Open Journal of Medical Microbiology
Vol.4 No.1(2014), Article ID:43022,9 pages DOI:10.4236/ojmm.2014.41005
Development and Evaluation of a Multiplex PCR for the Detection of Campylobacter concisus and Other Campylobacter spp. from Gastroenteritis Cases
Mohsina Huq1, Gena Gonis2, Taghrid Istivan1*
1Biotechnology and Environmental Biology, School of Applied Sciences, RMIT University, Melbourne, Australia
2Bacteriology Laboratory, Microbiology, Department of Laboratory Services, The Royal Children’s Hospital, Melbourne, Australia
Email: mohsina.huq@rmit.edu.au, gena.gonis@rch.org.au, *taghrid.istivan@rmit.edu.au
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/


Received 12 December 2013; revised 13 January 2014; accepted 20 January 2014
ABSTRACT
We developed and evaluated a multiplex PCR (m-PCR) for application in routine diagnostic laboratories to detect Campylobacter spp. in stool samples including C. concisus, C. jejuni, and C. coli. When this m-PCR was applied on spiked faecal samples, C. concisus, C. jejuni, and C. coli were specifically identified at 105 cells/gm of faeces. To compare the sensitivity of the m-PCR with conventional culture techniques, the same spiked stool samples were cultured on an antibiotic free Columbia blood agar using the filtration technique. The detection limit of conventional culture method was 105 cells/gm of stool for C. concisus and 106 cells/gm of stool for C. jejuni and C. coli. The m-PCR was applied to test 127 faecal samples from children with gastroenteritis and the results were compared with the conventional bacterial cultures data. By this m-PCR technique, C. jejuni was detected in 7 samples, C. coli in 2 samples, and C. concisus in 7 samples. However, the conventional culture results for these samples were 6 for C. jejuni, 2 for C. coli and only one sample was positive for C. concisus. In total, 19 samples were positive for Campylobacter spp. by m-PCR while only 9 samples were positive for Campylobacter spp. by culture. In conclusion, m-PCR is more sensitive than the culture technique to detect C. concisus and other fastidious campylobacters in faeces.
Keywords:Campylobacter concisus; Campylobacter spp.; Gastroenteritis; Faecal Spiking; Multiplex PCR

1. Introduction
The incidence of campylobacteriosis is gradually increasing, and Campylobacter spp. are now considered to be the leading cause of bacterial gastroenteritis worldwide [1] [2] . Campylobacter spp. colonise different parts of the human body and may cause many extra-intestinal diseases and severe long-term complications [3] . Although C. jejuni and C. coli are the most common etiological agents of campylobacteriosis [4] , there is evidence suggesting that other Campylobacter spp. such as C. concisus, C. upsaliensis, C. hyointestinalis, and C. fetus may also cause disease in humans [5] -[7] .
Campylobacter spp. are fastidious organisms and require a microaerophilic environment to grow. Most laboratory procedures are optimized to isolate the more common species: C. jejuni and C. coli. However, the other Campylobacter spp. may have additional growth requirements. For example, C. concisus is an emerging pathogen which requires hydrogen for growth and is a slow growing organism commonly found in the human oral cavity and was first described in 1981 [8] . It is usually associated with healthy than with diseased periodontal sites and in healthy sites it is usually found in shallow rather than in deeper sites [9] . Past studies have associated this species with gingivitis [10] , periodontitis [11] , and gastritis since 1989 [12] . More recently, it was isolated from foot ulcers [13] , other abscesses from different parts of the body [14] and intestinal biopsies and feacal samples of children with Crohn’s disease and patients with inflammatory bowel diseases (IBD) [15] [16] . An early molecular study reported that C. concisus produced cytotonic-like effects on CHO cells and induced intracytoplasmic vacuole formation that is similar to H. pylori [17] . Also a recent study in the United Kingdom showed a significantly higher incidence of C. concisus DNA throughout intestinal biopsies of adults with ulcerative colitis compared to those of healthy controls [18] . C. concisus requires a hydrogen enriched atmosphere for growth such as that recommended by the Cape Town Protocol [19] . If optimum growth conditions are not adhered to, it may result in a reduced isolation rate [20] . In a study in Japan, metagenomic analysis detected DNA sequences of the Campylobacter spp. genome including C. concisus, whilst the standard culture methods were negative [21] . In a recent study in The Netherlands, the detection frequency (DF) of the emerging pathogen C. concisus was found to be at least similar to the DF of C. jejuni when the species was confirmed by PCR product sequencing [22] . In a more recent study in Chile, a significant difference was reported between conventional culture methods and molecular methods used for the detection of Campylobacter spp. and Arcobacter spp. in faecal samples of patients with diarrhea and of controls. Emerging pathogens like C. concisus and C. ureolyticus were only detected by the molecular method used in the study [23] .
