Advances in Microbiology
Vol.3 No.5(2013), Article ID:35913,8 pages DOI:10.4236/aim.2013.35058
First TaqMan Assay to Identify and Quantify the Cylindrospermopsin-Producing Cyanobacterium Aphanizomenon ovalisporum in Water
1Institute IMDEA Water, Alcalá de Henares, Spain
2Departamento de Biología, Universidad Autónoma de Madrid, Madrid, Spain
Email: *rehab.elshehawy@imdea.org
Copyright © 2013 Elena Campo et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received July 8, 2013; revised August 8, 2013; accepted August 14, 2013
Keywords: Cyanobacteria; Aphanizomenon Ovalisporum; TaqMan Assay; PCR; Cylindrospermopsin
ABSTRACT
Cylindrospermopsin (CYN) is an alkaloid that causes hepatotoxicity, neurotoxicity and general cytotoxicity in vertebrates. It is currently gaining widespread attention after its reported appearance in water bodies around the world. A. ovalisporum is capable of CYN-production and can form toxic blooms when favorable environmental conditions are available. We have developed for the first time a two-step qPCR assay using Taqman probes to detect and quantify potential CYN-producing A. ovalisporum in water samples. The assay was sensitive enough to discriminate between CYN-producing and non-CYN-producing A. ovalisporum in a mixed background, and discriminate between A. ovalisporum and other nostocales as C. raciborskii and A. bergii. The detection limit of the assay falls in the log linear range of 102 and 105 gene copies per reaction and is thus within the sensitivity range of previously published assays for the detection of other toxic cyanobacteria species. Our assay allows for the first time to quickly assess water quality for the presence of potentially CYN-producing A. ovalisporum and can be easily used for the purposes of monitoring water bodies.
1. Introduction
Cyanobacteria grow in freshwaters worldwide. They dominate the phytoplankton communities of many lakes and reservoirs. They are capable of forming massive growth of blooms with adverse effects on fisheries, tourism and sanitation. Problems associated with blooms in water bodies are aggravated when toxic cyanobacterial species are present. It is known that some species can produce potent toxins that display hepatotoxicity, neurotoxicity, dermatotoxicity and a general cytotoxicity mode of action and therefore can adversely affect humans and cattle. It is predicted that the current rise in global temperature, in synergy with eutrophication, may not only favor the growth of cyanobacteria in general but also the presence of toxic genotypes, and may even lead to increased cellular toxicity [1]. Careful monitoring and sustainable management of water bodies, especially potableand recreational-waters, are urgently needed along with the continuous development of efficient monitoring tools and toxin risk assessment.
Cylindrospermopsin (CYN) is an alkaloid that causes hepatotoxicity, neurotoxicity and general cytotoxicity in vertebrates. It inhibits glutathione, cytochrome P450 and protein synthesis [2] and there is evidence that it may interfere with DNA synthesis [3]. Although historically it received less attention than other cyanotoxins, except in Australia [4] and Israel [5], it is now gaining widespread attention after its reported appearance in five continents (Africa [6], Asia [7], Europe [8], North and South America [9]). In Europe it had only been detected in very low concentrations until Quesada et al. [8] described a bloom of CYN-producing cyanobacteria in a Spanish water reservoir. Since then, there has been an increasing number of reports from Europe in the last few years [10,11]. CYN is released during exponential and stationary phase of cellular growth [12-14] and tends to accumulate due to limited photodegradation [15] and biodegradation [16]. This accumulation of CYN in water bodies explains why there is no clear correlation between the occurrence of CYN in water bodies and the presence of a CYN-producing species [9,17-19] representing remarkable problems for water management.
Most of the CYN producing strains belong to the genera Cylindrospermopsis, Aphanizomenon and Anabaena. While strains belonging to Cylindrospermopsis form a monophyletic group within the nostocales family of heterocystous cyanobacteria, the strains belonging to Aphanizomenon and Anabaena are polyphyletic [20,21]. Strains of Cylindrospermopsis in general, as well as the toxic strains, cluster according to their geographical origins [22,23]. Toxicity seems to be restricted to Asian and Australian strains [17,19,23].
Aphanizomenon ovalisporum (Forti), isolated for the first time from a lake near Istanbul, is considered a tropical invasive species in Europe [24]. A. ovalisporum is typically found in thermally stratified waters with optimal temperatures between 26˚C and 30˚C [25,26]. It is a filamentous nitrogen-fixing cyanobacterium reported in the Mediterranean region [5,8,25,27,28], North America and Australia [19,29]. Generally, A. ovalisporum filaments are distributed in the euphotic zones with the help of gas vesicles [30]. Under unfavorable conditions, as other nostocales, akinetes are formed, enabling species survival [31,32].
Due to the recent increase in the number of reports detecting CYN-producing genera and specifically CYNproducing A. ovalisporum, it is necessary to develop fast and sensitive tools for monitoring the presence and proliferation of A. ovalisporum in water bodies in addition to current analytical tools for measuring CYN. It is difficult to morphologically distinguish species of the genus Aphanizomenon, moreover, some species show morphological characteristics intermediate between Aphanizomenon and Anabaena [33,34]. A. ovalisporum shares high phylogenetic and morphologic features with Anabaena bergii [33, 34], which is nowadays considered a non-toxic species. Such high morphological similarities, supported by some phylogenetic analysis, led to the suggestion that both are the same species. However, Stüken et al. [34] have demonstrated that both are closely related but genetically, toxicologically and morphologically distinct taxa. The task of identifying A. ovalisporum is further complicated by the recent report on the presence of toxic and nontoxic strains of A. ovalisporum in Lake Kinneret, Israel, [28]. Accordingly, for the purpose of proper water management, it is important to accurately identify toxic cells of A. ovalisporum in order to undertake proper measures against the health risk associated to the production and release of CYN in the water. Here we report for the first time the development of a sensitive assay that is capable of discriminating CYN-producing A. ovalisporum in a mixed background.
