Advances in Microbiology
Vol.4 No.3(2014), Article ID:43004,7 pages DOI:10.4236/aim.2014.43023
Antimicrobial Resistance in Enterococcus sp Isolated from Soft Cheese in Southern Brazil
1Department of Food Microbiology, Technological Federal University of Paraná, Londrina, Brazil
2Department of Microbiology, State University of Londrina, Londrina, Brazil
Email: *lucianamaia@utfpr.edu.br
Copyright © 2014 Luciana Furlaneto-Maia et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In accordance of the Creative Commons Attribution License all Copyrights © 2014 are reserved for SCIRP and the owner of the intellectual property Luciana Furlaneto-Maia et al. All Copyright © 2014 are guarded by law and by SCIRP as a guardian.
Received October 22, 2013; revised November 22, 2013; accepted November 29, 2013
KEYWORDS
Enterococci; Dairy Products; Foods; Antimicrobial Resistance
ABSTRACT
The aim of this research was to study the incidence of antibiotic resistance in 56 Enterococcus strains isolated from dairy products. The identification of enterococci was detected by polymerase chain reaction (PCR) using specific primers to E. faecalis, E. faecium, E. gallinarum and E. casseliflavus, and antibiotic resistance was tested by the disk diffusion method. The most prevalent species was E. faecium with a rate of 58.33%, followed by 27.77% E. faecalis, 11.11% E. casseliflavus and 2.7% E. gallinarum. Distribution of resistance was found in different species. All isolates were susceptible to chloramphenicol, ampicillin, imipenem and amoxicillin/clavulanic acid. In addition, isolates resistant to tetracyclin, nalidixic acid, amikacin, erythromycin, vancomycin and cephalothin were detected. A total of 66.6% of E. faecium and 58.3% of E. faecalis strain were resistant to multiple drugs. The van(A) gene was detected in 100% of vancomycin resistant enterococci. Considering the results of our study, dairy enterococci can be considered a potential source for dissemination of antibiotic resistances.
1. Introduction
Enterococci are ubiquitous bacteria that have a predominant habitat in the digestive tract of humans and animals, but may be present in soil, water’s surface and in plants and vegetables. They can also be found in foods, especially in cheese [1], but their presence is unwanted in certain kinds of cheese and can cause them to be spoiled [2-4].
The reasons for the prevalence of enterococci in dairy products has long been considered a result of unhygienic conditions during milk collection and processing, together with their resistance to pasteurization temperatures and their adaptability to different substrates and growth conditions [5]. However, the presence of enterococci in cheese and other fermented foods is a matter of debate, since some enterococcal species are involved in clinical infections such as endocarditis, bacteraemia, urinary tract infections and neonatal sepsis [6]. One of the reasons for the rise of nosocomial infections related to enterococci might be their ability to develop resistance against a wide variety of antibiotics [7,8].
Their resistance to a wide variety of antibiotics can be acquired or found intrinsically. Intrinsic antibiotic resistance of enterococci to cephalosporins, beta-lactams, sulphonamids, low levels of clindamycin and aminoglycosides may have selectively allowed them to acquire further genes of resistance against many commonly used antimicrobial agents such as aminoglycosides, penicillins, tetracycline, chloramphenicol, and vancomycin [8,9]. Enterococcus sphave evolved an adaptive response to the antibiotic stress and have developed drug resistance [10].
In addition, these bacteria are able to acquire resistance determinants through gene transference by plasmids and transposons [11,12], and toacquire resistance to many antimicrobial agents, including aminoglycosides, chloramphenicol, B-lactams, macrolides, quinolones, tetracycline, vancomycin and teicoplanin [13,14].
The occurrence of antimicrobial resistance among enterococci is not only restricted to the nosocomial setting, and therefore resistant strains carried on sources such as food may also act as potential reservoirs of antimicrobial resistance genes. The food chain can be considered as the main route of transmission of antibiotic resistant bacteria between the animal and human population [15]. More specifically, fermented dairy products that are not heattreated before consumption provide a vehicle for antibiotic resistant bacteria with a direct link between the animal indigenous microflora and the human gastrointestinal tract.
In the present study, we investigated the incidence of antibiotic resistances among enterococci derived from soft cheese produced in the South of Brazil.
2. Materials and Methods
2.1. Strains and Phenotype Characterization
A total of 56 enterococci were isolated from samples of soft cheeses types, comercialized at several local street markets of Londrina city-Paraná State, and were collected over a period of one year from 2011 to 2012. Phenotype tests were used to separate the enterococci group and the non-enterococcalstrains. Colonies with typical enterococci morphology were identified to genus level through following methods: Gramstaining, catalase production and esculin hydrolysis tests incombination with resistance to bile salts and growth at 10˚C and 45˚C, according to Teixeira and Facklam [16] and molecular approach.
2.2. Isolation of Enterococcal DNA and Identification by PCR
Enterococcus spp. genomic DNA was extracted through the boiling method as described by Marques and Suzart [17]. The identification of enterococci was detected by polymerase chain reaction (PCR) using specific primers targeted [18-20] (Table 1).
