Advances in Microbiology
Vol. 2 No. 2 (2012) , Article ID: 19668 , 6 pages DOI:10.4236/aim.2012.22011
A Preliminary Molecular Typing by PCR Assays of Clostridium perfringens and Clostridium difficile Isolates from Dogs
1Department of Animal Health, Faculty of Veterinary Medicine, University of Parma, Parma, Italy
2Department of Pathology and Laboratory Medicine, Faculty of Medicine and Surgery, University of Parma, Parma, Italy
Email: *mariacristina.ossiprandi@unipr.it
Received February 3, 2012; revised February 22, 2012; accepted March 20, 2012
Keywords: Clostridium perfringens; Clostridium difficile; molecular typing; dogs; toxigenic/non-toxigenic
ABSTRACT
Clostridium perfringens and C. difficile have been associated with acute and chronic large and small bowel diarrhoea, and acute haemorrhagic diarrhoeal syndrome in dogs. The objective of this study was to investigate by toxin gene profile and PCR-ribotyping the molecular characteristics of 14 C. perfringens and 10 C. difficile isolates from 95 canine faeces (n = 36, diarrhoeic and n = 59, non-diarrhoeic). Concerning C. perfringens, 13 strains (92.9%) were type A, of which 3 (23.1%) also possessed the beta 2 toxin (CPB2)-encoding gene. One isolate (7.1%) was type D and possessed CPB2 gene. On the whole, 4 of the 14 strains (28.6%) tested cpb2-positive. Six C. difficile isolates (60.0%) demonstrated tcdA+/tcdB+ and cdtA+/cdtB+ genotype and tested positive for, in vitro, toxin production by EIA. Eight distinct ribotypes were observed. In conclusion, the PCR assays may provide useful and reliable tools for C. perfringens and C. difficile molecular typing in routine veterinary diagnostics.
1. Introduction
Clostridium perfringens and C. difficile are important enteropathogenic agents in veterinary medicine [1].
C. perfringens is one of the most widespread pathogen, inhabiting the gastrointestinal tract of human beings and animals as well as terrestrial and marine environments [2]. It has been associated with outbreaks of acute, often severe diarrhoea in humans, horses, dogs and cats. The elaboration of four major toxins, alpha (a), beta (b), iota (i), and epsilon (e), is the basis for typing the microorganism into five toxigenic phenotypes (A, B, C, D and E). The different toxinotypes cause different forms of enteritis and enterotoxaemia in various hosts [3-5]. Each type may also express a subset of at least 15 other established toxins, including C. perfringens enterotoxin (CPE), a wellcharacterized virulence factor whose production is coregulated with sporulation [6,7]. Virtually all strains isolated from dogs are type A, with only one published report documenting a type C infection in five cases of canine peracute lethal hemorrhagic enteritis [2]. Although several studies have shown an association between the immunodetection of CPE in faecal specimens and canine diarrhoea, the pathogenesis of C. perfringens-associated diarrhoea in the dog is not fully understood, because CPE is also detected in up to 14% of non-diarrhoeic dogs. Isolation of non-enterotoxigenic type A strains from a diarrhoeic specimen does not preclude an involvement of such strains in disease, because there is a plethora of other virulence factors not yet evaluated. One of these is the recently characterized C. perfringens b2 toxin, which has been associated with both necrotic enteritis in piglets and equine typhlocolitis [3,8].
C. difficile is the major cause of antibiotic-associated pseudomembranous colitis in human patients. It has also been associated with diarrhoea and enterocolitis in foals and adults horses, as well as diarrhoea in dogs [6].
Three toxins produced by C. difficile have been described: toxin A (TcdA, enterotoxin), toxin B (TcdB, cytotoxin), and an adenosine diphosphate (ADP)-ribosyltransferase (binary toxin, CDT). Diseases associated with C. difficile have primarily been attributed to the activity of TcdA and TcdB, and strains have historically been thought to produce both toxins (toxigenic isolates) or neither (non-toxigenic). There are increasing reports of variant strains isolated from human clinical cases of C. difficile-associated infection (CDI) that produce only TcdA or TcdB, however [2].
