International Journal of Intelligence Science
Vol.2 No.4(2012), Article ID:23697,3 pages DOI:10.4236/ijis.2012.24014
Molecular Environment of Reserving Information (M.E.R.I. Technology)*
1Nanotechnology Consultatory and Development Center (NCDC), Padova, Italy
2Isfahan University of Medical Sciences, Isfahan, Iran
3Department of Cardiology, Isfahan University of Medical Sciences, Isfahan, Iran
Email: #behjati@med.mui.ac.ir
Received June 15, 2012; revised August 18, 2012; accepted August 25, 2012
Keywords: Information Reserving; DNA Information; Molecular Environment; Nucleotides; DNA Macromolecules
ABSTRACT
The idea of application from DNA information organization for providing the next generation super-computers has been proposed by lessons learnt from nature. Thus, molecular environment of reserving information was considered as succeeding generation of a mother-board project containing some subcategories with various applications. MERI, constructed based on the binary coding system using DNA synthesizer machine, enables retrievable reading of molecular structures using DNA sequencer machine. It has high capacity for reservation of information about 1.212 × 1011 Kb/cm2 with long-term stability. It can be used in various fields of science and technology regarding its high capacity, small size and safety measures.
1. Introduction
Rapid pace of discovery in the field of science and technology, especially in electronic and computer sciences, made it the “age of information”. Parallel to these advances, application of these sciences in the field of boilogy is progressing. In this context, it has been recognized that E. coli can process life activated with huts 5 × 106 base pairs in the way that a supercomputer with million megabits is not capable to do part of them [1,2]. Thus, a new generation of these computers using especial type of these proteins is under evaluation in order to enhance rate and capacity of ordinary computers [3]. In our study, we introduce a new technology named MERI with various applications. MERI basically relies on DNA macromolecules. It contains 205 × 106 times (in Kb/cm2) capacity compared with CD. Its microscopic size (nanometer) and long-term stability are noteworthy. MERI technology is yet in its infancy but is able to direct us toward new generation of computers. To best of our knowledge, its application in transferring passwords in safe communication systems for military and security measures is noticed for the first the time by us. Regarding its small microscopic size, it covers security measures. Indeed, this technology can transfer messages born in each costume and at special time point to offsprings. Since this technology has various aspects in terms of applications, it can open new horizons for information reservation with high capacity in least occupied space for diverse science fields.
2. Methods and Materials
First, one binary code is attributed to each of the DNAbuilding nucleotides in Table 1.
A molecular equivalent is considered for digital messages. Using this predefined measures, tables are designed which enables determination of molecular equivalents applicable in Hexdecimal systems (Table 2).
Then, another table will be designed which determines molecular equivalents for alphabetic words based on binary codes (Table 3).
All of these steps are performed using home-made software. Molecular equivalents are constructed and molecules are constructed as single strands using DNA Synthesizer machine [4]. These stages are performed automatically and are accessible by ordering the desired DNA sequences to the machine. Synthesized DNA is derived using a vacuum centrifugation. Approximate number of DNA can be adjusted through dilation. For this purpose, DNA should be diluted in TE buffer or strill double-distilled water. In this way, DAN can be stored in +4˚C or –20˚C for short term and long term, respectively. DNA concentration was determined using calculation of optical density (OD) in 260 nm wavelength. Each unit of 260 OD equals 33 µgr/ml of single stranded DNA. Following formula is advised for determination of DNA concentration (µM): OD 260 dilution.
∑ Molar extinction coefficient of dNTPs A = 1520 T = 8400 G = 12,010 C = 7050.
DNA sequencing was performed automatically by Alf express DNA sequencer. After DNA sequencing, Table 3 was filled using home-made software to find the saved message by MERI.
Reservation capacity of MERI was calculated using following formula:
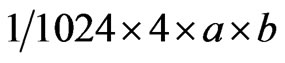
Width and length of each nucleotide were both 0.33 nm and the distance between toe nucleotide was 0.28 nm, thus:
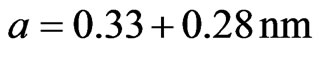
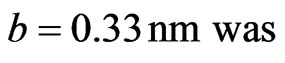
The calculated reservation capacity 1.212 × 1011 Kb/cm2.