The true incidence of these Campylobacter spp. as causative agents of disease cannot be accurately determined using standard culture techniques. Consequently, there is a demand for a rapid, sensitive and specific method for detecting these organisms. Therefore, the objective of our study was to optimize a four-gene multiplex PCR (m-PCR) for the detection of C. jejuni, C. coli and C. concisus from faecal samples from children with gastroenteritis and to evaluate the PCR results with conventional culture results from the Bacteriology Laboratory at The Royal Children’s Hospital (RCH), Melbourne, Australia.
2. Materials and Methods
The study was approved by the Human Research Ethics Committee of RMIT University, Melbourne (Approval number: 15/08) and the Royal Children’s Hospital Human Research Ethics Committee (Approval number: CA28021).
2.1. Bacterial Strains and DNA Preparation
A panel of 15 Campylobacter spp. strains and a non-Campylobacter strain were included as control strains in the study (Table 1). Campylobacter spp. strains were grown on Columbia agar base (Oxoid, Australia) supplemented with 5% defibrinated horse blood, and incubated at 35˚C for 3 - 5 days under microaerophilic conditions (6% O2, 10% CO2, 10% H2, and balanced N2). For PCR, bacterial DNA was extracted by the Wizard Genomic DNA purification kit (Promega BioSciences, USA) following the manufacturer’s instructions.
Cell densities of liquid cultures were estimated by measuring optical density using a spectrophotometer (Eppendorf, Australia) and were also confirmed by viable counts of 10-fold serially diluted cultures (in Brucella broth, Oxoid, Australia) plated on Columbia horse blood agar plates.
Table 1. List of strains used for selectivity testinga.
aATCC, American Type Culture Collection, Manassas, VA; NCTC, National Collection of Type Cultures, London, United Kingdom.
2.2. Spiking of Faecal Samples
Three blood-free, campylobacter-culture negative faecal samples were selected for the spiking experiment. Liquid campylobacter cultures were added to the faecal samples, resulting in final campylobacter concentrations of 107, 106, 105, 104, 103 and 102 cells/gm of faeces of C. coli, C. jejuni and C. concisus. Faecal DNA was extracted by QIAamp DNA Stool Kit (Qiagen, Australia), according to the manufacturer’s instructions. One hundred microliters of the spiked faecal samples were placed on a 0.65 micron filter paper (Sartorius Stedim, Australia) and left to adsorb for 20 minutes then the filter was removed, and the plates were streaked with a loop and incubated in appropriate conditions for 2 - 5 days following The Cape Town Protocol [19] . Any visible growth of campylobacters was identified by biochemical tests including oxidase, catalase, sodium hippurate hydrolysis, indoxyl acetate hydrolysis and light microscopy.
