Journal of Biosciences and Medicines
Vol.04 No.12(2016), Article ID:72720,8 pages
10.4236/jbm.2016.412018
A Complete and Accurate Short Sequence Alignment Algorithm for Repeats
Shuaibin Lian*, Tianliang Liu, Ke Gong, Xinwu Chen, Gang Zheng
School of Physics and Electronic Engineering, Xinyang Normal University, Xinyang City, China

Copyright © 2016 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: November 10, 2016; Accepted: December 11, 2016; Published: December 14, 2016
ABSTRACT
Eukaryotic genomes contain a significant fraction of repeats, which have very important biomedical function. Thus, aligning repeats from short sequences back to reference genome is the key step for further genome analysis. Unfortunately, the current aligning algorithms performed poorly in distinguishing repeats and nonrepeats. To this end, we proposed a new algorithm, named HashRepAligner, to address this problem. Finally, the cross comparison with other algorithms was performed, and the results indicated that HashRepAligner outperformed other aligners in terms of the detecting repeats.
Keywords:
Sequence Alignment, Next Generation Sequencing, Hash Index, Repeats Detection

1. Introduction
During the past twenty years, the new DNA sequencing technologies have significantly improved throughput and dramatically reduced the cost [1] . Currently, the available commercial next generation sequencing (NGS) platforms include MiSeq, and HiSeq from Illumina [2] , SOLiD and Ion Torrent from Life Technologies [3] , RS system from Pacific Bioscience, and Heliscope from Helicos Biosciences [4] [5] . These sequencing machines can sequence the whole genome in a shorter time, which inspired scientists to sequence large kinds of animals and plants [6] [7] . NGS can be characterized by parallel operation, higher throughput and much lower cost [8] , but share a common disadvantage of producing very short reads.
Furthermore, large researches indicated that repeats comprise a significant fraction of genomes, for example, ~20% of Caenorhabditis elegans and Caenorhabditis briggsae genomes [9] and ~50% of the human genome [10] have been identified as repeats. Most of them have some important biomedical functions and are closely related to some complex disease [11] [12] . Therefore, it is an important step to analyze genome functions from NGS data by aligning the sequencing data back to reference genome. Currently, there are two famous aligning tools, such as bowtie [13] and Soap [14] . Even though, each of them can align millions of reads in one hour, but their performance of detecting repeats also very poor.
In order to improve the completeness of aligning repeats, we proposed a new algorithm aiming for distinguishing repeats and non-repeats, named HashRepAligner, which is based on the combination of Hash index and sliding site math strategy. Hash- RepAligner has the following properties: 1) estimating the copy number of detected repeats; 2) in terms of completeness of aligning repeats, HashRepAligner outperforms others. Simulation data are used to assess the feasibility of HashRepAligner, while the real sequencing data are used to cross comparison with other two aligners, Soap and Bowtie. The results indicated that HashRepAligner outperformed others in terms of aligning repeats. Consequently, HashRepAligner is a complete and accurate repeats aligning tool.
2. Results
The principle of HashRepAligner is based on hash index and sliding site match. Hash index is used to speed the aligning process, while sliding site match is to use the matched number of every site to find the location of repeats, which can decrease the coverage bias and increase the confidence of aligning repeats.
HashRepAligner runs in key four steps: hash index construction, sliding site match, coverage depth estimation and boundary detection. The concrete steps and processes are detailed as follows.
