Advances in Microbiology
Vol.3 No.7(2013), Article ID:40134,6 pages DOI:10.4236/aim.2013.37072
Dominance of Enterobacteria among Histamine-Producing Bacteria Isolated from Indian Mackerel
Central Institute of Fisheries Education (CIFE) Seven Bungalows, Versova, Andheri (W), Mumbai, India
Email: *nayakbb@gmail.com
Copyright © 2013 Meena Tembhurne et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received September 29, 2013; revised October 29, 2013; accepted November 5, 2013
Keywords: Histamine; Scombroid Fish; Enterobacteria; 16SrDNA
ABSTRACT
Histamine fish poisoning (HFP) is a major illness occurring throughout the world due to the consumption of quality of deteriorated fish containing pre-formed histamine from bacterial activities. In the study reported here, the histamine-producing bacteria were isolated from the muscle, gills and the gut of 19 samples of Indian mackerel (Rastrelliger kanagurta) from Mumbai, India. The isolates from modified Niven’s medium (MNM) were confirmed for their ability to produce histamine by using 4 different pH-indicator media, followed by HPLC analyses. Out of 202 isolates, 63 isolates produced considerable amounts of histamine on at least 3 out of 4 media used in this study. The histamine formers were identified by biochemical tests followed by sequencing of their 16SrDNA gene, which showed that 89% of the isolates belonged to 13 different genera of the family Enterobacteriaceae. The non-enterobacterial histamine-producing bacteria belonged to the genera Staphylococcus, Alkaligenes, Shewanella and Psychrobacter.
1. Introduction
Histamine fish poisoning (HFP) occurs throughout the world due to the consumption of spoiled fish containing histamine [1,2]. Several species of bacteria are known to produce histamine via decarboxylation of the amino acid histidine by their enzyme histidine decarboxylase [3]. Histamine-producing bacteria may be introduced by contamination before, during or after processing of fish. HFP is more frequently associated with scombroid fish such as mackerel, tuna, bonito, seer fish etc., but of late, many non-scombroid fish such as sardines, pilchards, anchovies, herring and marline are known to contain high amounts of histamine in their muscle and are being reported as agents of HFP [3,4]. The family Enterobacteriaceae is primarily responsible for the decomposition of the scombroid fish. Among the enteric bacteria reported, Morganella morganii have been identified as prolific histamine producer [5]. Although only Morganella morganii, Klebsiella pneumoniae and Hafnia alvei have been isolated from fish incriminated in scombroid poisoning [6], a variety of other bacterial species including Proteus vulgaris, Proteus mirabilis, Enterobacter aerogenes, E. cloacae, Serratia planticola, S. liquefaciens and Citrobacter freundii have been identified as histamine formers in fish [7-10]. From salted sardine and fermented fish products, histamine-producing Staphylococcus spp., Vibrio spp., and Pseudomonas have been reported [11,12]. Several Vibrio spp. such as V. alginolyticus, V. harveyi, V. fischeri and Photobacteriaum leognathi have been reported to be histamine formers [13,14].
The source of the histamine formers in fresh fish could be the aquatic environment itself or they may be introduced during various stages of post-harvest handling. With the availability of more sensitive histamine detection methods, many new histamine-producing bacteria are being reported from fish. In the study discussed here, the diversity of histamine-producing bacteria was studied in Indian mackerel (Rastrelliger kanagurta), which is a popular staple fish in India and a common agent of histamine fish poisoning.
2. Materials and Methods
2.1. Fish Sample Collection and Preparation
During 2011-12, samples of fresh Indian Mackerel (Rastrelliger kanagurta Cuvier, 1817) were collected from the fish landing centers and the local retail markets of Mumbai, India, chilled immediately in ice and transported to the laboratory for analysis within two hours of collection. Ten-grams each of muscle with skin, gill and intestine were taken and homogenized with 90 ml of sterile physiological saline for 2 minutes using a stomacher (Seward Inc., UK). The homogenates were used as initial samples for bacteriological analysis.