2. Material and Methods
2.1. Cyanobacterial Cultures
Aphanizomenon ovalisporum strain (UAM 290 and UAM 537], Cylindrospermopsis raciborskii (UAM 520) Microcystis flos-aquae (UAM 297), Microcystis aeruginosa (UAM 281), Aphanizomenon gracile (UAM 529, 521), Anabaena lemmermanii (UAM 506), are part of the Universidad Autónoma de Madrid Culture Collection. Anabaena bergii AB2010/03 and Aphanizomenon ovalisporum AB2010/06 were kindly provided by Dr. Sukenik (Kinneret Limnological Laboratory, Israel), while Cylindrospermopsis raciborskii strain LEGE 97047 was kindly provided by Dr. V. Vasconcelos (Universidade do Porto, Portugal). All strains were grown in 100 ml flasks containing BG110 medium at pH 7 except for strain UAM 290, which was maintained at pH 9 (pH was adjusted using 1 mM NaOH). Light intensity was maintained at 60 µmol photons/m2s provided by cool white fluorescent lamps. The temperature was kept at 28˚C. All strains used in this work and their CYN production capability are listed in Table 1.
2.2. Field Samples
Samples were collected on three occasions during summer 2009 and on six occasions during summer 2011 from Alange reservoir (with a known presence of CYN-producing Aphanizomenon ovalisporum demonstrated by the isolation of Aphanizomenon ovalisporum strains UAM 536 and UAM537 by Cirés and collaborators, Cirés personal communication) in Badajoz province, Spain. This reservoir is used for recreational purposes including fishing and sailing as well as drinking water supply, irrigation and electricity. Water samples were collected every two meters along the water column (~27 m) using a hydrographic bottle (5 L) and immediately mixed in a container. Phytoplankton, including cyanobacteria, were retained on GF/F filters (Whatman, UK). One liter of water was filtered through each filter and filters were stored frozen (−20˚C) until further analysis.
2.3. DNA Extraction
Genomic DNA was extracted from 15 - 20 ml of cultures using DNeasy Plant Mini Kit (Qiagen) according to manufacturer’s instructions. Genomic DNA from environmental samples was extracted as follows: Cells were mechanically lysed by alternating between homogenization and freezing in liquid nitrogen. The DNA was subsequently extracted using the DNeasy Plant Mini Kit (Qiagen) in accordance to the manufacturer’s instructions. DNA concentration was measured using a Nanophotometer (Epoch, Biotech). The purity of the extracted DNA was determined by calculating the ratio of the ab-
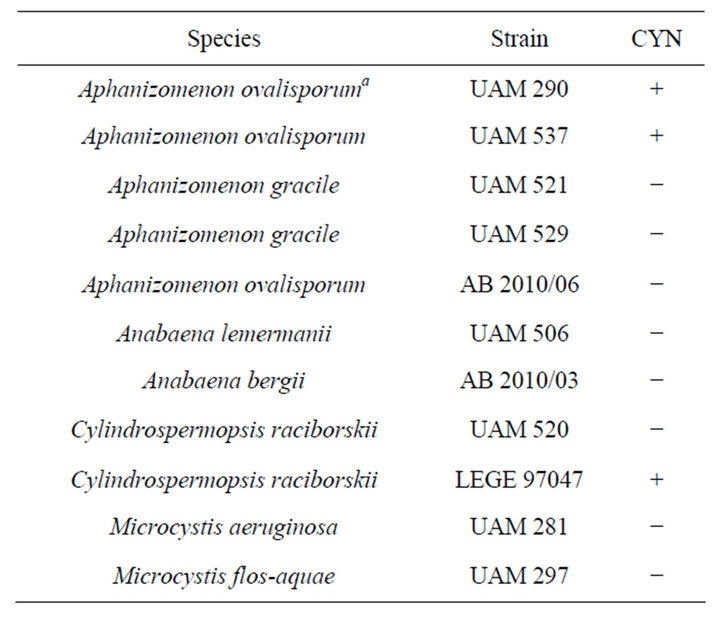
Table 1. List of strains used in this study. (a) reference strain used in this study, isolated by Wörmer et al., 2008. (CYN) ability to produce cylindrospermopsin.
sorbance measured at 260 nm (A260) to the absorbance measured at 280 nm (A280) using the Gen 5 data analysis software provided by the manufacturer (Epoch, Biotech). The purity (A260/A280) of DNA extracted in our study varied from 1.8 to 2.
2.4. Primers Design
Primers and probes designed for this work are listed in Table 2. Primers were designed using Primer 3 software (http://workbench.sdsc.edu/). Probes were designed using Primer Express 3.0 software (Applied Biosystems). Primers and probes were designed using all corresponding sequences available on the NCBI Genbank from A. ovalisporum (Accession numbers: FJ234844.1, FJ234857.1, FJ234845.1, FJ234843.1, FJ234858.1, FJ234859.1, GQ385961.1) and aligned against corresponding sequences available from C. raciborskii, Aphanizomenon sp. and A. bergii.
