Open Journal of Nephrology
Vol.3 No.2(2013), Article ID:33301,5 pages DOI:10.4236/ojneph.2013.32018
IGF2 ApaI A/G Polymorphism Evaluated in ESRD Individuals as a Biomarker to Identify Patients with New Onset Diabetes Mellitus after Renal Transplant in Asian Indians
1Department of Genetics and Molecular Medicine, Kamineni Hospital, Hyderabad, India
2Tissue Engineering Group, NOCERAL, Department of Orthopaedic Surgery, Faculty of Medicine, University of Malaya, Kuala Lumpur, Malaysia
3Center of Medical Genetics, Sir Ganga Ram Hospitals, New Delhi, India
4Department of Nephrology, Kamineni Hospital, Hyderabad, India
5Department of Biochemistry, Kasturba Medical College, Manipal University, Manipal, India
Email: *qhasan2000@yahoo.com
Copyright © 2013 Kiran Kumar Vattam et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received April 20, 2013; revised May 19, 2013; accepted May 27, 2013
Keywords: PTDM; IGF2 gene polymorphism; Diabetes mellitus; ESRD; New onset diabetes mellitus
ABSTRACT
Insulin like growth factors2 (IGF2) regulates pancreatic β-cell renewal and apoptosis, which in turn plays a role in altering insulin activity and glucose homeostasis. Polymorphisms in IGF2 gene have been associated with altered levels of IGF2. Hence, ApaI polymorphism in exon 9 of IGF2 (rs#680) gene was assessed in patients with end stage renal disease (ESRD) to identify individuals at risk of developing new onset diabetes mellitus (NODM) in Asian Indians. Isolated DNA was used for PCR&RFLP based genotyping of IGF2 ApaI polymorphism which was carried out in 364 individuals these included 140 patients who had undergone renal transplant, 42 of which developed new onset diabetes mellitus after renal transplant and 224 healthy control volunteers. In the present study NODM or post transplant diabetes mellitus (PTDM) showed a significant association with G allele and AG genotype when compared with the NonNODM ESRD patients after transplant (OR 2.081, 95% CI = 1.191 - 3.634, p = 0.01 and OR 3.188, 95% CI = 1.498 - 6.785, p = 0.002) ESRD patients with healthy controls also showed an association with G allele and AG genotype (OR 1.512, 95% CI = 1.060 - 2.155, p = 0.02 and OR 2.235, 95% CI = 1.453 - 3.438, p = 0.0002). IGF2 could be used as a biomarker to identify individuals at high risk of developing NODM, it would be a valuable asset in selecting appropriate immunosuppressive regimens for individuals undergoing transplant. Present study shows the importance of IGF2 ApaI polymorphism in assessing the risk of NODM in ESRD individuals in Asian Indians with ESRD.
1. Introduction
Insulin-like growth factors (IGFs) are regulators of processes like growth and metabolism. IGF1 and IGF2 contribute to pancreatic β-cell growth and development by regulating β-cell replication, renewal, and apoptosis [1]. Deregulation of balance between β-cell renewal and apoptosis due to alterations in IGF levels is potentially of great importance in the development of glucose intolerance, a major characteristic of diabetes. In addition, insulin-dependent glucose homeostasis may be affected by IGFs as they act via the insulin signaling pathway [2]. Defects in the IGF/insulin-signaling pathway affects birth weight and fat metabolism in both domestic animals and humans, which are known risk factor for development of Type 2 diabetes (T2D) [3]. IGF2 polymorphisms have been associated with weight gain, Body mass, obesity and adiposity [4].
Hence in the present study Apa1 polymorphism (rs# 680) has been evaluated in ESRD individuals who have undergone renal transplant to help identify individuals at a risk of developing new onset/Post transplant diabetes mellitus (PTDM).