Current diagnosis of C. difficile-associated diarrhoea is primarily based on detection of TcdA and/or TcdB in faecal specimens by EIA. Isolation of the microorganism alone is not sufficient for diagnosis, due to the presence of non-toxigenic strains. Toxigenic C. difficile has been isolated from dogs with chronic diarrhoea, and reports have documented a carriage rate of C. difficile ranging from 0% - 40% in diarrhoeic and non-diarrhoeic dogs [2, 9]. Toxigenic C. difficile can be isolated from up to 94% of neonate dogs in the absence of clinical signs of disease [2]. Clinical signs that have been associated with canine C. difficile infection range from asymptomatic carriage to a potentially fatal acute hemorrhagic diarrhoeal syndrome.
A simple and rapid method is needed to differentiate toxigenic and non-toxigenic strains of C. perfringens and C. difficile in animals. In this regard, the objective of the current study was to investigate the molecular characteristics of various strains of C. perfringens and C. difficile isolates from diarrhoeic and non-diarrhoeic dogs, through the use of toxin gene profiling and PCR-ribotyping.
2. Materials and Methods
2.1. Samples
Ninety-five faecal samples were collected over an 8 month period (July 2006-March 2007) from diarrhoeic (n = 36) and non-diarrhoeic (n = 59) dogs. Thirty-eight were shelter dogs (diarrhoeic n = 3, non-diarrhoeic n = 35), 47 were privately-owned dogs (diarrhoeic n = 26, non-diarrhoeic n = 21) belonging to students or staff of the Veterinary Medicine Faculty of Parma (Italy), and another 10 dogs were patients at the Faculty Veterinary Hospital (diarrhoeic n = 7, non-diarrhoeic n = 3). Assays were performed on specimens collected within 3 hours after natural voiding. After analysis, samples were immediately stored at −20˚C.
2.2. Faecal Culture
All faecal samples were cultured onto pre-reduced Schaedler agar plates (Oxoid, Basingstoke, Hampshire, England), and at the same time inoculated into cooked meat broth (Oxoid, England). Samples were also streaked onto pre-reduced selective medium containing cycloserinecefoxitin-fructose agar (CCFA) for C. difficile isolation. Plates were incubated anaerobically at 37˚C for 48 - 72 hours. After 3 days of incubation into cooked meat broth, the samples were subjected to heat shock for spore selection and then cultured onto Schaedler agar and/or CCFA. Clostridium identification was confirmed through the Rapid ID32A (bioMérieux SA, Marcy-l’Etoile, France).
2.3. Reference Strains
C. perfringens ATCC 12917 cpa+/cpe+ was utilized as positive control for duplex and multiplex PCRs. C. perfringens NCTC 8346, ATCC 373, and ATCC 27324 were used as cpa+/etx+, cpa+/cpb+/cpb2+ and cpa+/ iap+/cpe+/cpb2+ controls, respectively, for multiplex PCR. C. difficile VPI 10463 and 51377 were used as C. difficile tcdA+/tcdB+ and cdtA+/cdtB+ controls, respectively. A strain characterized as PCR ribotype 078 was utilized to compare the PCR-ribotyping banding patterns.
2.4. Rapid Immunoassays
For rapid, in vivo, detection of TcdA/B in faecal samples, a commercial microplate EIA was performed according to manufacturer instructions (ProSpecT Clostridium difficile Toxin A/B, Remel, Lenexa, Kansas, USA). The, in vitro, toxin production by C. difficile was detected by two distinct immunological tests (ProSpecT Clostridium difficile Toxin A/B, Remel, USA, and C. diff Quik Chek CompleteTM, TechLab, Princeton, USA) on isolates following 3 and 5 days of anaerobic growth into cooked meat broth. C. difficile VPI 10463 was used as TcdA+/ TcdB+ positive control.