2.3. Collection of Clinical Faecal Samples and m-PCR
To evaluate culture and molecular methods, a total of 140 faecal samples were collected from children with gastroenteritis, aged between 1 month to 18 years at the Royal Children’s Hospital (RCH), Melbourne from July to December, 2009. All samples were routinely cultured and identified at the RCH Bacteriology Laboratory on Columbia horse blood agar using the Cape Town Protocol [19] and on blood free Campylobacter media (mCCDA) (Modified Charcoal Cefoperazone Deoxycholate Agar) [24] . Plates were checked on days 3 and 5 for typical small colonies of Campylobacter spp. and were identified by biochemical characteristics as described earlier. Samples were also subjected to an in house Rota EIA and cell culture for Adenovirus, both of which were performed at RCH. Then a proportion of each faecal sample was tested by the m-PCR method developed for this study. DNA was extracted from the samples using QIAamp DNA Stool mini kit (Qiagen, Germany) according to the manufacturer’s instruction. The extracted DNA was stored at −20˚C until used in the m-PCR to amplify coding regions of three Campylobacter spp.; C. jejuni, C. coli and C. concisus and a universal coding region for Campylobacter spp. Primers selected for this m-PCR are from the hippuricase gene (hipO) of C. jejuni [25] , the aspartokinase gene of C. coli [26] , the gyrB gene of C. concisus [27] and a universal 16S rRNA gene sequence of Campylobacter spp. [5] serving as an internal positive control for the PCR (Table 2). The primers used in the present study were carefully selected so that the sizes of PCR products, produced by each of the primer sets, are distinguishable on a single gel. The primers’ sequences were checked by the Clone Manager suite of analysis tools (Sci Ed Central) to confirm that they do not form primer dimers, hair-pin loops and for crossreactivity.
The m-PCR system was optimized by serially diluting the Campylobacter spp. DNA templates and by gradient temperature cycles using a G-storm thermocycler (Gene works Pty Lt, Australia). All PCR reactions were performed in a total reaction volume of 25 µl containing 1X PCR buffer [10 mM Tris-HCl, 50 mM KCl; pH 8.3], 0.16 mM MgCl2, 0.2 mM dNTP each (Applied Biosystems, Australia), 1.25U recombinant Taq Polymerase (Roche Diagnostics, Australia), 0.1 µM of each primer, and 2.5 µl the DNA template. Thermocycler conditions were 94˚C for 2 min, followed by 30 cycles of 94˚C for 15 sec, 54˚C for 30 sec and 68˚C for 2 min, and a final extension of 7 min at 72˚C. PCR products were visualized by UV following electrophoresis on 1.5% agarose gel.
3. Results and Discussion
In our newly developed m-PCR method, PCR products with the expected size of 816 bp, 500 bp, 406 bp and 344 bp were detected for Campylobacter spp., C. coli, C. concisus and C. jejuni respectively using a mixture of the four sets of primers (Table 2) with DNA samples from the control strains (Figure 1). Blood-free campylobacter-negative faecal samples were spiked with 10-fold serial dilutions of our selected three campylobacter strains namely C. jejuni, C. coli or C. concisus cultures resulting in final concentrations of 102 - 107 cells/gm of, faeces to evaluate the detection limit of the m-PCR method in comparison with conventional culture techniques. Different amounts (40 - 200 ng/µl) of extracted DNA from the spiked faecal samples were tested by the m-PCR method for highest sensitivity and the optimal template volumes were found to be 100 ng/µl per reaction for m-PCR. C. jejuni, C. coli or C. concisus specific primers could produce an amplicon when it had a minimum concentration of 105 cells/gm in the spiked faecal sample (Figure 2). The detection limit of C. jejuni and C. coli in a spiked faecal sample was also reported to be at 105 cells per ml by Persson et al. when the researchers evaluated their developed multiplex PCR method [25] .
When the same spiked faecal samples were plated on Columbia horse blood agar plates using the filtration method following the Cape Town Protocol [19] before extracting DNA, the detection limit was 105 cells/gm of faeces for C. concisus and 106 cells/gm of faeces for C. jejuni, and C. coli. The motility and physical properties such as size and shape of C. concisus might be the explanation to be detected at a 10-fold lower concentration than C. jejuni and C. coli. The use of the filter paper to selectively allow Campylobacter spp. to pass through is considerably effective due to the complex bacterial and chemical nature of faeces. However, the the successful isolation from faecal samples depends on the viability of campylobacter spp. in the sample during processing. Campylobacters are microaerophilic in nature, and they tend to have a low survival rate if exposed to room temperature and atmospheric air [28] . Long transport time from sample collection to sample analysis reduces the viability of the bacteria in clinical samples. In addition, the progressive decrease in oxygen tension when gas
Table 2. Primer sets used in m-PCR for Campylobacter genus, C. jejuni, C. coli and C. concisus.