PTDM or new onset diabetes mellitus (NODM) is a serious metabolic complication that may follow organ transplantation in patients on immunosuppressive therapy [5]. According to American Diabetes Association (ADA) criteria, 13% of patients develop NODM by 3 months after transplant and 39% in the long term have abnormal glucose metabolism [6]. Although there are currently no clearly established risk factors for identifying individuals who develop NODM, a number of characteristics have been identified that appear to predispose patients to the development of NODM. These are above 40 years of age, African-American and Hispanic populations, patient with a positive family history for T2D, increased BMI, hepatitis C virus infection and the immunosuppressive regimen [7]. The pathophysiology of NODM closely mimics that of T2D, both pathologies are characterized by a combination of insulin hyposecretion and insulin resistance [8,9]. TCF7L2, SLC30A8 and KCNQ1 polymorphisms have been studied in NODM which were earlier reported to be associated with T2D [10]. Increased activity of the IGF signaling pathway has been implicated as a major contributor to diabetic nephropathy, conferring the typical morphological appearance to diabetic glomeruli by virtue of its ability to induce characteristic changes and affect glomerular hemodynamics. Increased glucose levels in diabetics elevate IGF levels in the kidney. However, the association of IGF2 ApaI polymorphism with NODM has not been studied in any population.
Hence the aim of the present study was to evaluate the association of IGF2 ApaI polymorphism with ESRD and development of NODM after renal transplant.
2. Materials and Methods
2.1. Subjects
The present study was carried out in 364 Asian Indian individuals from a cosmopolitan city Hyderabad, located in South India. 140 of which were unrelated non-diabetic end stage renal disease (ESRD) patients who had undergone renal transplant and were on immunosuppressive therapy for more than three months, these individuals were on routine follow up and monitored periodically for renal function 42 of these developed NODM based on the ADA [11] criteria.
The control group consisted of 224 healthy volunteers, above the age of 40 years without T2DM or related health problems and who had a normal FBS. The study was approved by the Institutional Ethics Committees.
2.2. Sample
2 ml of peripheral blood was obtained from all the individuals included in the study along with a detailed clinical and family history recorded in a well designed proforma.
2.3. Isolation of Genomic DNA
Genomic DNA was extracted from leukocytes by a salting out technique which is routinely used in our lab [12]. The extracted DNA samples were dissolved in 0.1X TE buffer (pH 8.0) and stored at −20˚C till further use. DNA concentration was adjusted to 100 ng/µl before carrying out polymerase chain reaction (PCR).
2.4. Genotype Analysis by PCR-RFLP
A three step PCR-RFLP method was followed for IGF2 exon 9 ApaI genotyping, which has been published by our group [13]. Specific primers: forward primer: 5’- CTTGGACTTTGAGTCAAATTGG-3’; reverse primer: 5’-GGTCGTGCCAATTACATTTCA-3’ were synthesized by Bioserve biotechnology, (Hyderabad, India) for PCR analysis. DNA was denatured at 95˚C for 5 min, amplified by 35 cycles of 94˚C for 30 sec, 55˚C for 30 sec, 72˚C for 45 sec and the final extension with 72˚C for 5 minutes, 2.5 units of Taq polymerase (cat#MME27, Banglore Genei, Banglore, India) were used per reaction. PCR products were digested for 2 hours with ApaI at 30˚C (10 units of ApaI enzyme, 8 μl PCR product and 1.5 μl buffer in a final volume of 15 μl); and electrophoresed on 2% agarose gel with ethidium bromide.
2.5. Statistics
Genotype and allele frequencies were calculated for the described SNP (Table 1). The groups were compared using the χ2 test to analyze the statistical significance of the difference in allelic distribution of various polymorphisms in patients and controls. Values of p < 0.05 were considered statistically significant. Odds ratio was performed using MedCalc for Windows, version 7.4.1.0 (MedCalc Software, Mariakerke, Belgium).
3. Results
The IGF2 ApaI genotype and allele frequencies were identified based on the bands obtained after restriction enzyme digestion of the 292 bp PCR product. The A (not digested by ApaI) and G alleles (digested by ApaI) are 292 bp and 229 bp, respectively, heterozygotes were A/G (292/229 bp) at the ApaI polymorphism (Figure 1).