2.2. Isolation of Histamine-Producing Bacteria
Homogenates of muscle with skin, gill and intestine were serially diluted and plated on modified Niven’s medium (MNM) by spread plate technique. The plates were incubated at 30˚C for 18 - 24 hours. The colonies with a pink halo around on modified Niven’s medium were selected as presumptive histamine-forming isolates and streaked on trypticase soy agar (TSA) plates containing 0.1% L-histidine. The isolates were maintained in glycerol broth at −80˚C. The confirmations of histidine decarboxylase activity of the purified bacterial groups were done by using four different media- 1) histidine decarboxylase broth (medium-I) according to the method of Falkow [15]. A control tube with only basal medium, i.e. without histidine, was inoculated with each isolate to rule out false positive results. The tubes were overlaid with sterile liquid paraffin and incubated at 30˚C and were observed after 48 h. Change of colour from purple to yellow and back to purple due to changes in pH was taken as positive result; 2) histidine decarboxylase broth containing inverted Durham’s tube (medium-II) as previously described [9]. A control tube with only basal medium, i.e. without histidine, was also simultaneously inoculated with the respective culture. The tubes were incubated at 30˚C for 24 - 48 hours. A tube with air bubble trapped inside the Durham’s tubes was considered as the positive result; 3) the isolates were stab-inoculated into histidine decarboxylase agar medium (medium III). The medium was prepared as per the composition of modified Niven’s medium (MNM) [9]. The medium was boiled and dispensed in 5 ml volumes in test tubes before sterilization. The purified cultures were stab inoculated into the sterile medium and incubated for 18 - 24 h at 30˚C. One control tube without histidine was also inoculated for each culture for better comparison. The change of color to deep red was taken as positive result; 3) streaking on Modified Niven’s Medium (MNM) (medium IV) followed by incubation at 30˚C for 24 hrs. Change of colour of the medium to deep red was taken as the positive reaction.