2.5. Real Time PCR
The DNA of cyrJ gene (encoding a sulfotransferase) was detected and quantified using cyrJ207 F/R primers and cyrJ207 P(FAM) probe. The DNA of rpoC11 gene (encoding the gamma subunit of the DNA-dependent RNA polymerase in cyanobacteria) was detected and quantified using the rpoc1148F/R primers and rpoc11830P (VIC) probe. Quantification was carried on an ABI7300 qPCR machine (Applied Biosystems). Real time PCR reactions were performed in 25 µl volumes containing 12.5 µl of QuantiFast TaqMan Probe PCR kit (Qiagen), 0.4 µM of each primer and probe and various concentrations of template DNA according to experiments (as explained below). The Real Time PCR programme was as
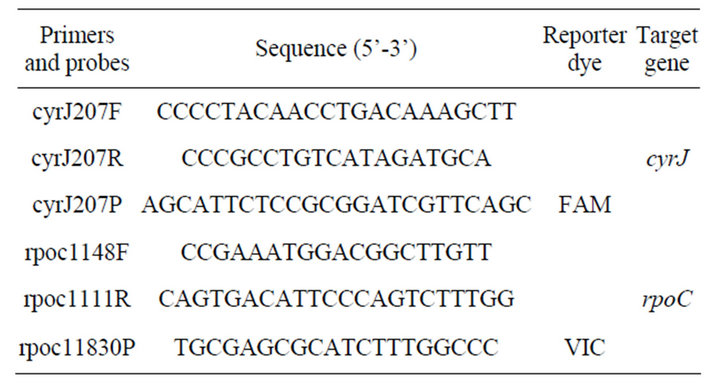
Table 2. Description of primers and probes used in Real time PCR reactions.
follows: a HotStar Taq DNA polymerase activation step at 95˚C for 15 min followed by 40 cycles of denaturation at 94˚C for 15s and annealing temperatures at 56˚C and 57˚C for cyrJ and rpoC1, respectively.
Specificity of the primers and probes was examined by testing them against a list of closely-related or distant strains (see Table 1 for list of strains used). From each strain listed, 2 ng/µl DNA was used and real time PCR reactions were run as described above.
To validate that the presence of closely or distinct related strains in the same reaction did not affect the efficiency of the amplification, 2 ng/µl DNA of A. ovalisporum UAM 290 was mixed with 2 ng/µl DNA from each of the strains listed in Table 3. Subsequently, tenfold serial dilutions were prepared from the mixture. Real Time PCR reactions were run as described above.
To quantify the numbers of copies of cyrJ and rpoC1 in field samples, standard curves were generated as described above using DNA from A. ovalisporum UAM290. Given that the genome size of A. ovalisporum is 4.5 Mb (Sukenik and Kaplan-Levy, Kinneret Limnological Laboratory, Israel, personal communication) and assuming that one genome contains only one copy of rpoC1 and cyrJ, the gene copy number was calculated as described in Vaitomaa et al. [35].
2.6. Determination of Extracellular Cylindrospermopsin Concentrations
Field water samples were filtered (GF/F Whatman, 0.7 um). Then the filtrate was pre-concentrated by a solid phase extraction according to Wörmer et al. [36] and used to determine extracellular cylindrospermopsin concentration by LC-MS/MS according to Dell’Aversano et al. [37] on a Varian 500 MS Ion Trap Mass Spectrometer (Agilent Technologies, USA).
3. Results
In this work we developed a two-step qPCR assay for the detection and quantification of toxic A. ovalisporum when present in a mixed background in laboratory or
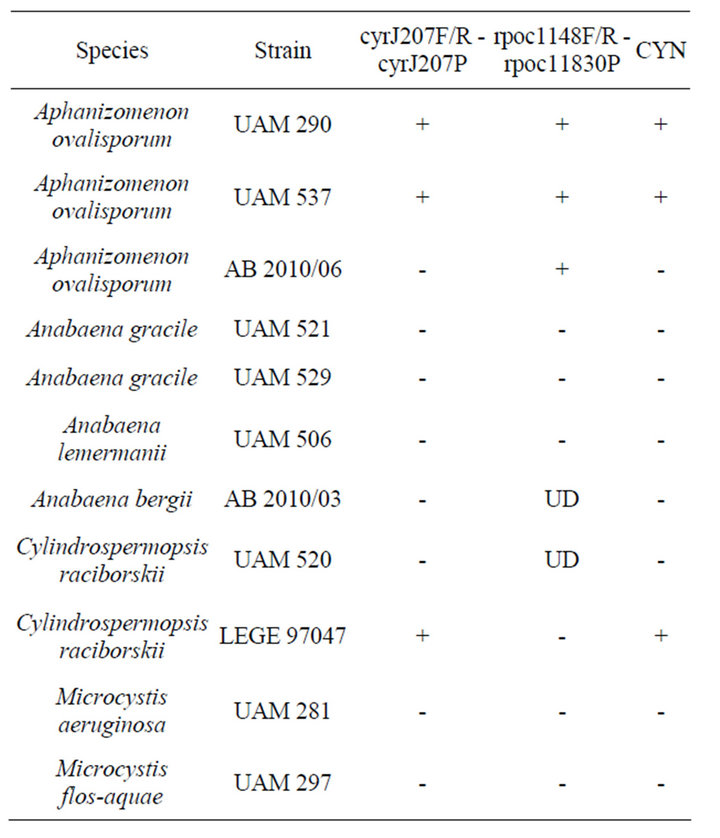
Table 3. Specificity of primers and probes used in Real time PCR runs. DNA from all strains was mixed in equal concentrations (see Material and Methods for description). (UD) under detection limit. (CYN) ability to produce cylindrospermopsin.
field samples. The first step is to report if potential CYNproducing cells are present in the sample and the second step is to identify if those cells belong to A. ovalisporum. For this purpose, we designed two TaqMan probes directed against cyrJ and rpoC1. The cyrJ gene was chosen as a molecular marker because in a recent study by Ballot et al. [28] the authors demonstrated that non-CYN producing A. ovalisporum lacked cyrJ, while harbored cyrA, cyrB and cyrC of the CYN gene cluster [28]. This makes cyrJ a good candidate to discriminate between CYNproducing and non-CYN producing A. ovalisporum.