2.5. Extraction of C. perfringens and C. difficile DNA
For each C. perfringens or C. difficile strain, a 100 ml suspension of cells in sterile water was vortexed, incubated at 100˚C for 5 and 10 min., respectively, and centrifuged at 12,000 g (Microliter Centrifuge, Hermle Z 233 M-2, Delchimica Scientific Glassware s.r.l.) for 2 min. Five ml of this preparation were used as the DNA template for all PCR assays. All PCRs were performed with a Techne TC-32 thermal cycler (Barloworld Scientific Ltd, Milano, Italy).
2.6. Duplex PCR for the C. perfringens Phospholipase C (PLC) and CPE Encoding Genes
All C. perfringens isolates and the ATCC 12917 reference strain were PCR-screened for the presence of PLC and CPE-encoding genes as previously described by Fach and Popoff [10]. Amplified products were subjected to 1.5% agarose gel electrophoresis (120 V, 1 h) and visualized by ethidium bromide staining and ultraviolet light exposure.
2.7. Multiplex PCR for the C. perfringens Toxins Encoding Genes
All C. perfringens isolates, along with the four reference strains, were PCR-subjected for the detection of a (cpa), b (cpb), e (etx), CPE (cpe), i (iap), and b2 (cpb2) toxin encoding genes, as described by Baums et al. [3]. The reaction products were subjected to agarose gel electrophoresis as mentioned above.
2.8. Duplex PCRs for the C. difficile TcdA/B and Binary Toxin Encoding Genes
All C. difficile isolates and the reference strains were PCR-screened for the presence of (a) TcdA/B-encoding genes (624-bp tcdA and 412-bp tcdB gene fragments), as previously described by Spigaglia and Mastrantonio [11], and (b) binary toxin genes (375-bp cdtA and 510-bp cdtB gene fragments), as described by Stubbs et al. [12]. The reaction products were subjected to agarose gel electrophoresis as above.
2.9. C. difficile PCR-Ribotyping
PCR-ribotyping was conducted with the primer pair RtFR1/RtFR2, as described by Bidet et al. [13,14]. The amplified products were analyzed by 3% gel electrophoresis (85 V, 5 h) and visualized as above.
3. Results
Sixty-two faecal samples were positive for Clostridium spp. presence (62/95 samples, 65.3%, confidence interval 95%: 55.3 to 74.3). Eighty-nine Clostridium spp. were isolated from the 62 positive faecal specimens. Frequently, more than one species of clostridia was observed in the same faecal sample. The completed results were published in a precedent work [15].
Overall, 14 dogs were positive for C. perfringens (14/ 95: 14.7%; I.C. 95.0%: 8.6 to 23.0). The isolation rate from diarrhoeic dogs (6/36: 16.7%) was similar to the rate from healthy dogs (8/59: 13.6%). The difference was statistically not significant at 95% level (P = 0.679, Upton’s Chi-square test). In one dog, affected by megaesophagus and treated with antibiotics for enteritis, C. difficile was also isolated [15].
None of the 14 strains were CPE-positive (plc+/cpe–) by duplex PCR. This result was confirmed by multiplex PCR assay (cpa+/cpe–). In particular, 13 isolates (13/14: 92.9%) were type A (cpa+), of which 3 (3/13: 23.1%) possessed the CPB2 toxin-encoding gene. Finally, 1 strain (1/14: 7.1%) was type D (cpa+/etx+) and possessed CPB2 gene (Figure 1). On the whole, 4 of the 14 strains (28.6%) tested cpb2-positive. Three of them (75.0%) were from diarrhoeic dogs, and 1 (25.0%) was from nondiarrhoeic dog. This difference was statistically not significant at 95% level (P = 0.486, Fisher’s Exact test).
Six type A strains (3 cpa+, and 3 cpa+/cpb2+) were isolated from faecal samples of dogs with enteritis. The other 7 type A isolates and the type D strain were from canine non-diarrhoeic faeces.