When the renal transplant patients were categorized as those who developed NODM and others who did not based on ADA criteria it was observed that 30% of the renal transplant recipients developed NODM. The IGF2 genotype in individuals with NODM was 26.1% AA and 73.8% AG, while non-PTDM group consisted of 56.1% AA and 43.8% AG. NODM cases showed a significant difference for the G allele and AG genotype
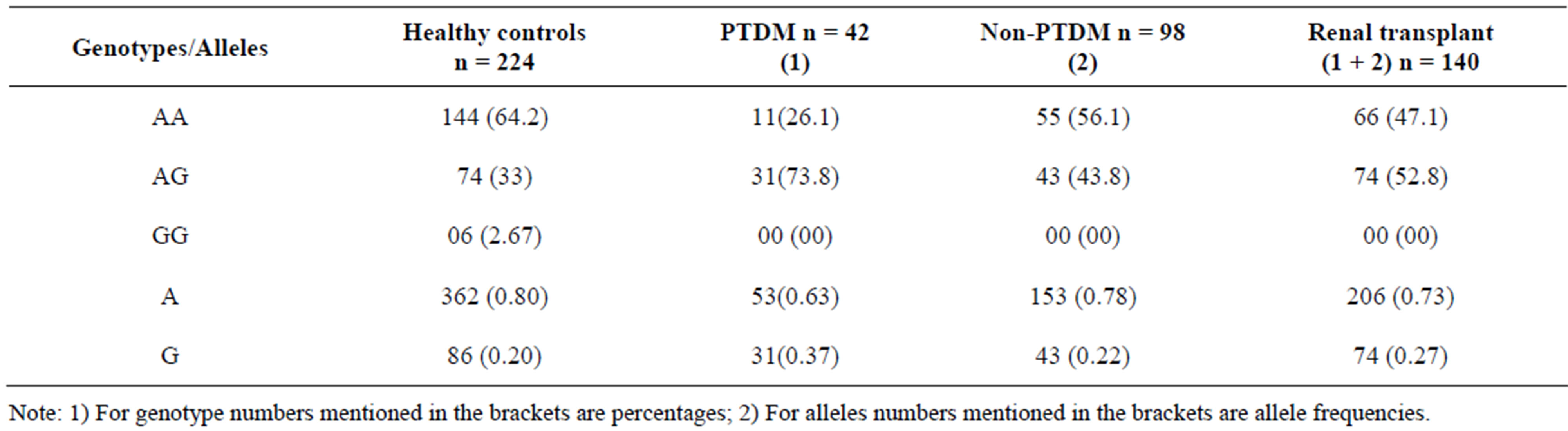
Table 1. Genotypes and alleles distributions of the IGF2 ApaI polymorphism in each of the cases studied along with matching controls.
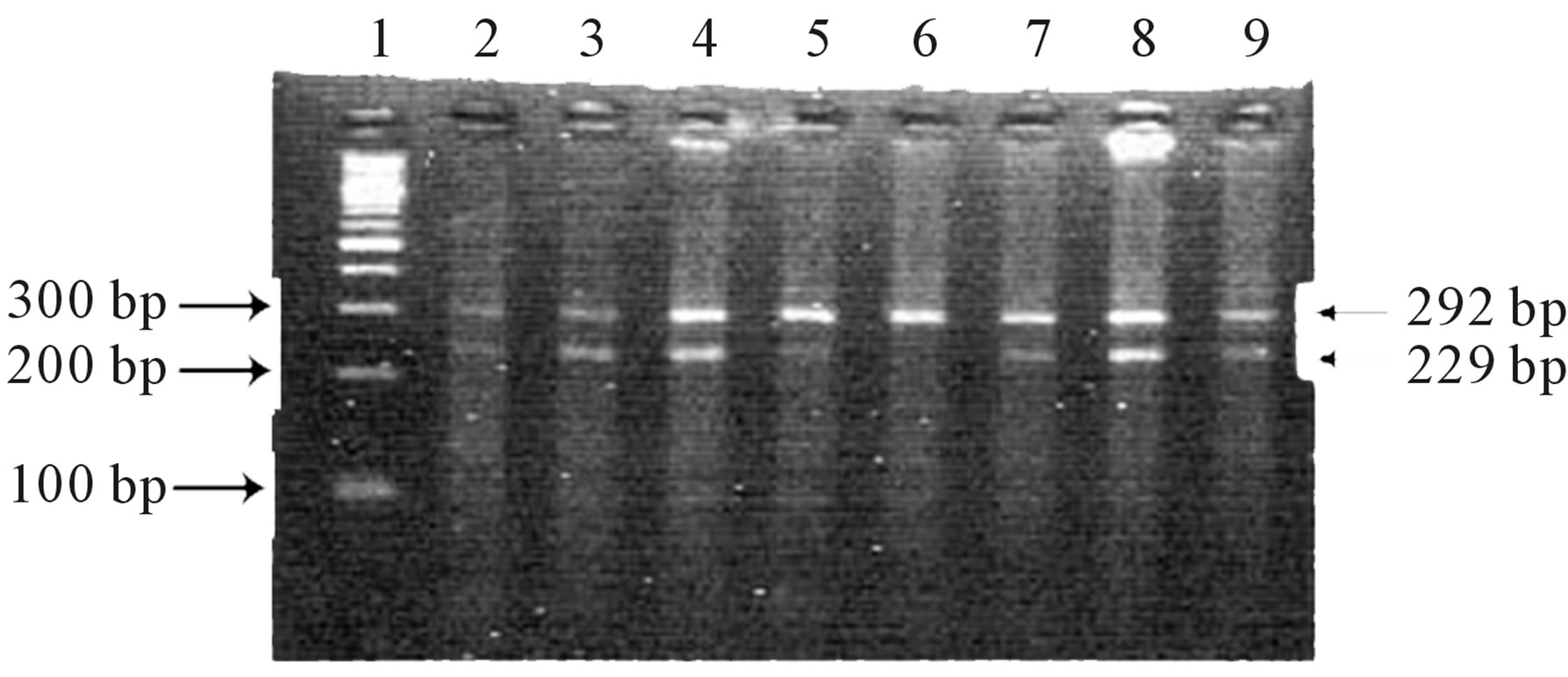
Figure 1. IGF2 PCR products were resolved by gel electrophoresis on 2% agarose gel stained with ethidium bromide after digestion with Apa I restriction enzyme. Lane 1: 100 bp DNA ladder, Lanes 2 - 5 & 7 - 9 showing 292/229 bp bands indicating heterozygous AG genotype, Lane 6: showing 292 bp band indicate homozygous AA genotype.
when compared to the ESRD patients (p = 0.01, p = 0.002) There was an association of NODM with G allele (OR 2.082, 95% CI = 1.191 and 3.634 AG genotype; OR 3.188, 95% CI = 1.498 - 6.785) (Table 2), which was significant. However, it was noteworthy that neither NODM nor the Non-NODM patients had the GG genotype, hence we compared ESRD patients who had undergone renal transplant with normal healthy controls. The G allele and AG genotype (p = 0.02, p = 0.0002) showed a significant association with ESRD (OR 1.512; 95% CI 1.060 - 2.155) and (OR 2.235, 95% CI = 1.453 - 3.438) respectively (Table 2).
4. Discussion
Several lines of evidence support that the susceptibility to NODM has a genetic component similar to Type 2 diabetes. Although no systematic studies have evaluated this, family studies suggest that NODM aggregates within families with a history of T2D [14]. Previously, it was shown that polymorphisms in the IGF1 and IGF2 genes are associated with features of the metabolic syndrome [15-18]. Gene variants in the IGF2 gene were found to be associated with IGF2 levels and BMI [16, 17]. Gomes et al. [19] (2006) studied the association between IGF2 ApaI polymorphism and the BMI, however, did not find any significant association.