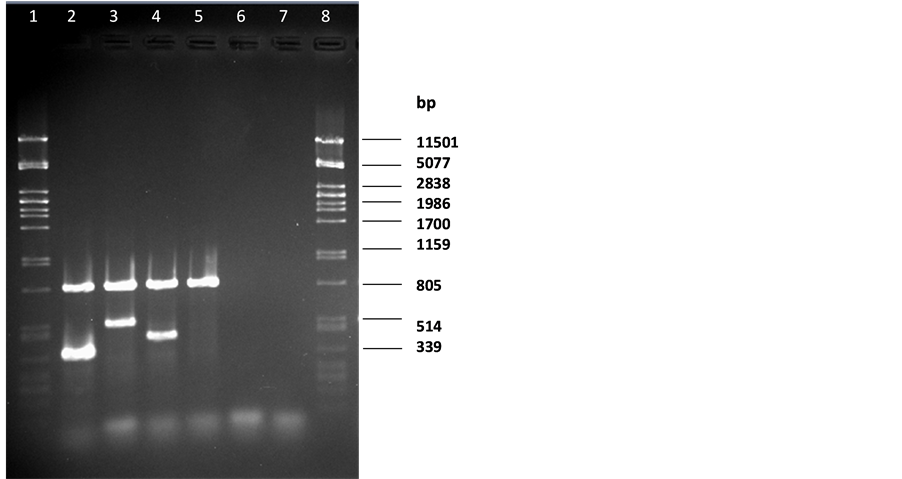
Figure 1. Multiplex PCR for Campylobacter spp. Lane 1 and 8: pstI digested lDNA marker; lane 2 C. jejuni DNA; lane 3 C. coli DNA; lane 4 C. concisus DNA; lane 5 C. mucosalis DNA; lane 6 E. coli; lane 7 reagent blank.
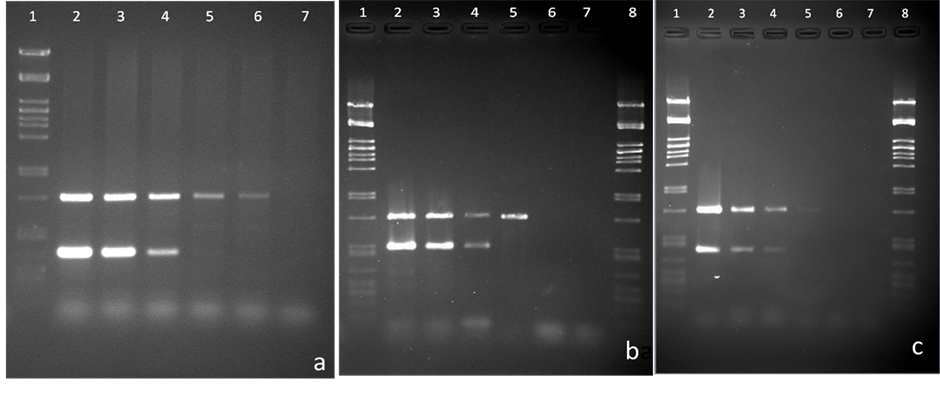
Figure 2. Sensitivity evaluation of the m-PCR method using 10-fold dilutions of bacterial DNA extracted from spiked faecal samples with C. jejuni in (a), C. coli in (b), and C. concisus in (c). Lanes 1&8: pstI digested lDNA marker. Bacterial cell concentrations (cells/gm) used: lane 2, 107; lane 3, 106; lane 4, 105; lane 5, 104; lane 6, 103; lane 7, 102.
generating kits are used may not favour adequate growth especially for the stressed bacteria in the sample. Furthermore the use of antibiotic mixtures in common selective media may inhibit the growth of certain Campylobacter spp. [29] .
To further test the routine diagnostic applicability of our m-PCR, it was compared to the conventional culturing system of 127 faecal samples at the Royal Children Hospital in Melbourne. A total of 140 faecal samples were collected and among these 13 samples were very small in volume and did not contain adequate DNA to run a PCR. Therefore, we excluded these 13 samples from the analysis. Results for positive samples by PCR or culture technique in addition to clinical symptoms and diagnosis are listed in Table 3. Ninteen samples were positive for Campylobacter spp. By m-PCR while only nine samples were positive for Campylobacter spp.