2.3. Confirmation of Histamine Production by HPLC
After growing the isolates in histidine broth, 1 ml of the culture was used for extraction in 10 ml of 10% trichloroacetic acid (TCA). The mixture was homogenized well and centrifuged at 3000 rpm at 4˚C for 10 minutes. The supernatant was collected and filtered through Whatman #1 filter paper and then through 0.22 µ membrane filter. The extracts were kept at −20˚C until assayed by HPLC for confirmation.
2.4. 16SrDNA Sequencing of Histamine-Forming Bacteria
The partial sequence of 16SrDNA gene of the histamineforming bacteria was derived by PCR amplification using previously described primers (10), followed by sequencing at Bioserve Biotechnologies, Secundeabad, India.
3. Results
3.1. Contribution of Histamine Formers to Total Bacterial Flora
The total bacterial count (TPC) and the histamine-forming bacterial counts (HFC) in different parts of fish are shown in Figure 1. The total viable count in gill, muscle
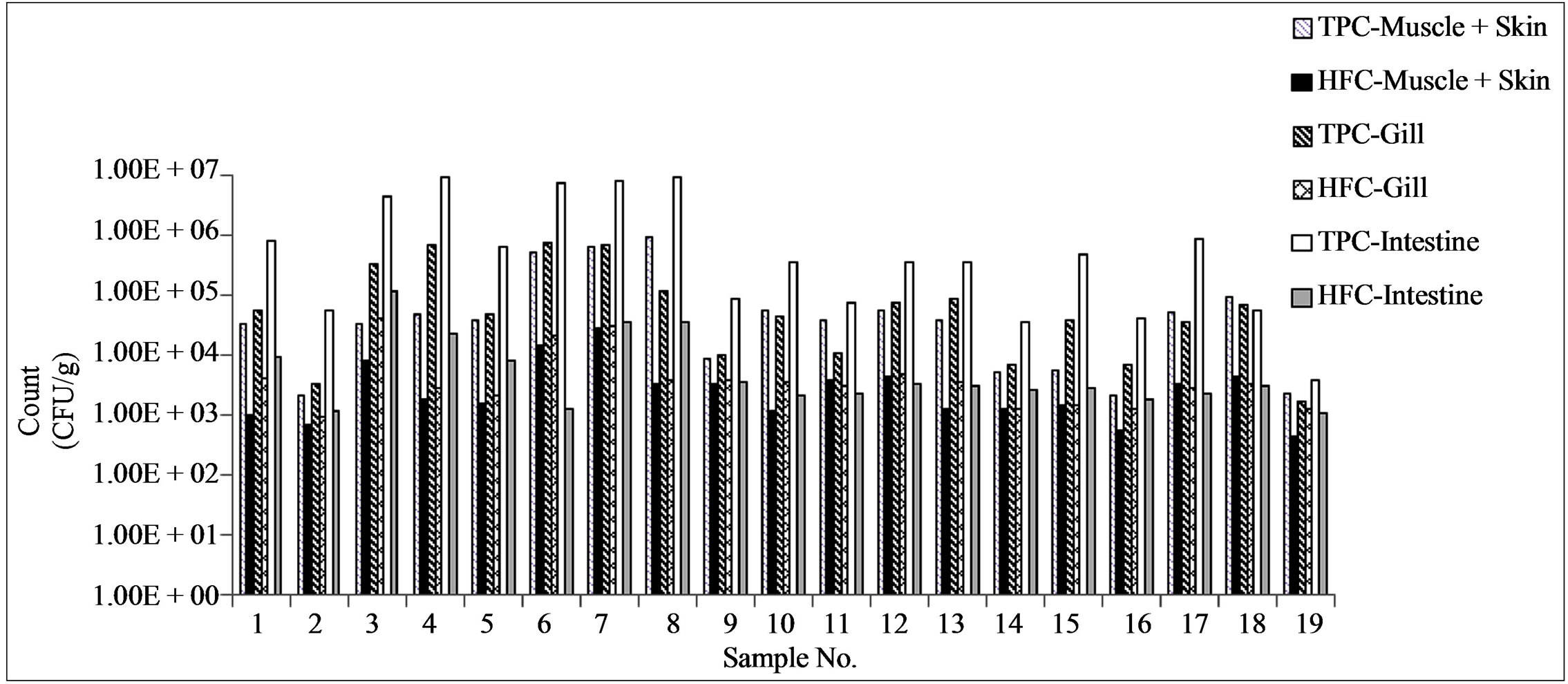
Figure 1. Distribution of histamine formers in different organs of the fish in relation to the total plate count. TPC = total plate count, HFC = Histamine former count.
with skin and intestine varied from 103 to 106 cfu/g. The total bacterial load was highest in the intestine (104 to 106 cfu/g), followed by gill (103 to 105) and muscle with skin (103 to 105 cfu/g). The histamine-forming bacteria count (HFC) varied from 102 to 104/g. The highest HFC was found in gills and intestine. HFC was as high as 20% of the total bacterial count (TPC) in muscle and gills compared to < 5% in the intestine (Figure 1).
3.2. Confirmation of Histamine Production on Multiple Media