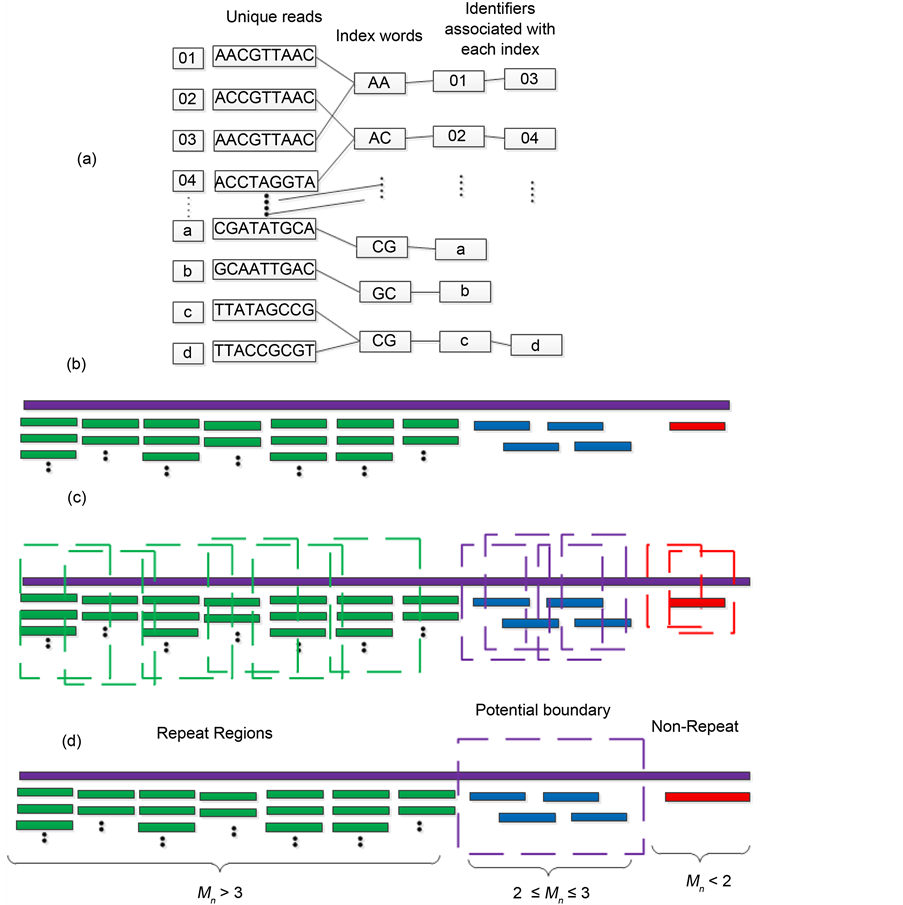
1) Constructing hash index (Figure 1(a)). In order to improve computing speed, an indirect hash structure was designed and adopted in this part. Firstly, the index key words are transformed into quaternary integers instead of the string itself. Secondly, the identifiers of the unique reads are recorded in decimal list. Thirdly, the mapping relations between unique reads and decimal list are constructed.
2) Sliding site match (Figure 1(b)). Based on the hash index, the short sequences are aligned back to the reference genome. For the repetitive seed, HashRepAligner align all of them as long as the keywords are matched according to the hash index.
3) Coverage depth estimation (Figure 1(c)). After aligning the short sequences back to the reference genome, the sliding window function was used to smooth the bias of data, and then coverage depth of each point in reference genome was computed.
4) Boundary detection (Figure 1(d)). According to the estimated coverage depth, read count are merged in a continuous interval. After merging process, the mean read counts 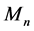 of the interval will be compared with mean sequencing depth
of the interval will be compared with mean sequencing depth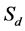 . If the
. If the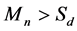 , this region will be considered as the repeats, while if
, this region will be considered as the repeats, while if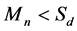 , this region will be considered as the non-repeats.
, this region will be considered as the non-repeats.
Figure 1. The graphic illustration of four steps of HashRepAligner. (a) Hash index construction. The first column is the corresponding identify of every reads. The second column is unique reads. The third column is index words of every reads. The subsequent column is the corresponding identity of every index words. (b) Sliding site match. The Violet line represents reference genome, the green, blue and red short line represent the reads. (c) Coverage depth estimation. The coverage information was estimated by using sliding window function. (d) Boundary detection. For example, if Sd = 2, the mean coverage depth of repeat region should meet Mn > 3, while the mean coverage depth of non-repeats region should meet Mn < 2.
3. Assessments
3.1. Metrics
In this part, we evaluated the performances of HashRepAligner in simulated Datasets and compared with others in real NGS datasets. We use some widely recognized metrics including Family, Total size, Family-accuracy, Size-accuracy, Repeat-accuracy, Copy- accuracy, Location-error to evaluate the performance. Some of them are widely recognized and used in reference [15] . Their definitions and effectiveness are as follows:
1) Family: Total size (T-size): the total size of detected repeats, which is used to evaluate the completeness of length of detected repeats, and which is defined as follows.
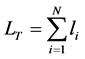 (3.1)
(3.1)
where 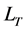 is the total length of all detected repeats,
is the total length of all detected repeats, 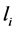 is the
is the 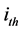 family of repeat, N is the number of family.
family of repeat, N is the number of family.
2) Family-accuracy (F-acc): the accuracy of detected repeats, which is used to evaluate the accuracy of detected repeats and it is defined as follows:
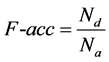 (3.2)
(3.2)
where 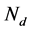 is the number of detected family,
is the number of detected family, 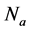 is the number of families.
is the number of families.
3) Size-accuracy (S-acc): the length accuracy of the detected repeat, which is defined as follows:
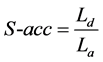 (3.3)
(3.3)
where 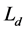 is total length of the detected repeat,
is total length of the detected repeat, 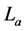 is total length of the actual repeat.
is total length of the actual repeat.
4) Repeat-accuracy (R-Acc): the global matching of the detected repetitive sequence and the actual repetitive sequence, which is defined as follows:
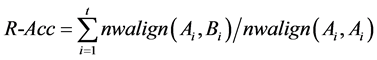 (3.4)
(3.4)
where R-Acc is the global matching value of the repetitive sequence, t is total copy number, nwalign is the global matching function of MATLAB software, Ai is the actual repetitive sequence, Bi is the detected repetitive sequence.