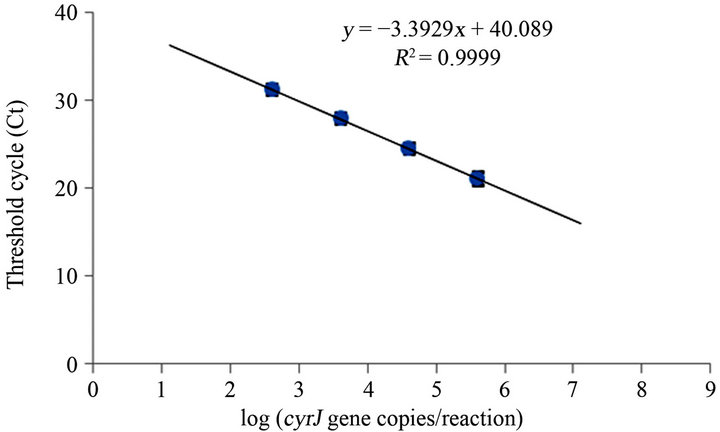
In order to test the efficiency of the two-step Real Time assay, standard curves were generated using DNA of A. ovalisporum strain UAM 290 as a reference strain and cyrJ207F/R - cyrJ207P or rpoc1148F/R - rpoc11830P, which amplify cyrJ and rpoC1, respectively (Figure 1).
The absolute values of the slopes of the standard curves were 3.39 ± 0.06 for cyrJ207F/R - cyrJ207P and 3.37 ± 0.07 for rpoc1148F/R - rpoc11830P, which correspond to amplification efficiency of 0.971 and 0.977, respectively (1 = 100%). The absolute values of the slopes are close to the ideal value (3.32), which indicates that primers and probes are binding to the target sequences and efficiently producing products during each PCR cycle. The detection limit of the assay falls in the log linear range of 102 and 105copies per reaction.
 (a)
(a)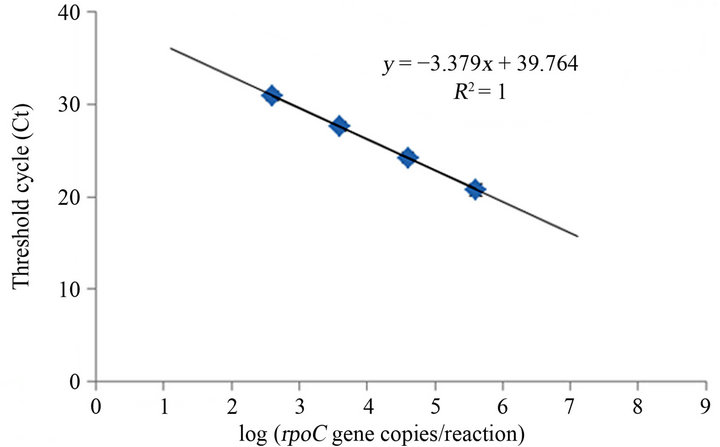 (b)
(b)
Figure 1. Standard curves generated using serial dilutions of DNA of A. ovalisporum with (a) cyrJ207F/R - cyrJ207P and (b) rpoc1148F/R - rpoc11830P. Error bars are hidden by the symbols. n = 3.
In order to test the specificity of the assay, equal concentrations of DNA of A. ovalisporum strain UAM 290 was mixed with equal DNA concentration of various strains of Aphanizomenon, Cylindrospermopsis, Anabaena and Microcystis (Table 3). As shown in Table 3, cyrJ207 F/R - cyrJ207P, successfully distinguished between CYNproducing and non CYN-producing strains. As shown in Table 3, rpoc1148F/R - rpoc11830P, successfully produced amplification with DNA from all A. ovalisporum strains and did not produce any amplification, or produced weak amplification under the detection limit of the assay, with DNA from other tested strains.
Accordingly, in order to detect CYN-producing A. ovalisporum in a mixed background and distinguish between toxic C. raciborskii and toxic A. ovalisporum, our developed two-step Real Time assay is applied by first using cyrJ207F/R - cyrJ207P (step 1) to detect the presence of potentially CYN-producing C. raciborskii or A. ovalisporum, and then by using rpoc1148F/R - rpoc 11830P (step 2) to discriminate between CYN-producing A. ovalisporum (positive results will be obtained) or CYN-producing C. raciborskii (negative results or products below detection limit will be obtained).
Such two-step assay may not be required for example in Europe, where toxic C. raciborskii has never been detected up till the writing of this work. In such case, only the first step will be needed.
In order to test that the efficiency of amplification is not affected when A. ovalisporum is present in a mixed background, as it is usually the case in the environment, DNA from all strains listed in Table 3 were mixed in equal proportions and used to spike an equal concentration of DNA from A. ovalisporum UAM 290. The efficiency of the amplification reactions as measured by the slope of the standard curves were 3.28 ± 0.04 and 3.26 ± 0.04 for cyrJ and rpoC1, respectively. This indicates that the efficiency of the assay is not affected by the presence of DNA of non-target species.
In order to validate the assay, qPCR reactions were run using DNA extracted from field samples collected in summer 2009 from a water reservoir in central Spain. The qPCR results were also compared with measurement of CYN concentrations in the water samples by LC-MS/ MS. As shown in Figure 2, the assay was able to detect and quantify the presence of A. ovalisporum in the samples. cyrJ gene copy numbers increased from August to September and then decreased again to fall below the detection limit of our qPCR assay. These data correlated well with the extracellular CYN concentrations as measured by LC-MS/MS. The fact that the gene copy numbers of rpoC1 are higher than those of cyrJ could indicate the presence of non-toxic cells of A. ovalisporum. This explanation merits further investigation because the presence of non-toxic A. ovalisporum in Europe is not yet demonstrated. Samples collected in summer 2011 did not appear to harbor A. ovalisporum filaments (following a careful microscopic examination) and qPCR assay tested negative (data not shown). This is not surprising giving the fact that blooms of A. ovalisporum in Spanish reservoirs are neither frequently encountered nor are an annual recurring phenomenon (A. Quesada, personal communication).