A total of 202 presumptive histamine-forming isolates from the Modified Niven’s Medium agar (Table 1) were further subjected to confirmatory tests for histidine decarboxylation using four different confirmatory media and also by HPLC. Sixty-three isolates were confirmed to be the histamine-forming bacteria, majority of which belonged to the family Enterobacteriaceae (Table 2). The results
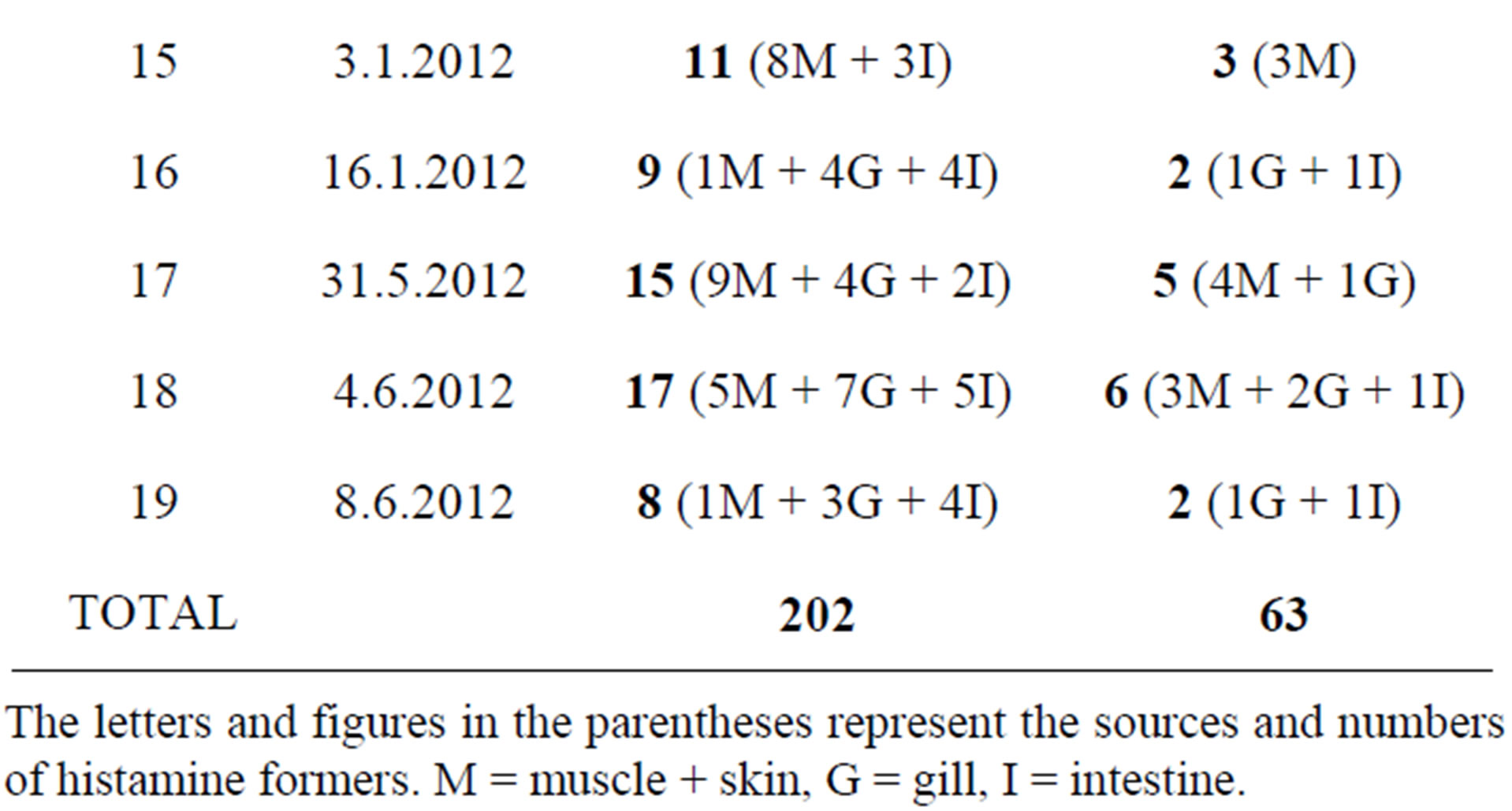

Table 1. Distribution of confirmed histamine formers in different organs of fish.
The letters and figures in the parentheses represent the sources and numbers of histamine formers. M = muscle + skin, G = gill, I = intestine.
of confirmatory tests are shown in Table 2. All confirmed histamine-forming bacteria were further identified by sequencing of partial 16SrDNA (Table 3). Of these, 35 were from muscle, 21 from gill and 7 were from intestine. Majority of the isolates such as K. variicola, K. pneumoniae, M. morganii, E. aerogenes, P. vulgaris, S. capitis and P. rustigianii produced histamine on all the four confirmatory media. While A. faecalis produced histamine on medium-II only, P. pulmonis produced histamine in all media except in medium-II.
Among different histamine-producing bacteria, M. morganii, P. vulgaris, P. rustigianii and all Klebsiella spp. showed rapid histidine decarboxylation on M-1. Decarboxylation by these bacteria was observed within 7 h as compared to 18 h by other bacteria. Others such as A. faecalis. P. pulmonis showed negative results on M-I. S. haliotis and S. capitis showed delayed decarboxylaion activity on M-I. P. mirabilis and P. penneri showed maximum variability in accumulation of gas inside Durham’s tube.
4. Discussion
Isolation and identification of histamine formers has always been a challenge due to diverse species of bacteria responsible for histamine formation in fish. Many media have been devised, which exploit the change of pH due

Table 2. Responses of histamine formers on different histidine decarboxylase media used in this study.