3. Results and Discussion
Modern biology was established by the discovery for double-stranded helix in 1953 by Francis H. C. crick, James D. Watson [5]. This discovery was the cornerstone for following astonishing advances in other biologic fields and life science. This progress is going on yet. In this regards, the hypothesis of application of macromolecules (proteins) in construction of computers back to the date 1994 [6]. Indeed, nowadays chips can save inherent properties of cells of alive creatures and analyze pathogenic agents carefully [7]. One molecule of DNA is constructed from assembly of nucleotides including:
Organic nitrogen, sugare (Deoxyribose or ribose) and phosphate group. Considering nucleotides (AdenineGuanine, Cytosine and Thymine) as informative symbols, one DNA molecule contains “n” nucleotides which will be equal to chain from symbols of a message with the length of “n”. Regarding the unlimited assembly of nucleotides from chemical point of views, each symbol of a message is statistically independent from other symbols. In addition, since all of these four nucleotides are similar in terms of cost and complexity, the probability of occurrence of these symbols is equal. The mean information bearing capacity of each symbol (entropy) equals to LogH24 = 2. Actually, one DNA molecule contains “2n” bits of information. One of the unique properties of MERI, is its high capacity. By now, the greatest information reservation capacity belongs to “Digital versatile Disk” which contains 4.7 Gbs in the same size with CD [8] Table 4. Samples of Gbs with 17 Gabs are made experimentally. This technology saves and process information with 0.4 µ distance on disk. Meanwhile, MERI’s capacity is 3 × 106, 20 × 106 and 5 × 109 times than that can be achieved by a DVD, CD and magnetic disk (Kbs/cm2). This high capacity relates to MERI’s property which carries information similar to nucleotides and functions as 2× bits of information, considering the length and width of nucleotide as 0.33 nm. Therefore, the reservation capacity of MERI is far higher than today’s technology.
Table 4 represents a comparison between MERI and

Table 1. Determination of molecular equivalent for binary codes.

Table 2. Determination of molecular equivalents applicable in Hexdecimal systems.
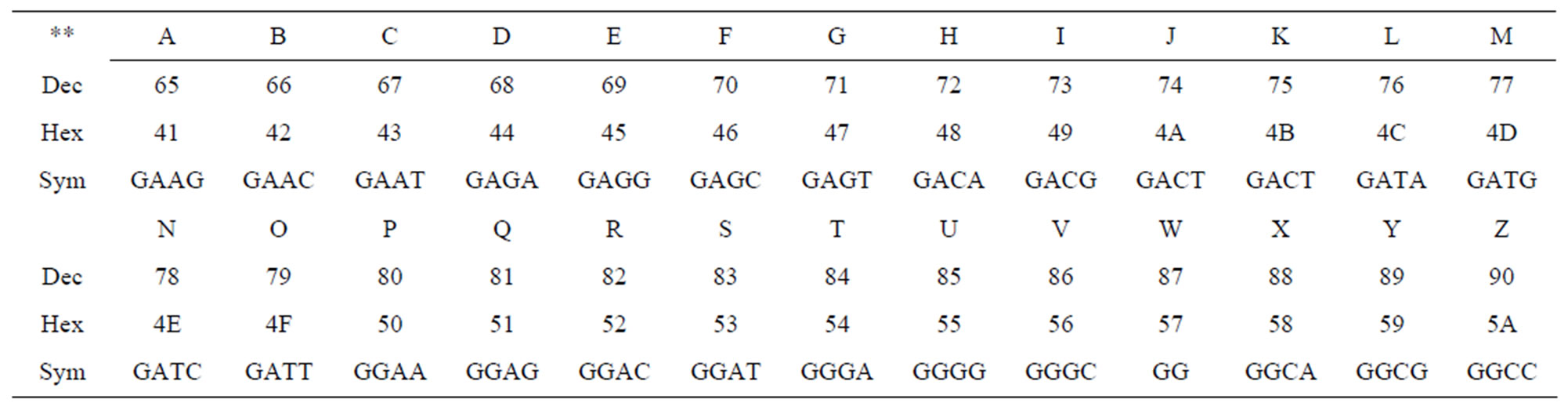
Table 3. Determination of molecular equivalents for alphabetic words.