Eight of 10 (80%) C. difficile culture-positive samples belonged to diarrhoeic dogs, 5 of which with enteritis
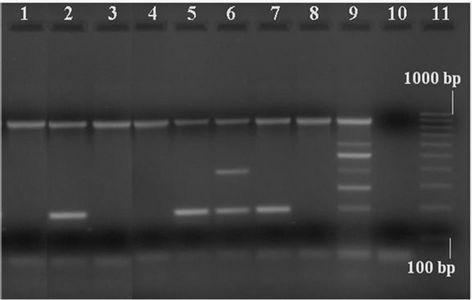
Figure 1. Detection of toxins encoding genes by multiplex PCR in Clostridium perfringens strains isolated from dogs. Lanes 1, 3, 4 and 8: type A strains (cpa+); lanes 2, 5 and 7: type A, cpb2+ strains; lane 6: type D, cpb2+ strain; lane 9: C. perfringens positive control (cpa+/cpb+/cpe+/etx+/iap+/cpb2+); lane 10: negative control (“0 DNA”); lane 11: molecular size markers (100 bp Molecular Ruler, Biorad, Italy).
after antibiotic therapy and 3 not treated with antibiotics since at least 6 months. The majority of C. difficile isolates (6/10, 60.0%) were toxigenic (tcdA+/tcdB+) and possessed cdtA and cdtB genes. All faeces tested EIAnegative. On the contrary, all PCR-positive strains were positive for, in vitro, toxin production when tested by both immunological tests. The isolation rates of C. difficile from diarrhoeic dogs (8/36, 22.2%) and non-diarrhoeic dogs (2/59, 3.4%) were statistically different (P = 0.006, Fisher’s Exact Test).
The proportion of toxigenic isolates (5/8, 62.5%) in diarrhoeic dogs was similar to the proportion (1/2, 50.0%) in non-diarrhoeic dogs. Such difference was not significant (P = 0.667, Fisher’s Exact Test).
Finally, the 10 C. difficile strains were subjected to ribotype analysis by comparing the primer-targeted amplicons of the intergenic spacer region localized between the 16S and the 23S rRNA genes. Eight ribotypes were noted (arbitrarily designated VETPR 1 - 8) (Figure 2). The observed ribotype distribution suggested wide diversity of C. difficile within the dog population. In particular, one ribotype (VETPR1) was predominant among the isolates, comprising 3/10 total strains (30.0%) (derived from 2 diarrhoeic and 1 non-diarrhoeic dogs) with a tcdA+/tcdB+ and cdtA+/cdtB+ genotype (Table 1). None of the observed ribotypes showed the ribotype 078.
4. Discussion
Detection of C. perfringens and C. difficile in canine faeces is important. It has been well documented that culture isolation of C. perfringens has not diagnostic value for canine C. perfringens-associated diarrhoea. Culture may be useful in procuring isolates for toxin neutralization tests and molecular techniques like PCR to
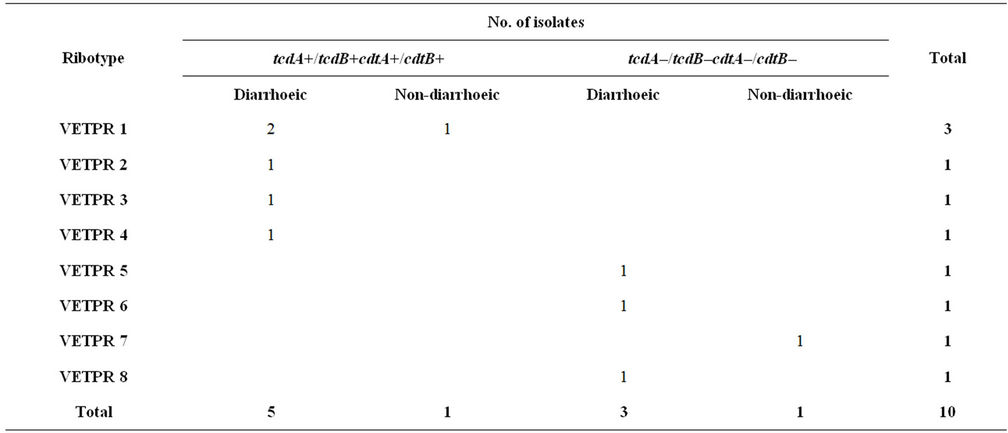
Table 1. Clostridium difficile PCR-ribotype prevalence versus toxin profile by PCR.