V = Variable, M-I; p = Histidine decarboxylase broth, M-II = Histidine decarboxylase broth containing inverted Durham’s tube, M-III = Stab culture in modified Niven’s agar medium, M-IV = streaked on modified Niven’s agar medium.
to histamine formation and consequently the change in the color of the medium. Glucose used in such media helps in lowering of pH to such a level that is conducive for histidine decarboxylase activity. An optimum pH of 5.0 - 5.5 for the activity is achieved due to the acid production from glucose and under such conditions, bromocresol purple seems to be the best pH indicator that gives lower false positive results. The presence of pyridoxal-5-phosphate, a cofactor, has a strong effect on the decarboxylase activity expression and is made an important ingredient of the medium for detecting amine formation [16]. Many differential media including Niven’s medium and modified Niven’s medium can give false-positive reaction due to production of other alkaline compounds which change the pH of the medium [17-20]. Nevertheless, they can be used for preliminary selection of strains possessing amino acid decarboxylase activity. Accordingly in the present work, the modified Niven’s medium (MNM) was used for initial selection of histamine formers. The presumptive positive colonies were selected from this medium and were further characterized using 4 different media as described in material methods. The methods followed in this study were found to be adequate to establish the histamine-forming ability of the bacteria. As observed in our study, the use of more than one medium increased the rate of isolation of histamine producers (Table 2).
In our study, a total of 202 isolates were screened and 63 were found to be confirmed histamine producers (Table 1). Most of them, apart from being histamine formers, also decarboxylated ornithine or lysine (data not shown), suggesting that they form at least one polyamine along with histamine. The roles of polyamines as potentiators of histamine poisoning are well documented [21].
The 16SrDNA gene based identification has become a convenient tool to precisely identify the bacteria [10,22]. Table 3 shows the BLAST identity of selected, representative isolates of histamine-forming bacteria isolated from mackerel. The majority of the confirmed histamineforming isolates belonged to the family Enterobacteriaceae. Enterobacteriaceae family members are often implicated in histamine poisoning and are also frequently isolated histamine-forming bacteria from fish. Some of the commonly isolated enterobacteria from scombroid fish implicated in histamine poisoning include M. morganii, K. pneumoniae, H. alvei P. vulgaris, E. aerogenes, P. mirabilis, C. freudii, E. coli, P. stuartii, K. oxytoca, E. agglomerans, E. cloacae, S. liquefaciens, S. marcescens, Edwardsiella spp. [6,13,19,23,24]. Several studies on Indian mackerel have isolated enterobacteria as major components of the histamine-forming bacterial population. A study by Ananthalakshmy et al. [25] reported Bacillus spp. as the major producer of histamine in Mackerel. In our study, a high prevalence of histamine-


Table 3. Species-wise distribution of histamine-formers based on partial sequencing of 16SrDNA gene.
M = Muscle + Skin; G = Gills; I = Intestine.
forming bacteria was observed in the gills and intestine of fish (Figure 1). In general, histamine-forming bacteria constitute normal skin, gill and intestinal microflora [26], but their numbers depend on the level of contamination and the temperature abuse of fish. Intestine of fish has been reported to be a major source of histamine-forming bacteria in many fish [9]. The histamine fish poisoning occurs when fish are subjected to temperature abuse leading to the proliferation of histamine-producing bacteria which produce and accumulate toxic levels of histamine in fish. Low levels of histamine may be produced by some bacteria at low temperatures [27]. It is possible that the isolation of histamine-forming bacteria in large numbers could also be due to the use of better and multiple isolation media.
5. Conclusion
Further study is required to determine the levels of histamine production. Further study is required to determine the levels of histamine production by the isolates obtained in the study to understand the relative contribution of individual species to the level of histamine in fish. Dominance of Enterobacteriaceae members among histamine forming isolates from all the organs sampled by us indicates that they may either be normal intestinal flora of mackerel or have come from post-harvest contamination in the markets and landing centres. Additionally, fish harvested from fecal contaminated water may also carry Enterobacteriaceae members as dominant flora. However, we assume post-harvest contamination as a stronger possibility. Further studies are required to establish the sources of the histamine formers in Indian mackerel.
6. Acknowledgements
The authors are grateful to Dr. W.S. Lakra, Director & Vice-Chancellor, CIFE Mumbai, for help and support.