Table 3. Conventional culture and m-PCR results with clinical symptoms of positive faecal samples.
aRota EIA, Rotavirus Enzyme Immunoabsorbent Assay; *Giardia cysts were also detected; ND, Not done.
by culture. For C. jejuni seven samples were positive by m-PCR while six samples were positive by culture method. It is worth mentioning that C. jejuni was isolated in the hospital from three other samples by culture; however these faecal samples did not have adequate DNA when measured by the spectrophotometer, following DNA extraction, and were excluded from the study as mentioned above. For C. coli, two samples were positive by both m-PCR and culture methods. Seven samples were positive for C. concisus by m-PCR while only one was detected by culture. In the m-PCR, all samples which produced PCR products for C. jejuni, C. coli and C. concisus, also produced a PCR product with the genus specific Campylobacter spp. Primers.
By applying the m-PCR in this study we were able to detect a co-infection of C. jejuni with C. coli in one of the tested samples, while only C. coli was detected in this sample by the conventional culture method. This could be due to overgrowth of C. coli, or that only one Campylobacter spp. representative colony was picked to perform biochemical tests. Co-infections with Campylobacter spp. have been reported previously [30] [31] and mixed infection of Campylobacter species were previously identified from stool samples by PCR and PCR-enzyme-linked immunosorbent assay [30] . Likewise, co-infections of different subspecies of C. jejuni in stool samples were previously identified by Pulsed-field gel electrophoresis and by flagellin gene typing [31] . Linton et al. (1997) also reported co-infection with C. jejuni and C. hyointestinalis by PCR method directly from stool samples [26] .
Interestingly in our study C. concisus DNA was detected in seven faecal samples while only one was isolated by the conventional culture method. As C. concisus is a fastidious hydrogen-requiring bacterium its isolation is difficult. Yet, only a small amount of its DNA can be detected by the more sensitive PCR method. Culture methods were previously compared to molecular techniques for detection of C. concisus and other fastidious campylobacters and were found to be less sensitive, therefore PCR would be the method of choice for the detection of these campylobacters [6] [29] . Furthermore, Giardia cysts were found in one of our samples that was positive for C. concisus by PCR only. There could be a co-infection of Giardia and C. concisus leading to gastroenteritis in this patient. Giardia lamblia is a prevalent enteric pathogen causing both asymptomatic carriage and diarrheal illness among children worldwide and there are some reports of co-infection of Campylobacter spp. with helminths and protozoans [32] .
It is worth noting that the faecal sample which was positive for C. concisus by both culture and PCR was from a patient previously diagnosed with Crohn’s disease. There are reports that C. concisus could be related to Crohn’s disease in children [33] and adults [34] . Several virulence markers have been previously identified in C. concisus such as secreted and cell-associated hemolytic activities and flagellum-mediated attachment [35] [36] . Furthermore, in this study four faecal samples which were negative by culture method produced a Campylobacter spp. genus specific PCR product by m-PCR. However, no specific PCR product was detected for any of C. coli, C. jejuni or C. concisus, indicating the presence of another Campylobacter spp. Yet, we could not determine the species because specific primers for other Campylobacter spp. were not included in the m-PCR, which is intended to be done in the future. This suggests that PCR is more sensitive than conventional culture method for detecting unusual Campylobacter spp. [6] [29] . It is also worth mentioning that Adenovirus was detected by conventional culture technique in one of these four samples, but no other infectious agents were detected in the other three samples. Molecular methods for detection of Campylobacter spp. are known to be more sensitive than culture [22] [23] [37] . Molecular tests are particularly advantageous for detection of non-cultivable organisms or those difficult to grow by conventional cultures. Detection of these organisms will help to identify them as potential causative agents of disease.
4. Conclusion
In conclusion, the present method offers a fast and robust identification of Campylobacter spp. in particular C. concisus, C. coli and C. jejuni present in faecal samples. The use of genus-specific primers also serves as an additional control in this method. Diagnostic PCR on template DNA extracted directly from the primary clinical source offers attractive advantages including reduced identification time and detection of non-viable and noncultivable bacteria contained in the sample. This m-PCR method with respect to sensitivity and specificity makes it suitable for application in diagnostic laboratories for the detection of Campylobacter spp. in clinical specimens.