In the present study ApaI IGF2 polymorphism was assessed out and it was identified that NODM showed an association with AG genotype and G allele (Table 2) when compared with controls i.e. normal healthy individuals and non-PTDM. GG genotype was completely absent even in the ESRD patients (includes both PTDM and non-PTDM) this was surprising as none of the ESRD cases included in the study were diabetic or had diabetic nephropathy, which has association with IGF2 in an earlier paper published by us Movva et al. [20] (2009). These ESRD patients showed a significant association with both AG genotype and G allele, indicating that both diabetic and non-diabetic renal disease were associated with IGF2 ApaI polymorphism. Similar studies in other populations are required to understand if this polymerphism has a role in ESRD in all ethnic groups or this is specific to the Asian Indian population.
NODM is a metabolic disorder that develops in response to a relative insulin deficiency in patients after organ transplantation treated with immunosuppressive drugs. From our study on 140 RT patients 30% were identified as PTDM. The onset of PTDM is most pronounced in the first few months after transplant and continue to present with PTDM at a steady rate after the first year post transplant. Our results suggest that IGF2 Apa I genotyping of patients is a possible method of predicting a patient’s risk for developing PTDM and would be a valuable asset in selecting appropriate immunosuppressive regimens for these individuals.
In conclusion our study for the first time shows that IGF2 ApaI G allele increases the risk of developing renal disease in Asian Indians and it can be used as a biomarke for identifying individuals at a high risk of developing NODM especially in case of RT recipients for appropriate management with immunosuppression to prevent the development of PTDM.
5. Acknowledgement
We acknowledge to the Dept of Nephrology of Kamineni
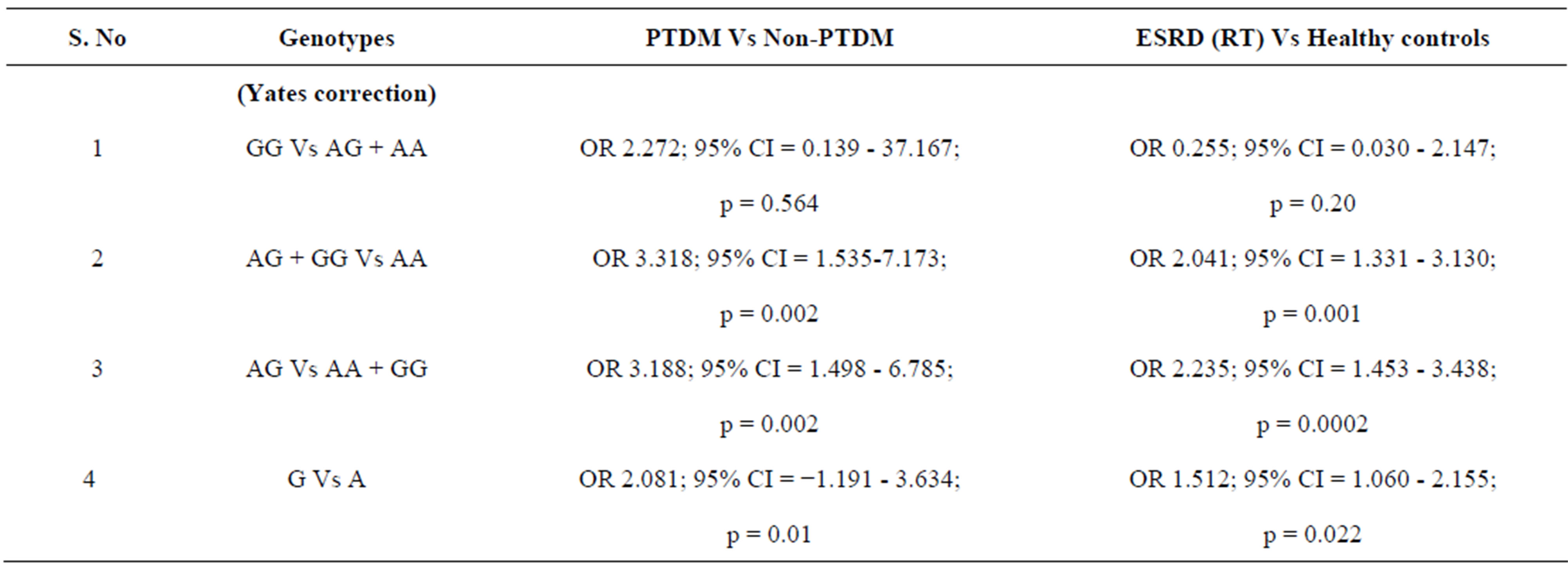
Table 2. Odds ratio and chi square test in IGF2 ApaI allele and genotype distribution in PTDM Vs non-PTDM and ESRD (RT) Vs Healthy controls.