Figure 2. PCR-ribotyping of Clostridium difficile strains isolated from dogs. Lanes 1 and 12: molecular size markers (100 bp DNA Ladder, Celbio, Milano, Italy); lanes 2-11: C. difficile isolates. In particular, lanes 2, 4 and 5: ribotype VETPR 1; lane 3: VETPR 2; lane 6: VETPR 3; lane 7: VETPR 4; lane 8: VETPR 5; lane 9: VETPR 6; lane 10: VETPR 7; lane 11: VETPR 8; lane 13: ribotype 078.
detect specific toxin genes, or molecular typing of strains to establish clonality in suspected outbreaks. Two commercially available immunoassays are currently used in veterinary diagnostic laboratories for CPE. It is important to note that the performance of these assays have not been validated in the dog, and there are concerns about their sensitivities and specificities [2]. Moreover, they not detect the CPB2 or other toxins.
The high rate of occurrence of cpb2-positivity among strains isolated from animals with enteritis would give strength to the hypothesis that CPB2 plays a role in pathogenesis of the disease [8,16]. On the contrary, the detection of strains harbouring cpb2 in healthy animals is not necessary itself a risk, although b2-toxigenic C. perfringens can become an emerging health threat when associated to enteric dysbiosis or immunosuppression [17].
In this work, the frequency of C. perfringens isolation from healthy and diarrhoeic dogs was similar. By multiplex PCR, 13 out of the 14 C. perfringens strains belonged to type A. This is in accord with literature [2]. Only one isolate tested type D. None strain resulted cpepositive, but a relatively high percentage of strains (4/14: 28.6%) were cpb2-positive. On the contrary, the type D isolate, positive for cpb2, came from a healthy dog.
We can not conclude that CPB2 is responsible for the enteritis in our strains because we didn’t verify the b2 protein expression in vitro, although we found a high revelation percentage of cpb2-positive diarrhoeic dogs. It may be important to consider the use of an additional method for the detection of CPB2 in cpb2-positive isolates, such as neutralization test. Preferably, detection of CPB2 should be performed directly from the tissue in enteritis cases where CPB2 may be expected to play a role [8].
Concerning C. difficile, the role that this microorganism plays in dogs is not well defined, and only a few studies evaluating the presence of toxins in diarrhoeic and non-diarrhoeic animals have been done [2].
The laboratory diagnosis of C. difficile-associated diarrhoea in the dog is controversial. The apparently high prevalence of EIA-positive, culture-negative canine specimens obtained with some commercial assays, never validated in the dog, is questionable, and may represent the consequence of false-positive results [2].
The results of this study confirmed the low sensitivity of EIA when performed directly on faecal specimens. This low sensitivity is not surprising, since none of the commercial EIA kits currently available has been validated in the dog. In contrast, the sensitivity and specificity for TcdA/B detection were higher when EIA was performed directly on isolates rather than on faecal samples. However, these results should be interpreted with caution, as toxins production, in vitro, does not automatically imply that toxin is produced and secreted in the intestinal tract [9].
Our significatively higher isolation rates from diarrhoeic dogs compared to non-diarrhoeic are in disagreement with previous reports [9]. However, it is important to underline that 5 out of the 10 C. difficile strains were isolated from dogs with enteritis consequent to antibiotic therapy which could have caused an overgrowth of C. difficile in intestine, thus predisposing the animals to enteritis.