5) Copy Accuracy (C-Acc): the accuracy of the copy numbers of detected repeats, which is defined as follows.
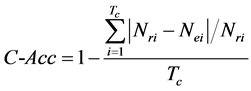 (3.5)
(3.5)
where 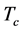 is the total copies of repeats.
is the total copies of repeats. 


6) Copy-accuracy (C-Acc): the accuracy of detected copy number, which is used to evaluate the accuracy of detected copy number and defined as follows:

where Cd is the detected total copy number, Ca is the actual total copy number.
7) Location-error (L-Err): the location error of the repeat, which defined as follows:

where Dsi is the starting location of ith detected repeat, Dei is the ending location of ith detected repeat, Asi is the starting location of ith real repeat, Aei is the ending location of ith real repeat. p is the total number of all repeats. All the repeats are detected by using repeat finding tool HashRepeatFinder [16] . In order to compute this metric, sequences similarity are computed with real repeats using swalign function in MATLAB201b.
For evaluating the accuracy, the metrics, such as Repeat accuracy, Copy accuracy and Location-error are computed by aligning the corresponding items back to the reference genome.
3.2. Simulation Study
We validated the performances of HashRepAligner in three kinds of simulated datasets containing interspersed repeats, tandem repeats and compound repeats, respectively. And then the effect of read depth, read length and the threshold value to HashRepAligner was evaluated, respectively. The detailed results were shown in Table 1.
Three sequences with length L = 500 kb, 300 kb, and 500 kb contain different types of repeats. Location of repeat and non-repeat is generated independently by HashRepAligner with basic parameters: read length Lr = 50, read depth = 2, the threshold value = 160 and step-size = 10. Repeat length smaller than 200 is removed.
From Table 1, three kinds of simulated datasets containing interspersed repeats, tandem repeats, and compound repeats were used to validate the performances of Hash- RepAligner. The repetitive contents contained in these three sequences represented a wide range of repeats with different copies and lengths. The Family-accuracy and Repeat-accuracy were almost up to 100% and 99%, respectively, which indicated that the family were all absolutely correct, and the error tolerance and Location-error of the repetitive sequence were lower than 2% and 15%. All of these indicate that HashRepAligner not only can find different kinds of repeats and non-repeats independently but also can seek out the starting and ending location of the repetitive sequence.
3.3. Cross Comparison
In this part, we use the real NGS dataset. A bacterial genome Rhodobacter sphaeroides (R.s) with genome size 4.6 Mb was downloaded from http://gage.cbcb.umd.edu/data/. All reads were error-corrected.
The Rhodobacter genome has two chromosomes and five plasmids. Thus even the bacteria had multiple chromosomes. Its repetitive structures were detected by HashRepeatFinder tool [16] . 23 families of repeats with total size 8.1 kb were detected. The following results can be concluded from Table 2. Firstly, there is 24, 20 and 26 family of
Table 1. The performances of finding different kinds of repeats.
Table 2. The performances of three tools in R.s.
repeats detected by HashRepAligner, Bowtie and Soap respectively. Their corresponding accuracy is 95.8%, 86.9% and 86.7%. Therefore, in terms of completeness, Hash- RepAligner outperformed others. Secondly, the total size of aligned repeats by three tools are 8.56 kb, 7.32 kb and 12.89 kb, the corresponding accuracy of which is 93.21%, 91.51% and 83.87% respectively. Therefore, in terms of size accuracy, HashRepAligner also outperformed others. Thirdly, HashRepAligner can estimate the copies of each aligned repeats with accuracy of 92.1%, but Bowtie and Soap cannot be used to estimate the copies of aligned items. Lastly, HashRepAligner has the minimum location error of aligned repeats.
4. Conclusions and Discussions
Genome repeats of eukaryotes occupy a significant fraction of the eukaryotes genomes. Most of them have played and are continuing to play critical roles in genome evolution. In order to align these repeats more completely and accurately, we proposed a short sequence aligning algorithm for repeats, named HashRepAligner, which is based on Hash index and sliding site match. In order to evaluate the performance, simulation study and cross comparison were conducted. The results indicated that 1) HashRepAligner can align the repeats more completely; 2) HashRepAligner also can estimate the copy numbers of each corresponding items; 3) HashRepAligner can find the starting and end location of the repetitive sequence. In one word, HashRepAligner is a complete and accurate ab repeat finding tool.