Hospitals for providing samples, we would like to thank Indian Council for Medical Research for funding the project (Sanction No. 5-3-8-39-2007; RHN), we would like to thank who all participated in the study.
REFERENCES
- L. M. Hart, A. Fritsche, I. Rietveld, J. M. Dekker, G. Nijpels, F. Machicao, M. Stumvoll, C. M. van Duijn, H. U. Häring, R. J. Heine, J. A. Maassen and T. W. van Haeften, “Genetic Factors and Insulin Secretion. Gene Variants in the IGF Genes,” Diabetes, Vol. 53, Suppl. 1, 2004, pp. 26-30. doi:10.2337/diabetes.53.2007.S26
- J. J. Kim and D. Accili, “Signalling through IGF-I and Insulin Receptors: Where Is the Specificity?” Growth Hormone and IGF Research, Vol. 12, No. 2, 2002, pp. 84-90. doi:10.1054/ghir.2002.0265
- C. N. Hales and D. J. Barker, “The Thrifty Phenotype Hypothesis,” British Medical Bulletin, Vol. 60, No. 1, 2001, pp. 5-20. doi:10.1093/bmb/60.1.5
- M. S. Sandhu, J. M. Gibson, A. H. Heald, D. B. Dunger and N. J. Wareham, “Low Circulating IGF-II Concentrations Predict Weight Gain and Obesity in Humans,” Diabetes, Vol. 52, No. 6, 2003, pp. 1403-1408. doi:10.2337/diabetes.52.6.1403
- M. Markell, “New-Onset Diabetes Mellitus in Transplant Patients: Pathogenesis, Complications and Management,” American Journal of Kidney Diseases, Vol. 43, No. 6, 2004, pp. 953-965. doi:10.1053/j.ajkd.2004.03.020
- B. L. Kasiske, J. J. Snyder, D. Gilbertson and A. J. Matas, “Diabetes Mellitys after Kidney Transplantation in the United States,” American Journal of Transplantation, Vol. 3, No. 2, 2003, pp. 178-185. doi:10.1034/j.1600-6143.2003.00010.x
- J. Davidson, A. Wilkinson, J. Dantal, F. Dotta, H. Haller, D. Herna´ndez, et al., “New-Onset Diabetes after Transplantation: International Consensus Guidelines,” Transplantation, Vol. 75, 2003, p. SS3. doi:10.1097/01.TP.0000069952.49242.3E
- S. E. Kahn, “The Relative Contributions of Insulin Resistance and Beta Cell Dysfunction to the Pathophysiology of Type 2 Diabetes,” Diabetologia, Vol. 46, No. 1, 2003, pp. 3-19.
- A. V. Ekstrand, J. G. Eriksson, R. C. Gronhagen, P. J. Ahonen and L. C. Groop, “Insulin Resistance and Insulin Deficiency in the Pathogenesis of Post-Transplantation Diabetes in MA,” Transplantation, Vol. 53, No. 3, 1992, pp. 563-569. doi:10.1097/00007890-199203000-00014
- S. K. Enu, S. K. Myoung, S. K. Yu, H. K. Chul and C. L. Hyun, “A Polymorphism in the Zinc Transporter Gene SLC30A8 Confers Resistance against Posttransplantation Diabetes Mellitus in Renal Allograft Recipients,” Diabetes, Vol. 57, No. 4, 2008, pp. 1043-1047.