4. Discussion
Toxic cyanobacteria in water bodies are of major concern to environmental authorities and water supply companies all over the world. In light of the conditions predicted to result from current levels of global warming, water managers may need to invest more resources in controlling toxic cyanobacterial blooms.
The frequent presence of toxic and non-toxic cells of the same species in one location and the lack of correlation between morphology and toxin production has complicated the microscopic identification of toxic species and created the need for a reliable and sensitive test to identify potentially toxin-producing cells in water bodies. To fulfill this need, PCR and chip-based technologies were developed rapidly. In this context, Real Time PCR
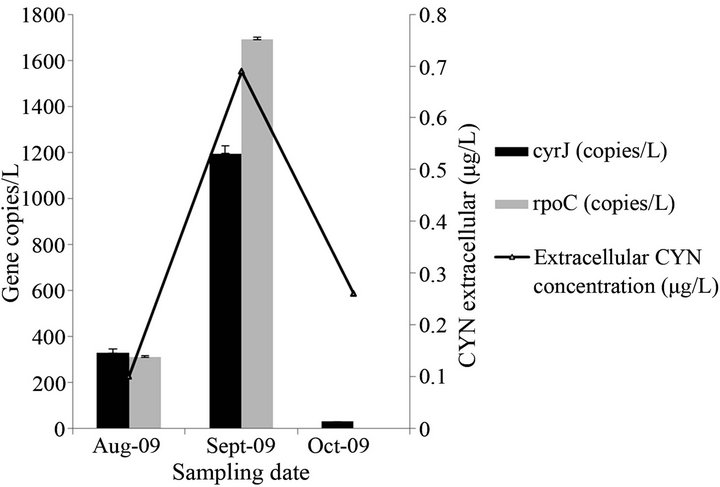
Figure 2. Copy number of cyrJ and rpoC and extracellular CYN concentration in field samples collected during summer 2009 from Alange reservoir, Spain. Error bars are marked. n = 2.
stands out as a powerful technique for the quantification of gene copy number, and hence quantification of potentially toxic cells, with its simple design, speed, and relatively inexpensive running costs.
A number of studies have developed Real Time PCR approaches for the detection and quantification of some of the genes belonging to the hepatotoxin synthetase clusters (mcy, nda and cyr) using SYBER-Green I and TaqMan technologies [38-40]. It was also demonstrated that the cylindrospermopsin synthetase gene C (cyrC) is useful for detecting and quantifying the toxic C. raciborskii in water [41,42]. In spite that A. ovalisporum is increasingly gaining attention after its reported appearance in many countries, there is no qPCR assay developed to specifically detect the CYN-producing A. ovalisporum cells when present in water samples. Although both C. raciborskii and A. ovalisporum are N2-fixing nostocales species and although both are tropical genera that are considered invasive in temperate regions [24], A. ovalisporum seems to present a competitive advantage over C. raciborskii and other nostocales due to its unique ability to acquire phosphate in oligotrophic waters [43], although such a feature still needs to be investigated in other cyanobacteria before withdrawing a definite conclusion. Furthermore A. ovalisporum presents higher growth rates at high temperatures than other nosotocales with optimum CYN production at 15˚C - 30˚C [26,44,24]. It is estimated that 25% - 35% of the produced CYN during typical growth phases will be released and such extracellular concentrations may rise to 50% at extreme temperatures or growth [44]. Altogether, it is thus important from the management perspective to be able to identify and discriminate toxin-producing A. ovalisporum when present in water bodies, especially in relation to the control of phosphate discharge, bloom proliferation during summer periods, overwintering strategies, and under the projected scenario of warmer temperatures due to climatic change.
Here we developed for the first time a two-step TaqMan Real Time PCR assay that is able to detect and discriminate between potential CYN-producing and nonCYN producing cells and also to discriminate between potential CYN-producing A. ovalisporum and C. raciborskii. In some cases, as is the case in Europe at the moment, only the first step is required to detect the presence of potential CYN-producing A. ovalisporum in water.
Our assay is similar in a way to the assay developed by Rantala et al. [45] in which several amplification steps are needed to detect potentially microcystinsand nodularinsproducing blooms. Our assay appeared to be equally as sensitive as the qPCR assay developed by Vaitomaa et al. [35] for the detection of microcystins genes and to the assay developed by Rasmussen et al. [41] for the detection of C. raciborskii. It is also one order of magnitude less sensitive than the assay developed by Al-Tebrineh et al. [39] and Koskenniemi et al. [46] for the detection of hepatotoxin-producing species.
The two-step assay described here proved to be specific and efficient to detect and quantify potential CYNproducing A. ovalisporum in a mixed background, rendering the assay suitable for application on environmental water samples for monitoring purposes. Indeed, when applying the assay on samples collected during summer of 2009 from a water reservoir in central Spain, the data obtained by qPCR correlated well with extracellular CYN concentrations as measured by LC-MS/MS.
5. Conclusion
We were able to develop a two-step qPCR assay for the detection and quantification of potential CYN-producing A. ovalisporum. The assay is sensitive, specific and offers the possibility of detecting this cyanobacterium in environmental samples. The combination of qPCR and LC-MS/MS techniques provides an accurate monitoring, detection and quantification tool that can be easily implemented within management strategies for the protection of human health from exposure to CYN produced by (the invasive cyanobacterium) A. ovalisporum, as well as for the sustainable management of water bodies in regards to cyanobacteria blooms. Finally, the strategy used to develop our qPCR can be implemented to develop similar assays for the specific detection of the CYNproducing Aphanizomenon flos-aquae, of which both toxic and non-toxic cells co-occur [12].
6. Acknowledgements
The authors acknowledge funding from Marie CurieCofund (AMAROUT) to RE.