The majority of C. difficile strains (60.0%) were toxigenic on the basis of results of the duplex PCR assays for the identification of TcdA/B and binary toxin genes. The carriage rates of toxigenic isolates in diarrhoeic dogs (62.5%) was similar than those in non-diarrhoeic dogs (50.0%). These findings are in agreement with those reported in previous studies [9,18].
None of our ribotypes showed the ribotype 078 that has emerged as hypervirulent genotype and predominant strain in pigs and calves [19]. The comparison of our ribotypes and tcd-profiles with additional C. difficile isolates from other sources could be useful to determine whether certain ribotypes are associated with variant toxin profiles in dogs, other animals and/or humans.
In conclusion, ideally, the application of PCR assays on C. perfringens and C. difficile isolates for the detection of toxins genes, combined with EIA tests for the demonstration of toxins production (in vivo and in vitro), should be implemented for diagnosing canine disease.
The results of this study highlight that the PCR assays may provide a useful and reliable tool for C. perfringens and C. difficile genotyping in routine veterinary diagnostics. The genotype, in many cases, could provide the final piece of information needed to establish a diagnosis [20].
5. Acknowledgements
The authors wish to thank Prof. Giuseppe Dettori, Department of Pathology and Laboratory Medicine, Faculty of Medicine and Surgery, University of Parma for his scientific contribute.
This work was supported by a grant of Local Funds for Research of University of Parma, FIL 2008.
REFERENCES
- S. L. Marks and E. J. Kather, “Antimicrobial Susceptibilities of Canine Clostridium difficile and Clostridium perfringens Isolates to Commonly Utilized Antimicrobial Drugs,” Veterinary Microbiology, Vol. 94, No. 1, 2003, pp. 39-45. doi:10.1016/S0378-1135(03)00061-0
- S. L. Marks and E. J. Kather, “Bacterial-Associated Diarrhea in the Dog: A Critical Appraisal,” Veterinary Clinics of North America: Small Animal Practice, Vol. 33, No. 5, 2003, pp. 1029-1060. doi:10.1016/S0195-5616(03)00091-3
- C. G. Baums, U. Shotte, G. Amtsberg and R. Goethe, “Diagnostic Multiplex PCR for Toxin Genotyping of Clostridium perfringens Isolates,” Veterinary Microbiology, Vol. 100, No. 1-2, 2004, pp. 11-16. doi:10.1016/S0378-1135(03)00126-3
- J. G. Smendley III, D. J. Fisher, S. Sayeed, G. Chakrabarti and B. A. McClane, “The Enteric Toxins of Clostridium perfringens,” Reviews of Physiology, Biochemistry and Pharmacology, Vol. 152, 2004, pp. 183-204. doi:10.1007/s10254-004-0036-2
- J. G. Songer and R. R. Meer, “Genotyping of Clostridium perfringens by Polymerase Chain Reaction Is a Useful Adjunct to Diagnosis of Clostridial Enteric Disease in Animals,” Anaerobe, Vol. 2, No. 4, 1996, pp. 197-203. doi:10.1006/anae.1996.0027
- S. L. Marks, “Bacterial Gastroenteritis in Dogs and Cats,” Proceedings of 28th World Congress of the World Small Animal Veterinary Association, Bangkok, 24-27 October 2003.
- S. B. Melville, R. E. Collie and B. A. McClane, “Regulation of Enterotoxin Production in Clostridium perfringens,” In: J. I. Rood, B. A. McClane, J. G. Songer and R. W. Titball, Eds., The Clostridia: Molecular Biology and Pathogenesis, San Diego, Academic Press, 1997, pp. 471- 485.