2.3. Antimicrobial Susceptibility Testing and Detection of Resistance Genes by PCR
The antibiotic discs (Laborclin®) were used to determine the susceptibility of the strains to ampicillin (AMP, 10 µg), nalidixic acid (NAL 30 µg), vancomycin (VAN, 30 µg), erythromycin(ERY, 15 µg), chloramphenicol (CLO, 30 µg), norfloxacin (NOR, 10 µg), tetracycline (TET, 30 µg), imipenem (IPM 10 µg), amikacin (AK 30 µg); cephalothin (CF 30 µg); ciprofloxacin (CIP 5 µg); amoxicillin/clavulanic acid (AMC 30 µg). The discs were placed onto Mueller-Hintonagar plates overlayed with the enterococcal culture with a cell concentration corresponding to 0.5 McFarland turbidity standard. After incubation at 37˚C for 18 - 24 h the diameter of inhibition haloes around the colonies was measured. Susceptibility or resistance was determined according to the recommendation of National Committee for Clinical Laboratory Standards guidelines (NCCLS, 2002) [21]. Minimal inhibitory concentrations (MICs) of vancomycin (VAN) were also determined by the broth microdilution according to methods described by (Clinical and Laboratory Standards Institute, 2009) [22]. E. faecalis ATCC 29212 and S. aureus ATCC 25923 were used as control strains.
The detection of resistance genes was conducted by PCR in all isolates of enterococci. The presence of gene vanA, aac(6’)-Ie-aph(2’’)-Ia, erm(B) and tet(L) was found for vancomycin, gentamicin, eritromicin and tetracycline, respectively (Table 2).
All PCR amplifications were performed in a final volume of 20 μl containing 1 ρmol of each primer (Forward and Reverse), 0.17 mMd NTPs, 2.5 mM MgCl2, 1 U of Taq DNA polymerase (Invitrogen®), buffer of Taq, and 10 µl template DNA. An initial cycle of denaturation (94˚C for 2 min) was followed by 30 cycles of denaturation (94˚C for 1 min), annealing at an appropriate temperature for 1 min and elongation (72˚C for 10 min). A Thermal Cycler (Techne-Tc3000) was used to carry out the PCR reactions. PCR products were analysed by gel electrophoresis in 1.2% agarose stained with ethidium bromide (0.5 g·ml−1), and observed under UV transillumination and photographed by L-PIX ST (LOCCUS®).
3. Results
A total of 56 soft cheese samples that were collected from local street market were investigated for the presence of enterococci, and a conventional cultivation showed that 100% of them were harbouring Enterococcus spp. Genus-specific PCR with detection of tuf gene was in accordance with these preliminary results. According to the presumptive phenotypic characterization followed by PCR of species-specific primers for E. faecium, E. faecalis, E. gallinarum, E. casseliflavus, revealed that the most prevalent species was E. faecium with a rateof 58.33%, followed by 27.77% E. faecalis, 11.11% E. casseliflavus and 2.7% E. gallinarum (Figure 1).
A summary of the resistence among the Enterococcus strains is reported in Table 3.
A total of 56 Enterococcus strains displayed resistance to at least one antibiotic tested and were resistant to antibiotic agents, some of which were resistant to multiple drugs.
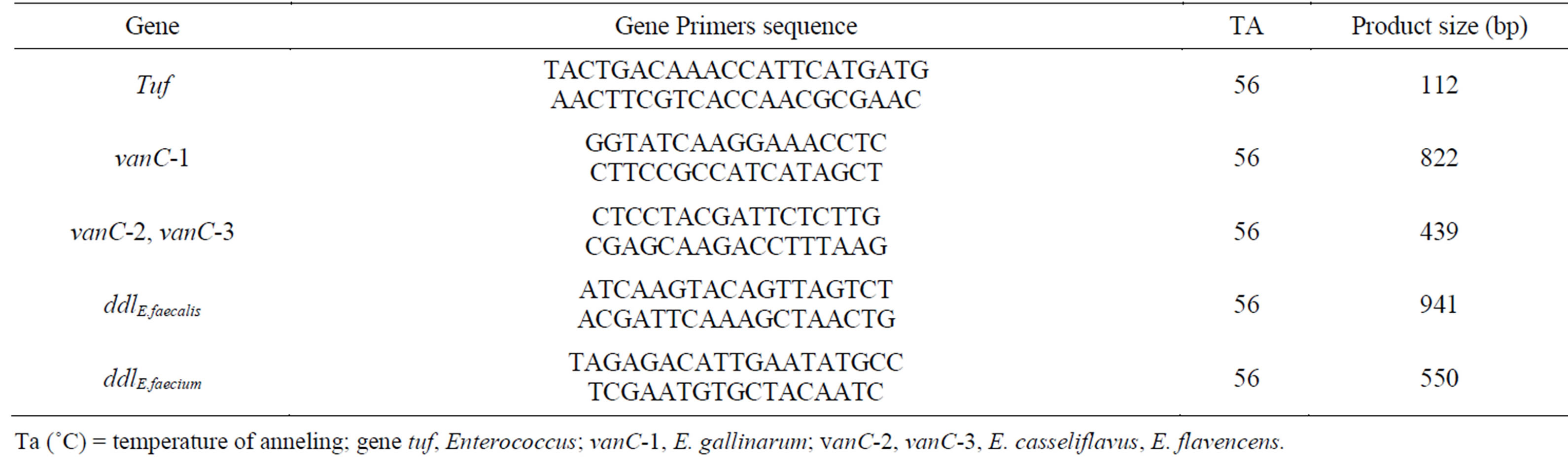
Table 1. List of primers and amplification conditions used in the present study.