REFERENCES
- B. A. Bartholomew, P. R. Berry, J. C. Rodhouse and R. J. Gilbert, “Scombrotoxic Fish Poisoning in Britain: Features of over 250 Suspected Incidents from 1976 to 1986,” Epidemiology and Infection, Vol. 99, No. 3, 1987, pp. 775-782. http://dx.doi.org/10.1017/S0950268800066632
- M. H. Silla, “Biogenic Amines: Their Importance in Foods,” International Journal of Food Microbiology, Vol. 29, No. 2-3, 1996, pp. 213-231. http://dx.doi.org/10.1016/0168-1605(95)00032-1
- S. L. Taylor, “Histamine Food Poisoning: Toxicology and Clinical Aspects,” Critical Reviews in Toxicology, Vol. 17, No. 2, 1986, pp. 91-128. http://dx.doi.org/10.3109/10408448609023767
- L. Lehane and J. Olley, “Histamine Fish Poisoning Revisited,” International Journal of Food Microbiology, Vol. 58, No. 1-2, 2000, pp. 1-37. http://dx.doi.org/10.1016/S0168-1605(00)00296-8
- J. E. Stratton and S. L. Taylor, “Scombroid Poisoning,” In: D. Ward and C. Hackney, Eds., Microbiology of Marine Food Products, Spectrum, AVI Publishers, New York, pp. 331-351.
- S. L. Taylor and M. W. Speckhard, “Isolation of Histamine Producing Bacteria from Frozen Tuna,” Marine Fisheries Review, Vol. 45, No. 1, pp. 35-39.
- A. R. Behling and S. L. Taylor, “Bacterial Histamine Production as a Function of Temperature and Time of Incubation,” Journal of Food Science, Vol. 47, No. 4, 1982, pp. 1311-1314. http://dx.doi.org/10.1111/j.1365-2621.1982.tb07675.x
- E. I. López-Sabater, J. J. Rodríguez-Jerez, A. X. Roig-Sagues and M. T. Mora-Ventura, “Determination of Histamine in Fish Using an Enzymic Method,” Food Additives and Contaminants, Vol. 10, No. 5, 1993, pp. 593-602. http://dx.doi.org/10.1080/02652039309374183
- D. H. Yoshinaga and H. A. Frank, “Histamine Producing Bacteria in Decomposing Skipjack Tuna Katsuwonuspelamis),” Applied and Environmental Microbiology, Vol. 44, No. 2, 1982, pp. 447-452.
- T. Iwamoto, K. Tani, K. Nakamura, Y. Suzuki, M. Kitagawa, M. Eguchi and M, Nasu, “Monitoring Impact of in Situ Biostimulation Treatment on Groundwater Bacterial Community by DGGE,” FEMS Microbiology Ecology, Vol. 32, No. 2, 2000, pp. 129-141. http://dx.doi.org/10.1111/j.1574-6941.2000.tb00707.x
- K. Yatsunami and T. Echigo, “Isolation of Salt Tolerant Histamine Forming Bacteria from Rice Bran Pickles of Sardine,” Bulletin of the Japanese Society of Scientific Fisheries, Vol. 57, No. 9, 1991, pp. 1723-1728. http://dx.doi.org/10.2331/suisan.57.1723
- K. Yatsunami and T. Echigo, “Occurrence of Halo Tolerant and Halophile Histamine-Forming Bacteria in Red Meat Fish Products,” Bulletin of the Japanese Society of Scientific Fisheries, Vol. 58, No. 3, 1992, pp. 515-520. http://dx.doi.org/10.2331/suisan.58.515
- H. A. Frank, J. D. Baranowski, M. Chongiriwatana, P. A. Brust and R. J. Premaratne, “Identification and Decarboxylase Activities of Bacteria Isolated from Decomposed Mahi Mahi (Coryphaenahippurus) after Incubation at 0˚C and 32˚C,” International Journal of Food Microbiology, Vol. 2, No. 6, 1985, pp. 331-340. http://dx.doi.org/10.1016/0168-1605(85)90023-6
- A. Ramesh and V. K. Venugopalan, “Densities and Characteristics of Histamine-Forming Luminous Bacteria of Marine Fish,” Food Microbiology, Vol. 3, No. 2, 1986, pp. 103-105. http://dx.doi.org/10.1016/S0740-0020(86)80033-8
- S. Falkow, “Activity of Lysine Decarboxylase as an Aid in the Identification of Salmonella and Shigella,” American Journal of Clinical Pathology, Vol. 29, No. 6, 1958, p. 598.