Acknowledgements
This study was funded by an ECR grant from RMIT University to Dr. Istivan. We thank RMIT University, Microbiology Teaching Unit for providing us the clinical isolates of Campylobacter spp. to use as controls. We also thank staff members in Bacteriology Laboratory at The Royal Children’s Hospital, Melbourne, for their assistance in sample collection.
Conflict of Interest Statement
None of the authors has any conflicts of interest associated with this study.
References
- Anonymous (2012) Monitoring the Incidence and Causes of Diseases Potentially Transmitted by Food in Australia: Annual Report of the OzFoodNet Network, 2010. Communicable Diseases Intelligence, 36, E213-E241.
- Eurosurveillance Editorial Team (2012) The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-Borne Outbreaks in 2010. Euro Surveillance, 17, 21. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20113
- Man, S.M. (2011) The Clinical Importance of Emerging Campylobacter Species. Nature Reviews Gastroenterology & Hepatology, 8, 669-685. http://dx.doi.org/10.1038/nrgastro.2011.191
- Skirrow, M.B. (1994) Diseases Due to Campylobacter, Helicobacter and Related Bacteria. Journal of Comparative Pathology, 111, 113-149. http://dx.doi.org/10.1016/S0021-9975(05)80046-5
- Linton, D., Owen, R.J. and Stanley, J. (1996) Rapid Identification by PCR of the Genus Campylobacter and of Five Campylobacter Species Enteropathogenic for Man and Animals. Research in Microbiology, 147, 707-718. http://dx.doi.org/10.1016/S0923-2508(97)85118-2
- Kulkarni, S.P., Lever, S., Logan, J.M., Lawson, A.J., Stanley, J. and Shafi, M.S. (2002) Detection of Campylobacter Species: A Comparison of Culture and Polymerase Chain Reaction Based Methods. Journal of Clinical Pathology, 55, 749-753. http://dx.doi.org/10.1136/jcp.55.10.749
- Samie, A., Obi, C.L., Barrett, L.J., Powell, S.M. and Guerrant, R.L. (2007) Prevalence of Campylobacter Species, Helicobacter pylori and Arcobacter Species in Stool Samples from the Venda Region, Limpopo, South Africa: Studies Using Molecular Diagnostic Methods. Journal of Infection, 54, 558-566. http://dx.doi.org/10.1016/j.jinf.2006.10.047
- Tanner, A.C.R., Lai, C.H., Listgarten, M.A., Visconti, R.A. and Socransky, S.S. (1981) Wolinella gen. nov., Wolinella succinogenes (Vibrio succinogenes Wolin et al.) comb. nov., and Description of Bateroides gracilis sp.nov., Wolinella recta sp. nov., Camylobacter concisus sp. nov., and Eikenella corrodens from Humans with Periodontal Disease. International Journal of Systematic Bacteriology, 31, 432-445. http://dx.doi.org/10.1099/00207713-31-4-432
- Macuch, P.J. and Tanner, A.C. (2000) Campylobacter Species in Health, Gingivitis, and Periodontitis. Journal of Dental Research, 79, 785-792. http://dx.doi.org/10.1177/00220345000790021301
- Socransky, S.S., Haffajee, A.D., Cugini, M.A., Smith, C. and Kent Jr., R.L. (1998) Microbial Complexes in Subgingival Plaque. Journal of Clinical Periodontology, 25, 134-144. http://dx.doi.org/10.1111/j.1600-051X.1998.tb02419.x
- Kamma, J.J., Nakou, M. and Manti, F.A. (1994) Microbiota of Rapidly Progressive Periodontitis Lesions in Association with Clinical Parameters. Journal of Periodontology, 65, 1073-1078. http://dx.doi.org/10.1902/jop.1994.65.11.1073
- Vandamme, P., Falsen, E., Pot, B., Hoste, B., Kersters, K. and De Ley, J. (1989) Identification of EF Group 22 Campylobacters from Gastroenteritis Cases as Campylobacter concisus. Journal of Clinical Microbiology, 27, 1775-1781.