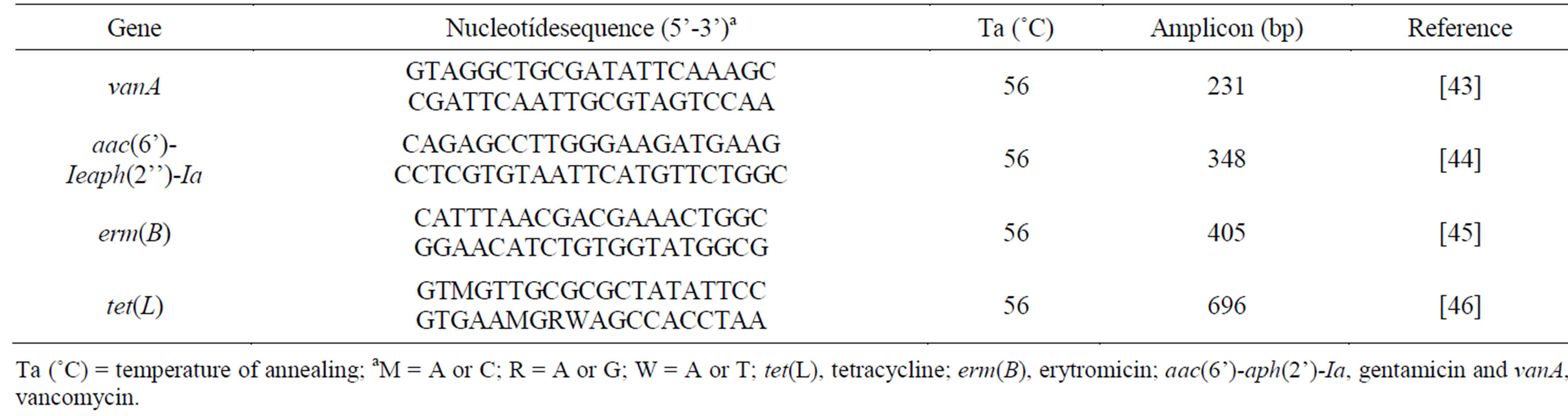
Table 2. Primers used in this study for detection of resistance genes by PCR-based method.
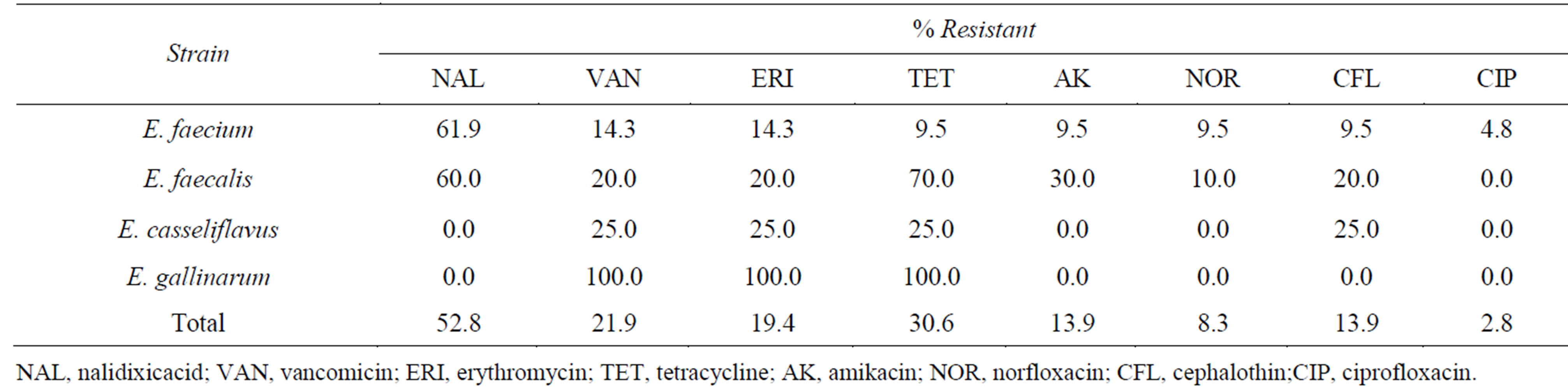
Table 3. Antibiotic resistant phenotypes of enterococcus species isolated from soft cheese.
The E. faecalis strains were mostly resistant to tetracycline (70%), followed by nalidixic acid (60%), amikacin (30%) and erythromycin, vancomicin and cephalothin (20%). E. faecium strains were mostly resistant to nalidixic acid (62%), vancomicin and erythromycin (14.3%), tetracycline, amikacin, norfloxacin and cephalothin (9.5%) and ciprofloxacin (4.8%). On the other hand, E. casseliflavus strains were resistant to vancomycin, erythromycin, tetracycline and cephalothin to 25% each one, and E. gallinarum strain were 100% resistant to vancomycin, erythromycin and tetracycline. None of the Enterococcus sp strains wasresistant to chloramphenicol, ampicillin, imipenem and amoxicillin/clavulanic acid. A total of 66.6% of E. faecium and 58.3% of E. faecalis strain were resistant to multiple drugs. All isolates were drug resistant for vancomycin with MICs of >32 to 64 µg·ml−1.