The alignment of repeats from sequencing data is difficult task for genome analysis and is still challenging many aligners, due to the complex repetitive structures and big datasets. Although a large number algorithms including Soap and Bowtie have been proposed to facilitate this problem, but this work is still not finished due to the following reasons. 1) Similarity: repeats can be classified as identical repeats and high similar repeats. For identical repeats, it is a little bit easy to detect as long as the length of repeat is determined. But for the similar repeats, it is difficult to unify the consensus sequences and detect them due to the uncertainty of similarity. Different researchers define different repeats similarity according to the different research task. 2) Types: interspersed repeats, tandem repeats and the compound repeats. The complexity of types of repeats is also the challenge of finding repeats. Eukaryotes genomes always contain different types of repeats. Notably, the compound repeats are almost everywhere. Different aligner has different advantages and specific applications. For the whole genome alignment, Soap or Bowite would be preferred. But for the repeat alignment, HashRepAligner should be preferred.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Acknowledgements
This work was financially supported by National Natural Science Foundation of China (Grant: 61501392), doctoral scientific research start-up funds of XYNU (No: 0201447). In addition, this study was financed in part by the Nanhu Scholars Program for Young Scholars of XYNU.
Cite this paper
Lian, S.B., Liu, T.L., Gong, K., Chen, X.W. and Zheng, G. (2016) A Complete and Accurate Short Sequence Alignment Algorithm for Repeats. Journal of Biosciences and Medicines, 4, 144-151. http://dx.doi.org/10.4236/jbm.2016.412018
References
- 1. Shendure, J., et al. (2004) Advanced Sequencing Technologies: Methods and Goals. Nature Reviews Genetics, 5, 335-344.
https://doi.org/10.1038/nrg1325 - 2. Bentley, D.R. (2006) Whole-Genome Re-Sequencing. Current Opinion in Genetics and Development, 16, 545-552.
https://doi.org/10.1016/j.gde.2006.10.009 - 3. Harris, T.D., Buzby, P.R., Babcock, H., et al. (2008) Single-Molecule DNA Sequencing of a Viral Genome. Science, 320, 106-109.
https://doi.org/10.1126/science.1150427 - 4. Metzker, M.L. (2010) Sequencing Technologies the Next Generation. Nature Reviews Genetics, 11, 31-46.
https://doi.org/10.1038/nrg2626 - 5. Mardis, E.R. (2008) The Impact of Next-Generation Sequencing Technology on Genetics. Trends in Genetics, 24, 133-141.
https://doi.org/10.1016/j.tig.2007.12.007 - 6. The 1000 Genomes Project Consortium (2010) A Map of Human Genome Variation from Population-Scale Sequencing. Nature, 467, 1061-1073.
- 7. Genome 10K Community of Scientists (2009) Genome 10K: A Proposal to Obtain Whole-Genome Sequence for 10,000 Vertebrate Species. Journal of Heredity, 100, 659-674.
https://doi.org/10.1093/jhered/esp086 - 8. Treangen, T.J. and Salzberg, S.L. (2012) Repetitive DNA and Next Generation Sequencing: Computational Challenges and Solutions. Nature Reviews Genetics, 13, 36-46.
- 9. Stein, L.D., Bao, Z., Blasiar, D., et al. (2003) The Genome Sequence of Caenorhabditis briggsae: A Platform for Comparative Genomics. PLoS Biology, 1, Article E45.
https://doi.org/10.1371/journal.pbio.0000045 - 10. International Human Genome Consortium (2001) Initial Sequencing and Analysis of the Human Genome. Nature, 409, 860-921.
https://doi.org/10.1038/35057062 - 11. Iafrate, A.J., Feuk, L., Rivera M.N., et al. (2004) Detection of Large-Scale Variation in the Human Genome. Nature Genetics, 36, 949-951.
https://doi.org/10.1038/ng1416 - 12. Feuk, L., Carson, A.R. and Scherer, S.W. (2006) Structural Variation in the Human Genome. Nature Reviews Genetics, 7, 85-97.
https://doi.org/10.1038/nrg1767 - 13. Langmead, B., Trapnell, C., Pop, M. and Salzberg, S.L. (2009) Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome. Genome Biology, 10, R25.
- 14. Li, R.Q., Li, Y.R., Kristiansen, K. and Wang, J. (2008) SOAP: Short Oligonucleotide Alignment Program. Bioinformatics Application Note, 24, 713-714.
https://doi.org/10.1093/bioinformatics/btn025 - 15. Saha, S., Bridges, S., Magbanua, Z.V. and Peterson., D.G. (2008) Empirical Comparison of Ab Initio Repeat Finding Programs. Nucleic Acids Research, 36, 2284-2294.
https://doi.org/10.1093/nar/gkn064 - 16. Lian, S.B., Chen, X.W., Wang, P., Zhang, X.L. and Dai, X.H. (2016) A Complete and Accurate Ab Initio Repeat Finding Algorithm. Interdisciplinary Sciences-Computational Life Sciences, 8, 75-83.
https://doi.org/10.1007/s12539-015-0119-6