- Johnson, C.C. and Finegold, S.M. (1987) Uncommonly Encountered, Motile, Anaerobic Gram-Negative Bacilli Associated with Infection. Reviews of Infectious Diseases, 9, 1150-1162. http://dx.doi.org/10.1093/clinids/9.6.1150
- De Vries, J.J., Arents, N.L. and Manson, W.L. (2008) Campylobacter Species Isolated from Extra-Oro-Intestinal Abscesses: A Report of Four Cases and Literature Review. European Journal of Clinical Microbiology & Infectious Diseases, 27, 1119-1123. http://dx.doi.org/10.1007/s10096-008-0550-2
- Zhang, L., Man, S.M., Day, A.S., Leach, S.T., Lemberg, D.A., Dutt, S., Stormon, M., Otley, A., O’Loughlin, E.V., Magoffin, A., Ng, P.H. and Mitchell, H. (2009) Detection and Isolation of Campylobacter Species Other than C. jejuni from Children with Crohn’s Disease. Journal of Clinical Microbiology, 47, 453-455. http://dx.doi.org/10.1128/JCM.01949-08
- Kalischuk, L.D. and Inglis, G.D. (2011) Comparative Genotypic and Pathogenic Examination of Campylobacter concisus Isolates from Diarrheic and Non-Diarrheic Humans. BMC Microbiology, 11, 53. http://dx.doi.org/10.1186/1471-2180-11-53
- Musmanno, R.A., Russi, M., Figura, N., Guglielmetti, P., Zanchi, A., Signori, R. and Rossolini, A. (1998) Unusual Species of Campylobacters Isolated in the Siena Tuscany Area, Italy. New Microbiology, 21, 15-22.
- Mukhopadhya, I., Thomson, J.M., Hansen, R., Berry, S.H., El-Omar, E.M. and Hold, G.L. (2011) Detection of Campylobacter concisus and Other Campylobacter Species in Colonic Biopsies from Adults with Ulcerative Colitis. PLoS One, 6, e21490. http://dx.doi.org/10.1371/journal.pone.0021490
- Le Roux, E. and Lastovica, A.J. (1998) The Cape Town Protocol: How to Isolate the Most Campylobacters for Your Dollar, Pound, Franc, Yen, etc. In: Lastovica, A.J., Newell, D.G. and Lastovica, E.E., Eds., International Workshop on Campylobacter, Helicobacter and Related Organisms, Institute of Child Heath, Cape Town, 30-33.
- Bessede, E., Delcamp, A., Sifre, E., Buissonniere, A. and Megraud, F. (2011) New Methods for Detection of Campylobacters in Stool Samples in Comparison to Culture. Journal of Clinical Microbiology, 49, 941-944. http://dx.doi.org/10.1128/JCM.01489-10
- Nakamura, S., Maeda, N., Miron, I.M., Yoh, M., Izutsu, K., Kataoka, C., Honda, T., Yasunaga, T., Nakaya, T., Kawai, J., Hayashizaki, Y., Horii T. and Iida, T. (2008) Metagenomic Diagnosis of Bacterial Infections. Emerging Infectious Diseases, 14, 1784-1786. http://dx.doi.org/10.3201/eid1411.080589
- De Boer, R.F., Ott, A., Guren, P., van Zanten, E., van Belkum, A. and Kooistra-Smid, A.M. (2013) Detection of Campylobacter Species and Arcobacter butzleri in Stool Samples by Use of Real-Time Multiplex PCR. Journal of Clinical Microbiology, 51, 253-259. http://dx.doi.org/10.1128/JCM.01716-12
- Collado, L., Gutierrez, M., Gonzalez, M. and Fernandez, H. (2013) Assessment of the Prevalence and Diversity of Emergent Campylobacteria in Human Stool Samples Using a Combination of Traditional and Molecular Methods. Diagnostic Microbiology and Infectious Disease, 75, 434-436. http://dx.doi.org/10.1016/j.diagmicrobio.2012.12.006
- Lopez, L., Castillo, F.J., Clavel, A. and Rubio, M.C. (1998) Use of a Selective Medium and a Membrane Filter Method for Isolation of Campylobacter Species from Spanish Paediatric Patients. European Journal of Clinical Microbiology & Infectious Diseases, 17, 489-492. http://dx.doi.org/10.1007/BF01691131
- Persson, S. and Olsen, K.E. (2005) Multiplex PCR for Identification of Campylobacter coli and Campylobacter jejuni from Pure Cultures and Directly on Stool Samples. Journal of Medical Microbiology, 54, 1043-1047.