- B. G. Marilia and A. CRoberta, “Post Transplant Diabetes Mellitus,” Diabetology & Metabolic syndrome, Vol. 1, No. , 2009, pp. 14.
- S. Chava, V. Mohan, N. Pasupuleti, M. MLatha, I. A. Khan, P. Upendram, A. Kumar, Y. R. Ahuja and Q. Hasan, “Evaluation of Aurora—A Gene Polymorphism and Esophageal Cancer Risk in a South Indian Population,” Genetic Testing and Molecular Biomarkers, Vol. 15, No. 3, 2011, pp. 185-189. doi:10.1089/gtmb.2010.0143
- J. S. Preetha, M. Sireesha, P. Nagarjuna, V. Bhavani, V. Sambasivan, R. A. You and Q. Hasan, “Regulation of IGF2 Transcript and Protein Expression by Altered Methylation in Breast,” Journal of Cancer Research and Clinical Oncology, Vol. 137, No. 2, 2011, pp. 339-345. doi:10.1007/s00432-010-0890-z
- M. A. Williams, C. Qiu, J. C. Dempsey and D. A. Luthy, “Familial Aggregation of Type 2 Diabetes and Chronic Hypertension in Women with Gestational Diabetes Mellitus,” The Journal of Reproductive Medicine, Vol. 48, No. 12, 2003, pp. 955-962.
- N. Vaessen, P. Heutink, J. A. Janssen, J. C. Witteman, L. Testers, A. Hofman, S. W. Lamberts, B. A. Oostra, H. A. Pols and C. M. Vanduijn, “A Polymorphism in the Gene for IGF1: Functional Properties and Risk for Type 2 Diabetes and Myocardial Infarction,” Diabetes, Vol. 50, No. 3, 2001, pp. 637-642. doi:10.2337/diabetes.50.3.637
- S. D. O’Dell, G. J. Miller, J. A. Cooper, P. C. Hindmarsh, P. J. Pringle, H. Ford, S. E. Humphries and I. N. Day, “Apa I Polymorphism in Insulin-Like Growth Factor II (IGF2) Gene and Weight in Middle-Aged Males,” International Journal of Obesity, Vol. 21, No. 9, 1997, pp. 822-825. doi:10.1038/sj.ijo.0800483
- T. R. Gaunt, J. A. Cooper, G. J. Miller, I. N. Day and S. D. O’Dell, “Positive Associations between Single Nucleotide Polymorphisms in the IGF2 Gene Region and Body Mass Index in Adult Males,” Human Molecular Genetics, Vol. 10, No. 14, 2001, pp. 1491-1501. doi:10.1093/hmg/10.14.1491
- O. Ukkola, G. Sun and C. Bouchard, “Insulin-Like Growth Factor 2 (IGF2) and IGF-Binding Protein 1 (IGFBP1) Gene Variants Are Associated with Overfeeding-Induced Metabolic Changes,” Diabetologia, Vol. 44, No. 12, 2001, pp. 2231-2236. doi:10.1007/s001250100034
- M. V. Gomes, M. R. Soares, N. A. Pasqualim, C. R. Marcondes, R. B. Lôbo and E. S. Ramos, “Association between Birth Weight, Body Mass Index and IGF2/ApaI Polymorphism,” Growth Hormone and IGF Research, Vol. 15, No. 5, 2005, pp. 360-362. doi:10.1016/j.ghir.2005.06.016
- S. Movva, S. Venkatasubramanian, K. K. Vattam, R. Sistla, Y. R. Ahuja and Q. Hasan, “Relevance of Insulin-Like Growth Factor 2 in the Etiopathophysiology of Diabetic Nephropathy: Possible Roles of Phosphatase and Tensin Homolog on Chromosome 10 and Secreted Protein Acidic and Rich in Cysteine as Regulators of Repair,” Journal of Diabetes, Vol. 1, No. 2, 2009, pp. 118- 124. doi:10.1111/j.1753-0407.2009.00025.x
NOTES
*Corresponding author.