- S. Bover-Cid and W, Holzapfel, “Improved Screening Procedure for Biogenic Amine Production by Lactic Acid Bacteria,” International Journal of Food Microbiology, Vol. 53, No. 1, 1999, pp. 33-41. http://dx.doi.org/10.1016/S0168-1605(99)00152-X
- V. Arribas, M. C. Polo, F. Jorganes and R. Mun˜oz, “Screening of Biogenic Amine Production by Lactic Acid Bacteria Isolated from Grape Must and Wine,” International Journal of Food Microbiology, Vol. 84, No. 1, 2003, pp. 117-123. http://dx.doi.org/10.1016/S0168-1605(02)00391-4
- J. Baranowski, “Assay for Histamine Decarboxylase Activity,” In: B. S. Pan and D. James, Eds., Histamine in Marine Products: Production by Bacteria, Measurement and Prediction of Formation, FAO Fishery Technical Paper 252, 1985, FAO, Rome, pp. 10-13.
- J. J. Rodriguez-Jerez, E. L. Lopez-Sabater, M. M. Hernandez-Herrero and M. T. Mora-Ventura, “Histamine, Putrescine and Cadaverine Formation in Spanish Semipreserved Anchovies as Affected by Time/Temperature,” Journal of Food Science, Vol. 59, No. 5, 1994, pp. 993- 997. http://dx.doi.org/10.1111/j.1365-2621.1994.tb08175.x
- A. X. Roig-Sagués, M. M. Hernández-Herrero, E. I. Ló- pez-Sabater, J. J. Rodríguez-Jerez and M. T. Mora-Ventura, “Evaluation of Three Decarboxylating Agar Media to Detect Histamine and Tyramine-Producing Bacteria in Ripened Sausages,” Letters in Applied Microbiology, Vol. 25, No. 5, 1997, pp. 309-312. http://dx.doi.org/10.1046/j.1472-765X.1997.00223.x
- J. M. Hungerford, “Scombroid Poisoning: A Review,” Toxicon, Vol. 56, No. 2, 2010, pp. 231-243. http://dx.doi.org/10.1016/j.toxicon.2010.02.006
- M. Maifreni, F. Frigo, I. Bartolomeoli, N. Innocente, M. Biasutti and M. Marino, “Identification of the Enterobacteriaceae in Montasio Cheese and Assessment of Their Amino Acid Decarboxylase Activity,” Journal of Dairy Research, Vol. 80, No. 1, 2013, pp. 122-127. http://dx.doi.org/10.1017/S002202991200074X
- L. Ababouch, M. E. Afilal and H. Benabedlijelil, “Quantitative Changes in Bacteria, Amino Acids and Biogenic Amines in Sardine (Sardinapllchardus) Stored at Ambient Temperature (25˚C - 28˚C) and in Ice,” International Journal of Food Science and Technology, Vol. 26, No. 3, 1991, pp. 297-306. http://dx.doi.org/10.1111/j.1365-2621.1991.tb01166.x
- C. F. Niven, M. B. Jeffery and D. A. Corlett, “Differential Plating Medium for Quantitative Defection of Histamine Producing Bacteria,” Applied and Environmental Microbiology, Vol. 41, No. 1, pp. 321-322.
- V. K. Ananthalakshmy, A. Ramesh and A. V. K. Venugopalan, “Bacterial Production of Histamine in Some Tropical Fish,” Microbios, Vol. 63, No. 255, 1990, pp. 71-77.
- S. H. Arnold and W. D. Brown, “Histamine (?) Toxicity from Fish Products,” Advances in Food Research, Vol. 24, 1978, pp. 113-154. http://dx.doi.org/10.1016/S0065-2628(08)60157-3
- M. Okuzumi, S. Okuda and M. Awano, “Isolation of Psychrophilic and Halophilic Histamine-Forming Bacteria from Scomber japonicus,” Bulletin of the Japanese Society for the Science of Fish, Vol. 47, No. 12, 1981, pp. 1591-1598. http://dx.doi.org/10.2331/suisan.47.1591
NOTES
*Corresponding author.