In our study, the presence of resistance genes erm(B), tet(L), vanA and aac(6’)-Ie-aph(2’’) were 86.7%, 23.3%, 80.0% and 66.7%, respectively. Several isolatesharboured resistance genes to more than oneantibiotic. The significant ones were tet(L)+/erm(B)+ to E. faecalis (42.8%) and erm(B)+/aac(6’)-Ie-aph(2’’)-Ia+/vanA+ to E. faecium (69.6%).
The presence of the vanA gene was detected in twenty three isolates of E. faecalis and fifty-four of E. faecium, corresponding to 42.8% and 96.6% of the isolates, respectively. The van(A) gene was detected in 100% of vancomycin resistant enterococci, however, five isolates harboured the van(A) gene and presented vancomycin
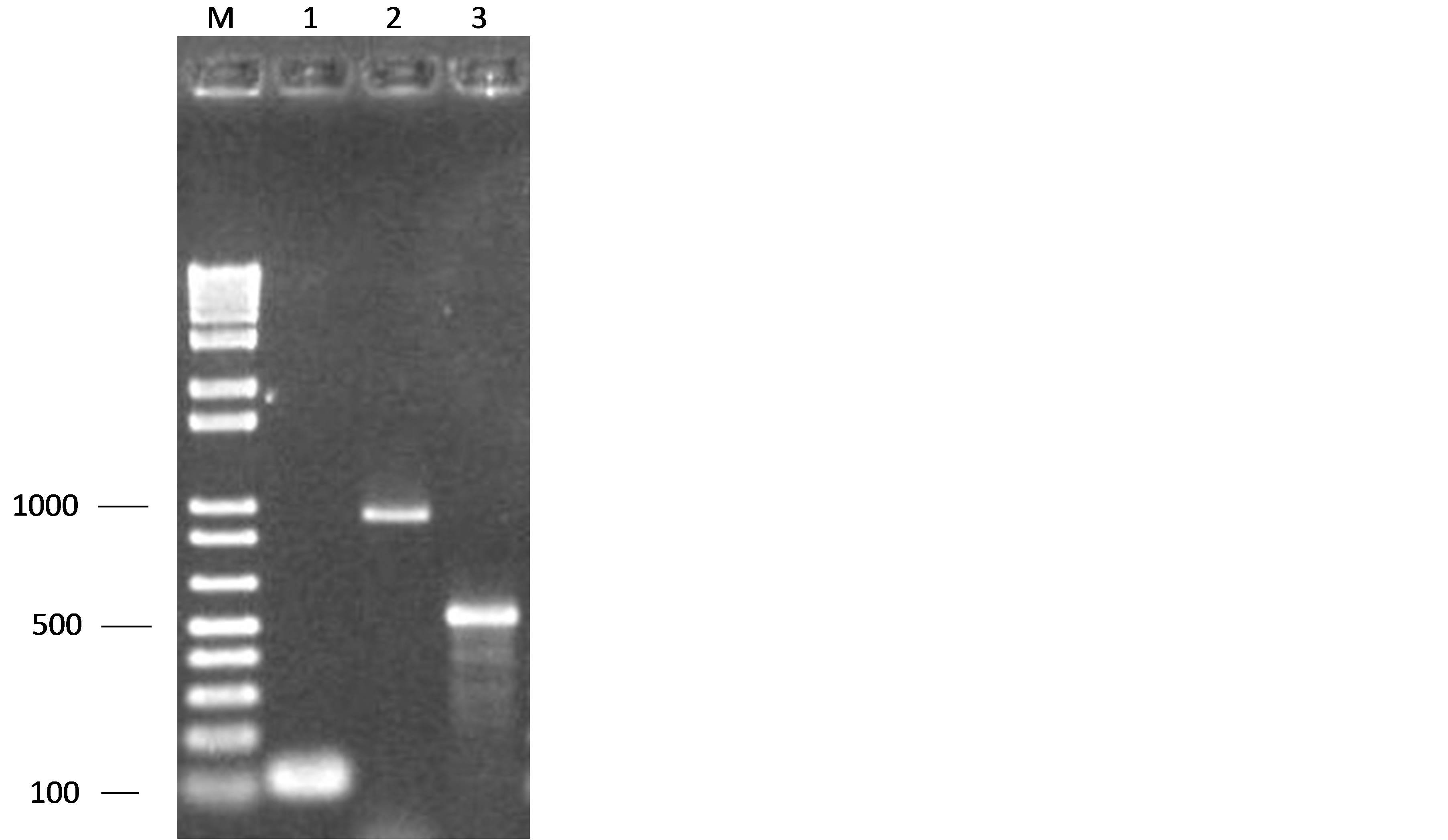
Figure 1. Amplification gel pictures characteristic of polymerase chain reaction (PCR) amplification of Enterococcus sp gene. Lanes: 1 Enterococcus spp. (112 pb); 2 E. faecalis (941 pb); 3 E.faecium (550 pb); M Ladder 1 kb plus (Invitrogen).
susceptibility phenotype.
4. Discussion
In this study, enterococci were isolated from soft cheese, the most prevalent species being E. faecium, followed by E. faecalis, E. casseliflavus and E. gallinarum. A higher prevalence of enterococci in processed foods may be attributed to their resistance to heat, extreme salinity and harsh conditions of foods [1-3,23].