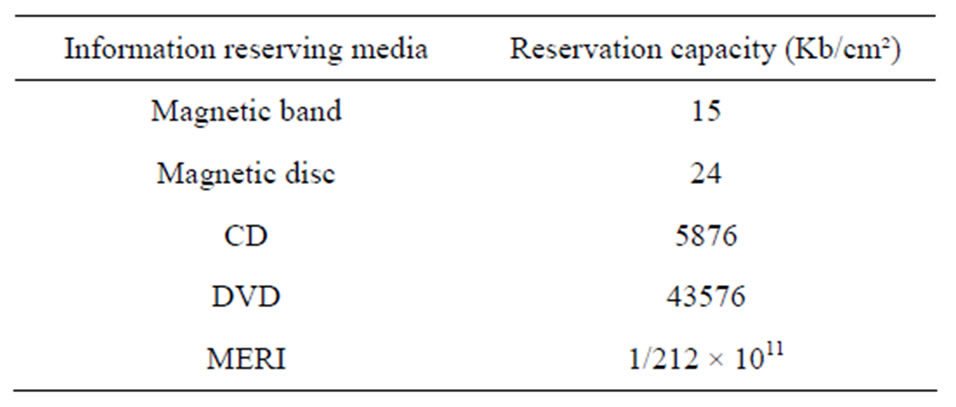
Table 4. Comparison of MERI capacity with different information reserving medias.
very small compared with a CD in the similar state. This microscopic size let them to be applied in transferring keys between sender and receiver through safe canals in safe communication systems through classic coding systems. In classic coding systems, coding or decoding functions or algorithms act in inverse manner under the control of a constant key. The secure key is defined by the agreement of sender and receiver and the security of the coding system is toughly dependent in the security of the key [8]. Even if some one is aware about the coding algorithm, no one is able to evaluate the original from coded text due to inaccessibility to the key. In digital communication systems, coding and decoding systems are composed from complex mathematical algorithms. In these systems, the key is a short binary code (around 100 bits). Key transferring between sender and receiver should be through a safe canal which can be a secure person or meeting in one secure place between sender and receiver. These approaches bring their own limitations. Alternative approaches are cutting the key into pieces and sending them through various canals. Key coding with one predefined key is another way. Key reservation and saving is of paramount value and is performed using various methods as keeping in mind or using magnetic cards known as smart card or Reomkey. Generally, considering the importance of key in coding systems, it’s transferring and reservation should be done under tight physical protection. Application of MERI for this purpose facilitates secure reservation and transferring of key in various physical ways. Due to its microscopic sizes (millennium of mm) it is possible to send key invisible. MERI will bring great advance in safe communication systems and political and military applications. We have designed some coding systems including standard DES system in different sizes (46 and 128 bits). Another unique property of MERI is its stable longterm durability in its best condition. This property is attributed to the excellent chemical stability of DNA molecule. Thus, MERI technology can be considered as a bridge to transferring information to the future human offspring. By now, using MERI at semi-commercial level is possible. We are extending this technology to a cheaper and more capacious media.
REFERENCES
- R. Weiss, S. Basu, S. Hooshangi, A. Kalmbach, D. Karig, R. Mehreja and I. Netravali, “Genetic Circuit Building Blocks for Cellular Computation, Communications, and Signal Processing,” Weiss-Natural Computing, 2003, in press.
- M. L. Simpson, G. S. Saylor and J. T. Fleming, “Applegate, Whole-Cell Biocomputing,” Trends in Biotechnology, Vol. 19, No. 8, 2001, pp. 317-323.
- S. Burbec, “Complexity and the Evolution of Computing: Biological Principles for Managing Evolving Systems,” Computing Systems, 2007, pp. 2004-2007.
- J. Watson and F. Crick, “Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid,” Nature, Vol. 171, 1953, pp. 737-738.
- L. M. Adleman, “Molecular Computation of Solutions to Combinatorial Problems,” Science, Vol. 266, No. 5187, 1994, pp. 1021-1024.
- J. Wise, T. Stadthagen, M. Schmidhuber, M. Brischwein, J. Ressler, U. Raeder and H. Grothe, “Intelligent Mobile Lab for Metabolics in Environmental Monitoring,” Analytical Letters, Vol. 39, 2006, pp. 1759-1771.
- Super Video Compact Disc, “A Technical Explanation (PDF),” Philips System Standards and Licensing, 1998, p. 2.
- T. Wu, “The SRP Authentication and Key Exchange System,” Network Working Group, Stanford University, Stanford, 2000.
NOTES
*Conflict of interest: The authors declare no commercial or financial conflict of interest.
#Corresponding author.