REFERENCES
- R. El-Shehawy, E. Gorokhova, F. Fernández-Piñas and F. F. del Campo, “Global Warming and Hepatotoxin Production by Cyanobacteria: What Can We Learn from Experiments?” Water Research, Vol. 46, No. 5, 2012, pp. 1420-1429. doi:10.1016/j.watres.2011.11.021
- L. Pearson, T. Mihali, M. Moffit, R. Kellman and B. Neilan, “On the Chemistry, Toxicology and Genetics of the Cyanobacterial Toxins Microcystin, Nodularin, Saxitoxin and Cylindrospermopsin,” Marine Drugs, Vol. 8, No. 5, 2010, pp. 1650-1680. doi:10.3390/md8051650
- G. R. Shaw, A. A. Seawright, M. R. Moore and P. K. S. Lam, “Cylindrospermopsin, a Cyanobacerial Alkaloid: Evaluation of Its Toxicologic Activity,” Therapeutic Drug Monitoring, Vol. 22, No. 1, 2000, pp. 89-92. doi:10.1097/00007691-200002000-00019
- I. Ohtani, R. E. Moore and M. T. C. Runnegar, “Cylindrospermopsin—A Potent Hepatotoxin from the BlueGreen Algae Cylindrospermopsis raciborskii,” Journal of the American Chemical Society, Vol. 114, No. 20, 1992, pp. 7941-7942. doi:10.1021/ja00046a067
- U. Pollinger, O. Hadas, Y. Z. Yaconi, T. Zohari and T. Berman, “Aphanizomenon ovalisporum (Forti) in Lake Kinneret, Israel,” Journal of Plankton Research, Vol. 20, No. 7, 1998, pp. 1321-1339. doi:10.1093/plankt/20.7.1321
- C. Berger, N. Ba, M. Gugger, M. Bouvy, F. Rusconi, A. Couté, M. Troussellier and C. Bernard, “Seasonal Dynamics and Toxicity of Cylindrospermopsis raciborskii in Lake Guiers (Senegal, West Africa),” FEMS Microbiology Ecology, Vol. 57, No. 3, 2006, pp. 355-366. doi:10.1111/j.1574-6941.2006.00141.x
- K. Harada, I. Ohtani, K. Iwamoto, M. Suzuki, M. F. Watanabe and K. Terao, “Isolation of Cylindrospermopsin from a Cyanobacterium Umezakia natans and Its Screening Method,” Toxicon, Vol. 32, No. 1, 1994, pp. 73-84. doi:10.1016/0041-0101(94)90023-X
- A. Quesada, E. Moreno, D. Carrasco, T. Paniagua, L. Wörmer, C. De Hoyos and A. Sukenik, “Toxicity of Aphanizomenon ovalisporum (Cyanobacteria) in a Spanish Water Reservoir,” European Journal of Phycology, Vol. 41, No. 1, 2006, pp. 39-45. doi:10.1080/09670260500480926
- W. W. Carmichael, S. Azevedo, J. S. An, R. J. R. Molica, E. M. Jochimsen, S. Lau, K. L. Rinehart, G. R. Shaw and G. K. Eaglesham, “Human Facilities from Cyanobacteria: Chemical and Biological Evidence for Cyanotoxins,” Environmental Health Perspectives, Vol. 109, No. 7, 2001, pp. 663-668. doi:10.1289/ehp.01109663
- L. Bláhová, M. Oravec, B. Maršálek, L. Šejnohová, Z. Šimek and L. Bláha, “The First Occurrence of the Cyanobacterial Alkaloid Toxin Cylindrospermin in the Czech Republic as Determined by Immunochemical and LC/MS Methods,” Toxicon, Vol. 53, No. 5, 2009, pp. 519-524. doi:10.1016/j.toxicon.2009.01.014
- L. Brient, M. Lengronne, M. Bormans and J. Fastner, “First Occurrence of Cylindrospermopsin in Freshwater in France,” Environmental Toxicology, Vol. 24, No. 4, 2009, pp. 415-420. doi:10.1002/tox.20439
- K. Preußel, A. Stüken, C. Wiedner, I. Chorus and J. Fastner, “First Report on Cylindrospermopsin Producing Aphanizomenon flos-aquae (Cyanobacteria) Isolated from Two German Lakes,” Toxicon, Vol. 47, No. 2, 2006, pp. 156- 162. doi:10.1016/j.toxicon.2005.10.013
- P. R. Hawkins, E. Putt, I. R. Falconer and A. Humpage, “Phenotypical Variation in a Toxic Strain of the Phytoplankter, Cylindrospermopsis raciborskii (Nostocales, Cyanophyceae) during Batch Culture,” Environmental Toxicology. Vol. 16, No. 6, 2001, pp. 460-467. doi:10.1002/tox.10005
- M. L. Saker and D. J. Griffiths, “The Effect of Temperature on Growth and Cylindrospermopsin Content of Seven Isolates of Cylindrospermopsis raciborskii (Nostocales, Cyanophyceae) from Water Bodies in Northern Australia,” Phycologia, Vol. 39, No. 4, 2000, pp. 349-354. doi:10.2216/i0031-8884-39-4-349.1
- L. Wörmer, M. Huerta-Fontela, S. Cirés, D. Carrasco and A. Quesada, “Natural Photodegradation of the Cyanobacterial Toxins Microcystin and Cylindrospermopsin,” Environmental Science & Technology, Vol. 44, No. 8, 2010, pp. 3002-3007. doi:10.1021/es9036012
- L. Wörmer, S. Cires, D. Carrasco and A. Quesada, “Cylindrospermopsin Is Not Degraded by Co-Occurring Natural Bacterial Communities during a 40-Days Study,” Harmful Algae, Vol. 7, No. 2, 2008, pp. 206-213. doi:10.1016/j.hal.2007.07.004
- J. Fastner, R. Heinze, A. R. Humpage, U. Mischke, G. K. Eaglesham and I. Chorus, “Cylindrospermopsin Occurrence in Two German Lakes and Preliminary Assessment of Toxicity and Toxin Production of Cylindrospermopsis raciborskii (Cyanobacteria) Isolates,” Toxicon, Vol. 42, No. 3, 2003, pp. 313-321. doi:10.1016/S0041-0101(03)00150-8
- J. Rücker, A. Stücken, B. Nixdorf, J. Fastner, I. Chorus and C. Wiedner, “Concentrations of Particulate and Dissolved Cylindrospermopsin in 21 Aphanizomenon-Dominated Temperate Lakes,” Toxicon, Vol. 50, No. 6, 2007, pp. 800-809. doi:10.1016/j.toxicon.2007.06.019
- M. Yilmaz, E. J. Phlips, N. J. Szabo and S. Badylak, “A Comparative Study of Florida Strains of Cylindrospermopsis and Aphanizomenon for Cylindrospermopsin Production,” Toxicon, Vol. 51, No. 1, 2008, pp. 130-139. doi:10.1016/j.toxicon.2007.08.013
- C. Lyra, S. Suomalainen, M. Gugger, C. Vezie, P. Sundman, L. Paulin and K. Sivonen, “Molecular Characterization of Planktic Cyanobacteria of Anabaena, Aphanizomenon, Microcystis and Planktothrix Genera,” International Journal of Systematic and Evolutionary Microbiology, Vol. 51, 2001, pp. 513-526.