Enterococci are considered as opportunistic pathogens which can cause a variety of infections in patients that have severe underlying diseases or that are immunocompromized. The major responsible for human enterococcal infections is E. faecalis followed by E. faecium and with a much lower incidence by other enterococcal species [2,6,8]. Furthermore, Enterococci are found in a variety of artisanal cheese made from raw or pasteurized milk from goats, sheep, water buffaloes, and cows [24].
A perusal of the literature revealed that E. faecalis and E. faecium were the most frequently isolated species from enterococci isolated from an artisanal cheese [25-29]. This explaination could be by the fact that these foods are manipulated by hands suggesting a contamination during the manufacturing process.
Antimicrobial susceptibility tests showed resistance phenotypes to a range of antibiotics widely administered in humans. A major concern is the presence of food strains harbouring multiple antibiotic resistance (from 3 to 8 out of the 12 antibiotics tested). Indeed, 100% of the vancomycin resistant enterococci showed resistance also to other clinically relevant antibiotics such as erythromycin (100%), tetracycline (100%), amikacin (50%), norfloxacin (50%), cephalothin (75%) and nalidixic acid (50%) thus leaving few therapeutic options. Results showed low level and high level resistant to vancomycin antibiotic at concentration of 64 to 128 μg·ml−1. Enterococcus sp strains isolated from soft cheese did not show chloramphenicol, ampicillin and amoxicilin-resistence, a clinical relevant antibiotic since ampicillin remains the drug of choice for the treatment of enterococcal infections.
Cariolato et al. [30] demonstrated only one isolate of Enterococcus from cheese was resistant to only two antibiotics (nalixidic acid and streptomycin). All the other ones were sensitive. Mannu et al. [31] showed lower occurrence of dairy strains resistant to clinically relevant antibiotics among food enterococci, suggesting that enterococci from milk and cheese cannot be considered the main potential sources for dissemination of antibiotic resistances. Martín-Platero et al. [32] showed that no more than 13% of the isolates of Enterococcus sp from cheese demonstrated any antimicrobial activity.
On the other hand, Flórez et al. [33], Cariolato et al. [30], Riboldi et al. [28] and Frazzon et al. [34], showed that in colonial cheese type was detected the presence of Enterococcus resistant to one of ampicillin gentamycin, streptomycin, ampicillin, vancomycin and tetracycline. High incidence of enterococci resistance to tetracycline chloramphenicol and erythromycin was found among isolated raw milk cheese.
Data on the incidence of vancomycin resistance within dairy enterococci remain controversial, though several papers indicate very lowor no presence of vanA and vanB resistance genes in enterococci isolated from cheese [20,23,27,35].
However, since in our study we did not apply any enrichment procedure, our data is not comparable with some results obtained in other foods, such as meat products, in which higher numbers of vancomycin resistant enterococci were isolated after a step of selective enrichment in the presence of vancomycin [36,37].
The association of resistence gene with conjugative elements is an important factor for spreading resistance in enterococci, which corroborate with the outlook that Enterococcus species are important sources of antibiotic resistance genes by horizontal gene transfer [38]. The vanA gene cluster, which confers resistance to vancomycin, is located on transposon Tn1546 [39].
Riboldi et al. [28] suggested that the emergence of Enterococcus spvancomycin-resistant (VRE) strain in food samples would be explained by the massive use of antibiotic in agriculture (e.g., avoparcin as animal growth promoters). Vancomycin-resistant indicates that Enterococcus is a controversial species that should not be used for probiotic applications [40].
Emergence of enterococci antimicrobial resistance and its spreading in food suggest a situation of risk for the community, and also a possible correlation between strains present in hospitals with those isolated from food must be considered [41-46].
Some studies have shown that the same resistance gene was found in bacteria isolated from both food samples and patients [37,41]. The food chain can be considered as the main route of transmission of antibiotic resistant bacteria between the animal and human population. More specifically, dairy products that are not heat-treated before consumption provide a vehicle for antibiotic resistant bacteria with a direct link between the animal indigenous microflora and the human gastrointestinal tract [15].
5. Conclusions
We have described here the first report of antibiotic resistant enterococci isolated from foods in the North of Paraná, in Southern Brazil. However, further studies are necessary in order to understand the possible role played by potential virulence factors and antibiotic resistances of dairy isolates in relation to their ecological habitat as well as the transferability of this virulence and resistance traits among strains in the cheese environment. Our results suggest that dairy enterococci can be considered as a potential source for dissemination of antibiotic resistances.
Finally, one reason that could explain the emergence of antibiotic resistant enterococci in food samples would be the use of antibiotic in agriculture, and some studies have shown that the same resistance gene was found in bacteria isolated from both food samples and patients.
Further studies are necessary in order to understand the possible role played by antibiotic resistances of dairy isolates in relation to their ecological habitat as well as the transferability of these resistance traits among strains in the cheese environment. However, this finding is the most relevant in this study and serves as a warning to authorities of public health since vancomycin is the last antibiotic alternative used for treatment of enterococal nosocomial infections.
Acknowledgments
L.F. Maia thanks the National Council for Scientific and Technological Development (CNPq) and K.R. Rocha thanks the Brazilian Graduate Educational Federal Agency (CAPES) for the fellowship grants. Authors thank CNPq (process n. 502230/2009-6) and The State of Paraná Research Foundation (Fundação Araucária) (process n. 185/2012) for financial support.