- D. M. Bueschel, B. H. Jost, S. J. Billington, H. T. Trinh and J. G. Songer, “Prevalence of cpb2, Encoding Beta2 toxin, in Clostridium perfringens Field Isolates: Correlation of Genotype with Phenotype,” Veterinary Microbiology, Vol. 94, No. 2, 2003, pp. 121-129. doi:10.1016/S0378-1135(03)00081-6
- N. Chouicha and S. L. Marks, “Evaluation of Five Enzyme Immunoassays Compared with the Cytotoxicity Assay for Diagnosis of Clostridium difficile-Associated Diarrhoea in Dogs,” Journal of Veterinary Diagnostic Investigation, Vol. 18, No. 2, 2006, pp. 182-188. doi:10.1177/104063870601800207
- P. Fach and M. R. Popoff, “Detection of Enterotoxigenic Clostridium perfringens in Food and Faecal Samples with a Duplex PCR and the Slide Latex Agglutination Test,” Applied and Environmental Microbiology, Vol. 63, No. 11, 1997, pp. 4232-4236.
- P. Spigaglia and P. Mastrantonio, “Molecular Analysis of the Pathogenicity Locus and Polymorphism in the Putative Negative Regulator of Toxin Production (TcdC) among Clostridium difficile Clinical Isolates,” Journal of Clinical Microbiology, Vol. 40, No. 9, 2002, pp. 3470- 3475. doi:10.1128/JCM.40.9.3470-3475.2002
- S. Stubbs, M. Rupnik, M. Gilbert, J. Brazier, B. Duerden and M. Popoff, “Production of Actin-Specific ADP-Ribosyltransferase (Binary Toxin) by Strains of Clostridium difficile,” FEMS Microbiology Letters, Vol. 186, No. 2, 2000, pp. 307-312. doi:10.1111/j.1574-6968.2000.tb09122.x
- P. Bidet, F. Barbut, V. Lalande, B. Burghoffer and J. C. Petit, “Development of a New PCR-Ribotyping Method for Clostridium difficile Based on Ribosomal RNA Gene Sequencing,” FEMS Microbiology Letters, Vol. 175, No. 2, 1999, pp. 261-266. doi:10.1111/j.1574-6968.1999.tb13629.x
- P. Bidet, V. Lalande, B. Salauze, B. Burghoffer, V. Avesani, M. Delmée, A. Rossier, F. Barbut and J. C. Petit, “Comparison of PCR-Ribotyping, Arbitrarily Primed PCR, and Pulsed-Field Gel Electrophoresis for Typing Clostridium difficile,” Journal of Clinical Microbiology, Vol. 38, No. 7, 2000, pp. 2484-2487.
- L. Zerbini and M. C. Ossiprandi, “Prevalence of Clostridium spp. in Diarrhoeic and Healthy Dogs,” In: Annali della Facoltà di Medicina Veterinaria dell’Università di Parma, Parma, Vol. 27, 2007, pp. 143-156.
- S. Thiede, R. Goethe and G. Amtsberg, “Prevalence of β2 Toxin Gene of Clostridium perfringens Type A from Diarrhoeic Dogs,” Veterinary Record, Vol. 149, No. 9, 2001, pp. 273-274. doi:10.1136/vr.149.9.273
- U. Schotte, U. Truyen and H. Neubauer, “Significance of b2-Toxigenic Clostridium perfringens Infections in Animals and Their Predisposing Factors—A Review,” Journal of Veterinary Medicine, Vol. 51, No. 10, 2004, pp. 423-426. doi:10.1111/j.1439-0450.2004.00802.x
- R. M. Batt and C. Rutgers, “Bacteria and Intestinal Disease in Dogs,” GDBA Technical Review No 11, UK, 1997.
- K. Keel, J. S. Brazier, K. W. Post, S. Weese and J. G. Songer, “Prevalence of PCR Ribotypes among Clostridium difficile Isolates from Pigs, Calves, and Other Species,” Journal of Veterinary Medicine, Vol. 45, No. 6, pp. 2007, pp. 1963-1964.
- F. A. Uzal and J. G. Songer, “Diagnosis of Clostridium perfringens Intestinal Infections in Sheep and Goats,” Journal of Veterinary Diagnostic Investigation, Vol. 20, No. 3, 2008, pp. 253-265. doi:10.1177/104063870802000301
NOTES
*Corresponding author.