- Linton, D., Lawson, A.J., Owen, R.J. and Stanley, J. (1997) PCR Detection, Identification to Species Level, and Fingerprinting of Campylobacter jejuni and Campylobacter coli Direct from Diarrheic Samples. Journal of Clinical Microbiology, 35, 2568-2572.
- Matsheka, M.I., Lastovica, A.J. and Elisha, B.G. (2001) Molecular Identification of Campylobacter concisus. Journal of Clinical Microbiology, 39, 3684-3689. http://dx.doi.org/10.1128/JCM.39.10.3684-3689.2001
- Holler, C., Witthuhn, D. and Janzen-Blunck, B. (1998) Effect of Low Temperatures on Growth, Structure, and Metabolism of Campylobacter coli SP10. Applied and Environmental Microbiology, 64, 581-587.
- Lawson, A.J., Shafi, M.S., Pathak, K. and Stanley, J. (1998) Detection of Campylobacter in Gastroenteritis: Comparison of Direct PCR Assay of Faecal Samples with Selective Culture. Epidemiology & Infection, 121, 547-553. http://dx.doi.org/10.1017/S0950268898001630
- Lawson, A.J., Logan, J.M., O’Neill, L., Desai, G.M. and Stanley, J. (1999) Large-Scale Survey of Campylobacter Species in Human Gastroenteritis by PCR and PCR-Enzyme-Linked Immunosorbent Assay. Journal of Clinical Microbiology, 37, 3860-3864.
- Richardson, J.F., Frost, J.A., Kramer, J.M., Thwaites, R.T., Bolton, F.J., Wareing, D.R. and Gordon, J.A. (2001) Coinfection with Campylobacter Species: An Epidemiological Problem? Journal of Applied Microbiology, 91, 206-211. http://dx.doi.org/10.1046/j.1365-2672.2001.01377.x
- Jensen, L.A., Marlin, J.W., Dyck, D.D. and Laubach, H.E. (2009) Prevalence of Multi-Gastrointestinal Infections with Helminth, Protozoan and Campylobacter spp. in Guatemalan Children. The Journal of Infection in Developing Countries, 3, 229-234.
- Man, S.M., Zhang, L., Day, A.S., Leach, S.T., Lemberg, D.A. and Mitchell, H. (2010) Campylobacter concisus and Other Campylobacter Species in Children with Newly Diagnosed Crohn’s Disease. Inflammatory Bowel Diseases, 16, 1008-1016. http://dx.doi.org/10.1002/ibd.21157
- Mahendran, V., Riordan, S.M., Grimm, M.C., Tran, T.A., Major, J., Kaakoush, N.O., Mitchell, H. and Zhang, L. (2011) Prevalence of Campylobacter Species in Adult Crohn’s Disease and the Preferential Colonization Sites of Campylobacter Species in the Human Intestine. PLoS One, 6, e25417. http://dx.doi.org/10.1371/journal.pone.0025417
- Istivan, T.S., Smith, S.C., Fry, B.N. and Coloe, P.J. (2008) Characterization of Campylobacter concisus Hemolysins. FEMS Immunology and Medical Microbiology, 54, 224-235. http://dx.doi.org/10.1111/j.1574-695X.2008.00467.x
- Kaakoush, N.O., Deshpande, N.P., Wilkins, M.R.T., Burgos-Portugal, J.A., Raftery, M.J., Day, A.S., Lemberg, D. A. and Mitchell, H. (2011) The Pathogenic Potential of Campylobacter concisus Strains Associated with Chronic Intestinal Diseases. PLoS One, 6, e29045. http://dx.doi.org/10.1371/journal.pone.0029045
NOTES

*Corresponding author.