REFERENCES
- G. Giraffa, “Enterococci from Foods,” FEMS Microbiology Reviews, Vol. 26, No. 2, 2002, pp. 163-171. http://dx.doi.org/10.1111/j.1574-6976.2002.tb00608.x
- C. M. A. P. Franz, W. H. Holzapfel and M. E. Stiles, “Enterococci at the Crossroads of Food Safety?” International Journal of Food Microbiology, Vol. 47, No. 1-2, 1999, pp. 1-24. http://dx.doi.org/10.1016/S0168-1605(99)00007-0
- G. Giraffa, “Functionality of Enterococci in Dairy Products,” International Journal of Food Microbiology, Vol. 88, No. 2-3, 2003, pp. 215-222. http://dx.doi.org/10.1016/S0168-1605(03)00183-1
- R. Malek, A. El-Attar, M. Mohamed, S. Anwar, M. ElSoda and C. Béal, “Technological and Safety Properties Display Biodiversity among Enterococci Isolated from Two Egyptian Cheeses, ‘Ras’ and ‘Domiati’,” International Journal of Food Microbiology, Vol. 153, No. 3, 2012, pp. 314-322. http://dx.doi.org/10.1016/j.ijfoodmicro.2011.11.019
- G. Giraffa, D. Carminati and E. Neviani, “Enterococci Isolated from Dairy Products: A Review of Risks and Potential Technological Use,” Journal of Food Protection, Vol. 60, 1997, pp. 732-738.
- D. R. Schaberg, D. H. Culver and R. P. Gaynes, “Major Trends in the Microbial Etiology of Nosocomial Infection,” American Journal of Medicine, Vol. 91, No. 3, 1991, pp. 72S-75S. http://dx.doi.org/10.1016/0002-9343(91)90346-Y
- A. Lombardi, D. Cariolato and C. Andrighetto, “Occurrence of Virulence Factors and Antibiotic Resistances in Enterococcus faecalis and Enterococcus faecium Collected from Dairy and Human Samples in North Italy,” Food Control, Vol. 19, No. 9, 2008, pp. 886-892. http://dx.doi.org/10.1016/j.foodcont.2007.08.019
- L. M. Mundy, D. F. Sahm and M. Gilmore, “Relationships between Enterococcal Virulence and Antimicrobial Resistances,” Clinical Microbiology Reviews, Vol. 13, No. 4, 2000, pp. 513-522. http://dx.doi.org/10.1128/CMR.13.4.513-522.2000
- S. Radu, H. Toosa, R. A. Rahim, A. Reezal, M. Ahmad, A. N. Hamid, G. Rusul and M. Nishibuchi, “Occurrence of the vanA and vanC2/C3 Genes in Enterococcus Species Isolated from Poultry Sources in Malaysia,” Diagnostic Microbiology and Infectious Disease, Vol. 39, No. 3, 2001, pp. 145-153. http://dx.doi.org/10.1016/S0732-8893(01)00222-X
- A. K. Oli, R. Sungar, N. Shivshetty, R. Hosamani1 and K. C. Revansiddappa, “Study of Scanning Electron Microscope of Vancomycin Resistant Enterococcus faecalis from Clinical Isolates,” Advances in Microbiology, Vol. 2, No. 2, 2012, pp. 93-97. http://dx.doi.org/10.4236/aim.2012.22012
- R. Leclercq, E. Derlot, J. Duval and P. Courvalin, “Plasmid Mediated Resistance to Vancomycin and Teicoplanin Resistance in Enterococcus faecium,” New England Journal of Medicine, Vol. 319, 1988, pp. 157-161. http://dx.doi.org/10.1056/NEJM198807213190307
- B. D. Shepard and M. S. Gilmore, “Antibiotic-Resistant Enterococci: The Mechanisms and Dynamics of Drug Introduction and Resistance,” Microbes and Infection, Vol. 4, No. 2, 2002, pp. 215-224. http://dx.doi.org/10.1016/S1286-4579(01)01530-1
- T. J. Eaton and M. J. Gasson, “Molecular Screening of Enterococcus Virulence Determinants and Potential for Genetic Exchange between Food and Medical Isolates,” Applied and Environmental Microbiology, Vol. 67, No. 4, 2001, pp. 1628-1635. http://dx.doi.org/10.1128/AEM.67.4.1628-1635.2001
- G. Huys G, K. D’Haene, J. C. Collard and J. Swings, “Prevalence and Molecular Characterization of Tetracycline Resistance in Enterococcus Isolates from Food,” Applied and Environmental Microbiology, Vol. 70, No. 3, 2004, pp. 1555-1562. http://dx.doi.org/10.1128/AEM.70.3.1555-1562.2004
- W. Witte, “Medical Consequences of Antibiotic Use in Agriculture,” Science, Vol. 279, No. 5353, 1998, pp. 996- 997. http://dx.doi.org/10.1126/science.279.5353.996
- L. M. Teixeira and R. R. Facklam, “Enterococcus,” In: P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller and R. H. Yolken, Eds., Manual of Clinical Microbiology, 9th Edition, American Society for Microbiology, Washington DC, 2007, pp. 422-433.