- M. Gugger, C. Lyra, P. Henriksen, A. Couté, J. F. Humbert and K. Sivonen, “Phylogenetic Comparison of the Cyanobacterial Genera Anabaena and Aphanizomenon,” International Journal of Systematic and Evolutionary Microbiology, Vol. 52, No. 5, 2002, pp. 1867-1880. doi:10.1099/ijs.0.02270-0
- M. Gugger, R. Molica, B. Le Berrre, P. Dufour, C. Bernard and J. F. Humbert, “Genetic Diversity of Cylindrospermopsis Strains (Cyanobacteria) Isolated from Four Continents,” Applied and Environmental Microbiology, Vol. 71, No. 2, 2005, pp. 1097-1100. doi:10.1128/AEM.71.2.1097-1100.2005
- S. Haande, T. Rohrlack, A. Ballot, K. Roberg, R. Skulberg, M. Beck and C. Wiedner, “Genetic Characterisation of Cylindrospermopsis raciborskii (Nostocales, Cyanobacteria) Isolates from Africa and Europe,” Harmful Algae, Vol. 7, No. 5, 2008, pp. 692-701. doi:10.1016/j.hal.2008.02.010
- A. Sukenik, R. N. Kaplan-Levy, J. M. Welch and A. F. Post, “Akinete Development in the Harmful Cyanobacterium Aphanizomenon ovalisporum,” Journal of Phycology, Vol. 48, Special Issue, 2012, pp. S26-S26.
- S. Gkelis, M. Moustaka-Gouni, K. Sivonen and T. Lanaras, “First Report of the Cyanobacterium Aphanizomenon ovalisporum Forti in Two Greek Lakes and Cyanotoxin Occurrence,” Journal of Plankton Research, Vol. 27, No. 12, 2005, pp. 1295-1300. doi:10.1093/plankt/fbi085
- G. Mehnert, F. Leunert, S. Cirés, K. D. Jöhnk, J. Rücker, B. Nixdorf and C. Wiedner, “Competitiveness of Invasive and Native Cyanobacteria from Temperate Freshwaters under Various Light and Temperature Conditions,” Journal of Plankton Research, Vol. 32, No. 7, 2010, pp. 1009- 1021. doi:10.1093/plankt/fbq033
- G. Bazzichelli and N. Abdelahad, “Caractérisation Morphométrique et Statistique de deux Populations d’Aphanizomenon du Group Aphanizomenon ovalisporum Forti des Lacs de Nemi et Albano (Italie),” Archiv für Hydrobiologie, Vol. 73, 1994, pp. 1-21.
- A. Ballot, J. Ramm, T. Rundberget, R. Kaplan-Levy, O. Hadas, A. Sukenik and C. Wiedner, “Occurrence of NonCylindrospermopsin Producing Aphanizomenon ovalisporum and Anabaena bergii in Lake Kinneret (Israel),” Journal of Plankton Research, Vol. 33, No. 11, 2011, pp. 1736-1746. doi:10.1093/plankt/fbr071
- G. R. Shaw, A. Sukenik, A. Livne, R. K. Chiswell, M. J. Smith, A. A. Seawright, R. Norris, G. K. Eaglesham and M. R. Moore, “Blooms of the Cylindrospermopsin Containing Cyanobacterium, Aphanizomenon ovalisporum (Forti), in Newly Constructed Lakes, Queensland, Australia,” Environmental Toxicology, Vol. 14, No. 1, 1999, pp. 167-177. doi:10.1002/(SICI)1522-7278(199902)14:1<167::AID-TOX22>3.0.CO;2-O
- G. Shalev-Malul, J. Lieman-Hurtwitz, Y. Viner-Mozzini, A. Sukenik, A. Gaathon, M. Lebendiker and A. Kapland, “An AbrB-Like Protein Might Be Involved in the Regulation of Cylindrospermopsin Production by Aphanizomenon ovalisporum,” Environmental Microbiology, Vol. 10, No. 4, 2008, pp. 988-999. doi:10.1111/j.1462-2920.2007.01519.x
- O. Hadas, R. Pinkas, E. Delphine, A. Vardi, A. Kaplan and A. Sukenik, “Limnological and Ecophysiological Aspects of Aphanizomenon ovalisporum Bloom in Lake Kinneret, Israel,” Journal of Plankton Research, Vol. 21, No. 8, 1999, pp. 1439-1453. doi:10.1093/plankt/21.8.1439
- S. Cirés, L. Wörmer, C. Wiedner and A. Quesada, “Temperature-Dependent Dispersal Strategies of Aphanizomenon ovalisporum (Nostocales, Cyanobacteria): Implications for the Annual Life Cycle,” Microbial Ecology, Vol. 65, No. 1, 2013, pp. 12-21. doi:10.1007/s00248-012-0109-8
- J. Komárek and L. Kovacik, “Trichome Structure of 4 Aphanizomenon Taxa (Cyanophyceae) from Czechoslovakia, with Notes on the Taxonomy and Delimitation of the Genus,” Plant Systematics and Evolution, Vol. 164, No. 1-4, 1989, pp. 47-64.