- E. B. Marques and S. Suzart, “Enterococcus faecalis Strains Isolated in Londrina, Brazil,” Journal of Medical Microbiology, Vol. 53, No. 11, 2004, pp. 1069-1073. http://dx.doi.org/10.1099/jmm.0.45654-0
- S. Dukta-Malen, S. Evers and P. Courvalin, “Detection of Glycopeptide Resistance Genotypes and Identification to the Species Level of Clinically Relevant Enterococci by PCR,” Journal of Clinical Microbiology, Vol. 33, 1995, pp. 24-27.
- D. Ke, F. J. Picard, F. Martineau, P. H. R. Ménard, M. Ouellette and M. G. Bergeron, “Development of a PCR Assay for Rapid Detection of Enterococci,” Journal of Clinical Microbiology, Vol. 37, 1999, pp. 3497-3503.
- C. Andrighetto, E. Knijff, A. Lombardi, S. Torriani, M. Vancanneyt and K. Kersters, “Phenotypic and Genetic Diversity of Enterococci Isolated from Italian Cheeses,” Journal of Dairy Research, Vol. 68, No. 2, 2001, pp. 303- 316. http://dx.doi.org/10.1017/S0022029901004800
- National Committee for Clinical Laboratory Standards, “Performance Standards for Antimicrobial Disk Susceptibility Testing,” Twelfth Informational Supplement (M100- S12), ACCLS, Wayne, 2002.
- Clinical and Laboratory Standards Institute, “M07-A.B. Methods for Dilution Antimicrobial Susceptility Tests for Bacteria that Grow Aerobically; Approved Standard: 8th Edition,” CLSI, Wayne, 2009.
- D. Jurkovic, L. Krizkova, R. Dusinsky, A. Belicova, M. Sojka and J. Krajcovic, “Identification and Characterization of Enterococci from Bryndza Cheese,” Letters in Applied Microbiology, Vol. 42, 2006, pp. 553-559.
- R. Gelsomino, M. Vancanneyt, T. M. Cogan, S. Condon and J. Swings, “Source of Enterococci in a Farmhouse Raw-Milk Cheese,” Applied and Environmental Microbiology, Vol. 68, No. 7, 2002, pp. 3560-3565. http://dx.doi.org/10.1128/AEM.68.7.3560-3565.2002
- G. Suzzi, M. Caruso, F. Gardini, A. Lombardi, L. Vannini, M. E. Guerzoni, C. Andrighetto and M. T. Lanorte, “A Survey of the Enterococci Isolated from an Artisanal Italian Goat’s Cheese (Semicottocaprino),” Journal of Applied Microbiology, Vol. 89, No. 2, 2000, pp. 267-274. http://dx.doi.org/10.1046/j.1365-2672.2000.01120.x
- S. Cosentino, M. B. Pisano, A. Corda, M. E. Fadda and C. Piras, “Genotypic and Technological Characterization of Enterococci Isolated from Artisanal Fiore Sardo Cheese,” Journal of Dairy Research, Vol.71, No. 4, 2004, pp. 444- 450. http://dx.doi.org/10.1017/S002202990400041X
- H. Drahovská, L. Slobodníková, D. Kocíncová, M. Seman, R. Konceková and J. Turna, “Antibiotic Resistance and Virulence Factors among Clinical and Food Enterococci Isolated in Slovakia,” Folia Microbiologica, Vol. 49, No. 6, 2004, pp. 763-768. http://dx.doi.org/10.1007/BF02931562
- G. P. Riboldi, J. Frazzon, P. A. D’Azevedo and A. P. G. Frazzon, “Antimicrobial Resistance Profile of Enterococcus spp Isolated from Food in Southern Brazil,” Brazilian Journal of Microbiology, Vol. 40, 2009, pp. 125- 128.
- P. Nieto-Arribas, S. Seseña, J. M. Poveda, R. Chicón, L. Cabezas and L. Palop, “Enterococcus Populations in Artisanal Manchego Cheese: Biodiversity, Technological and Safety Aspects,” Food Microbiology, Vol. 28, No. 5, 2011, pp. 891-899. http://dx.doi.org/10.1016/j.fm.2010.12.005
- R. Cariolato, C. Andrighetto and A. Lombardi, “Occurrence of Virulence Factors and Antibiotic Resistances in Enterococcus faecalis and Enterococcus faecium Collected from Dairy and Human Samples in North Italy,” Food Control, Vol. 19, 2008, pp. 886-892.
- L. Mannu, A. Paba, D. E. Daga, R. Comunian, S. Zanetti and L. Dupre, “Comparison of the Incidence of Virulence Determinants and Antibiotic Resistance between Enterococcus faecium Strains of Dairy, Animal and Clinical Origin,” International Journal of Food Microbiology, Vol. 88, No. 2-3, 2003, pp. 291-304. http://dx.doi.org/10.1016/S0168-1605(03)00191-0
- A. M. Martín-Platero, E. Valdivia, M. Maqueda and M. Martínez-Bueno, “Characterization and Safety Evaluation of Enterococci Isolated from Spanish Goats’ Milk Cheeses,” Internecional Journal of Food Microbiology, Vol. 132, 2009, pp. 24-32.