- A. Stüken, R. J. Campbell, A. Quesada, A. Sukenik, P. K. Dadheech and C. Wiedner, “Genetic and Morphologic Characterization of Four Putative Cylindrospermopsin Producing Species of the Cyanobacterial Genera Anabaena and Aphanizomenon,” Journal of Plankton Research, Vol. 31, No. 5, 2009, pp. 465-480. doi:10.1093/plankt/fbp011
- J. Vaitomaa, A. Rantala, K. Halinen, L. Rouhiainen, P. Tallberg, L. Mokelke and K. Sivonen, “Quantitative Real Time PCR for Determination of Microcystin Synthetase E Copy Numbers for Microcystis and Anabaena in Lakes,” Applied and Environmental Microbiology, Vol. 69, No. 12, 2003, pp. 7289-7297. doi:10.1128/AEM.69.12.7289-7297.2003
- L. Wörmer, D. Carrasco, S. Cirés and A. Quesada, “Advances in Solid Phase Extraction of the Cyanobacterial Toxin Cylindrospermopsin,” Limnology and Oceanography: Methods, Vol. 7, 2009, pp. 568-575. doi:10.4319/lom.2009.7.568
- C. Dell’Aversano, G. K. Eaglesham and M. A. Quilliam, “Analysis of Cyanobacterial Toxins by Hydrophilic Interaction Liquid Cromatography-Mas Spectrometry,” Journal of Chromatography, Vol. 1028, No. 1, 2004, pp. 155- 164. doi:10.1016/j.chroma.2003.11.083
- L. A. Pearson and B. A. Neilan, “The Molecular Genetics of Cyanobacterial Toxicity as a Basis for Monitoring Water Quality and Public Health Risk,” Current Opinion in Biotechnology, Vol. 19, No. 3, 2008, pp. 281-288.
- J. Al-Tebrineh, M. M. Gehringe, R. Akcaalan and B. A. Neilan, “A New Quantitative PCR Assay for the Detection of Hepatotoxigenic Cyanobacteria,” Toxicon, Vol. 57, No. 4, 2011, pp. 546-554. doi:10.1016/j.toxicon.2010.12.018
- H. Hautala, U. Lamminmäki, L. Spoof, S. Nybom, J. Meriluoto and M. Vehniäinen, “Quantitative PCR Detection and Improved Sample Preparation of MicrocystinProducing Anabaena, Microcystis and Planktothrix,” Ecotoxicology and Environmental Safety, Vol. 87, 2013, pp. 49-56. doi:10.1016/j.ecoenv.2012.10.008
- J. P. Rasmussen, S. Giglio, P. T. Monis, R. J. Campbell and C. P. Saint, “Development and Field Testing of a Real-Time PCR Assay for Cylindrospermopsin-Producing Cyanobacteria,” Journal of Applied Microbiology, Vol. 104, No. 5, 2008, pp. 1503-1515. doi:10.1111/j.1365-2672.2007.03676.x
- P. T. Orr, J. P. Rasmussen, M. A. Burford, G. K. Eaglesham and S. M. Lennox, “Evaluation of Quantitative Real Time PCR to Characterize Spatial and Temporal Variations in Cyanobacteria, Cylindrospermopsis raciborskii (Woloszynska) Seenaya and Subba Raju and Cylindrospermopsin Concentrations in Three Subtropical Australian Reservoirs,” Harmful Algae, Vol. 9, No. 3, 2010, pp. 243-254. doi:10.1016/j.hal.2009.11.001
- Y. Bar-Yosef, A. Sukenik, O. Hadas, Y. Viner-Mozzini and A. Kaplan, “Enslavement in the Water Body by Toxic Aphanizomenon ovalisporum, Inducing Alkaline Phosphatase in Phytoplanktons,” Current Biology, Vol. 20, No. 17, 2010, pp. 1557-1561. doi:10.1016/j.cub.2010.07.032
- S. Cirés, L. Wörmer, J. Timon, C. Wiedner and A. Quesada, “Cylindrospermopsin Production and Release by the Potentially Invasive Cyanobacterium Aphanizomenon ovalisporum under Temperature and Light Gradients,” Harmful Algae, Vol. 10, No. 6, 2011, pp. 668-675. doi:10.1016/j.hal.2011.05.002
- A. Rantala, E. Rizzi, B. Castiglioni, G. de Bellis and K. Sivonen, “Identification of Hepatotoxin-Producing Cyanobacteria by DNA-Chip,” Environmental Microbiology, Vol. 10, No. 3, 2008, pp. 653-664. doi:10.1111/j.1462-2920.2007.01488.x
- K. Koskenniemi, C. Lyra, P. Rajaniemi-Wacklin, J. Jokela and K. Sivonen, “Quantitative Real-Time PCR Detection of Toxic Nodularia Cyanobacteria in the Baltic Sea,” Applied and Environmental Microbiology, Vol. 73, No. 7, 2007, pp. 2173-2179. doi:10.1128/AEM.02746-06
NOTES
*Corresponding author.