- A. B. Flórez, S. Delgado and B. Mayo, “Antimicrobial Susceptibility of Lactic Acid Bacteria Isolated from a Cheese Environment,” Canadian Journal Microbiology, Vol. 51, No. 1, 2005, pp. 51-58. http://dx.doi.org/10.1139/w04-114
- A. P. G. Frazzon, G. P. Riboldi, J. Frazzon and P. A. D’Azevedo, “Antimicrobial Resistance Profile of Enterococcus spp Isolated from Food in Southern Brazil Brazilian,” Journal of Microbiology, Vol. 40, 2009, pp. 125- 128.
- S. Morandi, M. Brasca, C. Andrighetto, A. Lombardi and R. Lodi, “Technological and Molecular Characterisation of Enterococci Isolated from North-West Italian Dairy Products,” International Dairy Journal, Vol. 16, No. 8, 2006, pp. 867-875. http://dx.doi.org/10.1016/j.idairyj.2005.09.005
- C. Novais, T. M. Coque, M. J. Costa, J. C. Sousa, F. Baquero and L. V. Peixe, “High Occurrence and Persistence of Antibiotic-Resistant Enterococci in Poultry Food Samples in Portugal,” Journal of Antimicrobial Chemotherapy, Vol. 56, No. 6, 2005, pp. 1139-1143. http://dx.doi.org/10.1093/jac/dki360
- B. Robredo, K. V. Singh, F. Baquero, B. E. Murray and C. Torres, “Vancomycin-Resistant Enterococci Isolated from Animals and Food,” International Journal of Food Microbiology, Vol. 54, No. 3, 2005, pp. 197-204. http://dx.doi.org/10.1016/S0168-1605(99)00195-6
- L. Rizzotti, F. La Gioia, F. Dellaglio and S. Torriani, “Molecular Diversity and Transferability of the Tetracycline Resistance Gene Tet(M), Carried on Tn916-1545 Family Transposons, in Enterococci from a Total Food Chain,” Antonie Van Leeuwenhoek, Vol. 96, No. 1, 2009, pp. 43- 52. http://dx.doi.org/10.1007/s10482-009-9334-7
- T. L. Poole, M. E. Hume, L. D. Campbell, H. M. Scott, W. Q. Alali and R. B. Harvey, “Vancomycin-Resistant Enterococcus faecium Strains Isolated from Community Wastewater from a Semiclosed Agri-Food System in Texas, Antimicrobial Agents and Chemotherapy, Vol. 49, No. 10, 2005, pp. 4382-4385. http://dx.doi.org/10.1128/AAC.49.10.4382-4385.2005
- L. Chang, Z. Zhuo-Yang, D. Ke, Y. Jian-Ping and G. Xiao-Kui, “Antibiotic Resistance of Probiotic Strains of Lactic Acid Bacteria Isolated from Marketed Foods and Drugs,” Biomedicala and Environmental Sciences, Vol. 22, No. 5, 2009, pp. 401-412. http://dx.doi.org/10.1016/S0895-3988(10)60018-9
- S. M. Donabedian, L. A. Thal, E. Hershberger, M. B. Perri, J. W. Chow, P. Bartlett, R. Jones, K. Joyce, S. Rossiter, K. Gay, J. Johnson, C. Mackinson, E. Debess, J. Madden, F. Angulo and M. J. Zervos, “Molecular Characterization of Gentamicin-Resistant Enterococci in the United States: Evidence of Spread from Animals to Humans through Food,” Journal of Clinical Microbiology, Vol. 41, No. 3, 2003, pp. 1109-1113. http://dx.doi.org/10.1128/JCM.41.3.1109-1113.2003
- R. Joshua, R. Hayes, L. Linda, J. Peggy, Y. Kyung, D. David and D. G. White, “Prevalence and Antimicrobial Resistance of Enterococcus Species Isolated from Retail Meats”, Applied and Environmental Microbiology, Vol. 69, No. 12, 2003, pp. 7153-7160.
- I. M. Bell, J. C. Paton and J. Turnidge, “Emergence of Vancomycin-Resistant Enterococci in Australia: Phenotypic and Genotypic Characteristics of Isolates,” Journal of Clinical Microbiology, Vol. 36, 1998, pp. 2187-2190.
- S. B. Vakulenko, S. M. Donabedian, A. M. Vorkresenskiy, M. J. Zervos, S. A. Lerner and J. W. Chow, “Multiplex PCR for Detection of Aminoglycoside Resistance Genes in Enterococci,” Antimicrobial Agents Chemotherapy, Vol. 47, No. 4, 2003, pp. 1423-1426. http://dx.doi.org/10.1128/AAC.47.4.1423-1426.2003
- L. B. Jensen, A. Peter, L. Dons, R. N. Jones, A. M. Hammerum and F. M. Aarestrup, “ Molecular Analysis of the Tn1546 in Enterococcus faecium Isolated from Animals and Humans,” Journal Clinical Microbiology, Vol. 36, 1998. pp. 437-442.
- D. Gevers, M. Danielsen, G. Huys and J. Swings, “Molecular Characterization of tet(M) Genes in Lactobacillus Isolates from Different Types of Fermented Dry Sausage,” Applied Environmental Microbiology, Vol. 69, No. 2, 2003, pp. 1270-1275. http://dx.doi.org/10.1128/AEM.69.2.1270-1275.2003
NOTES
*Corresponding author.

