Open Journal of Ecology
Vol.3 No.1(2013), Article ID:27698,4 pages DOI:10.4236/oje.2013.31005
Biodiversity and secretion of enzymes with potential utility in wastewater treatment

1Departamento de Genética, Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais, Belo Horizonte, Brazil;
*Corresponding Author: kalapothakis@gmail.com
2Phoneutria Biotecnologia e Serviços Ltda, Belo Horizonte, Brazil
Received 1 December 2012; revised 31 December 2012; accepted 8 January 2013
Keywords: Lipase; Esterase; Wastewater; Sewage Treatment; Lipolytic Microorganisms; Amylase; Protease; Cellulase
ABSTRACT
The main organic contaminants in municipal wastewater are proteins, polysaccharides, and lipids, which must be hydrolyzed to smaller units. A high concentration of oil and grease in wastewater affects biological wastewater treatment processes by forming a layer on the water surface, which decreased the oxygen transfer rate into the aerobic process. Microbial proteases, lipases, amylases, and celullases should play essential roles in the biological wastewater treatment process. The present study aimed to isolate lipaseand other hydrolytic enzyme-producing microorganisms and assess their degradation capabilities of fat and oil wastewater in the laboratory. We also evaluated microbial interactions as an approach to enhance lipolytic activity. We place emphasis on lipase activity because oil and grease are not only environmental pollutants, but also form an undesirable tough crust on pipes of sewage treatment plants. Thirty-five lipolytic microorganisms from sewage were identified and assessed for hydrolytic enzyme profiles. Lipases were characterized in detail by quantification, chain length affinity, and optimal conditions for activity. The good stability of isolated lipases in the presence of chemical agents, thermal stability, wide range of pH activity and tolerance, and affinity for different lengths of ester chains indicates that some of these enzymes may be good candidates for the hydrolysis of organic compounds present in wastewater. A combination of enzymes and fermenting bacteria may facilitate the complete hydrolysis of triglycerides, proteins, and lignocellulose that normally occur in the wastes of industrial processes. This study identifies enzymes and microbial mixtures capable of digesting natural polymeric materials for facilitating the sewage cleaning process.
1. INTRODUCTION
For more than a century, biological wastewater treatment has been used to minimize anthropogenic damage to the environment. Oil and Grease (O&G) are the major problems and contaminants in biological wastewater treatment processes. Because of their nature, O&G form a layer on the water surface and decrease the oxygen transfer rate into an aerobic process [1]. These contaminants are mainly discharged from restaurants, food industries, and households [1,2]. Proteins and polysaccharides must also be hydrolyzed to smaller units by extracellular enzymes in a municipal wastewater treatment plant [3,4].
The composition and activity of the microbial community within a wastewater treatment plant play a substantial role in the efficiency and robustness of the purification process [5]. The efficiency of conventional biological processes in wastewater treatment is reduced by the high concentrations of O&G in effluents [6]. In the activated sludge process, high levels of O&G lead to a reduction of biological activity of the flocs due to the difficulty of oxygen and substrate to penetrate the floc due to the oil film formation around it [7]. Moreover, in the case of anaerobic digestion, excessive amounts of O&G inhibit the action of acetogenic and methanogenic bacteria [6,8,9]. The Brazilian National Council on the Environment (CONAMA) established the maximum level of mineral oil concentration allowed for effluent in water bodies at 20 mg/l, and the maximum level of vegetable oils and animal fats to 50 mg/l in Article 34, Resolution number 357 established on March 17, 2005 [10].
Traditional approaches to treat oily effluents include gravity separation, dissolved air flotation (DAF), deemulsification, coagulation, and flocculation. Free oil is removed from wastewater by gravity separation; however, this process cannot remove small oil droplets and emulsions. Oil that adheres to the surface of solid particles can be removed by particle settling [11]. DAF uses solubilized air to increase buoyancy of the smaller oil droplets and improve separation. In addition, emulsified oil is removed by chemical or thermal de-emulsifying processes, or both [11]. Wastewater containing emulsified oil is heated to reduce the viscosity, accentuate density differences, and weaken the interfacial films stabilizing the oil phase. Thereafter, acidification and the addition of a cationic polymer neutralize the negative charges, and elevation of pH to an alkaline level induces flocculation of inorganic salts. Flocs with adsorbed oil are separated and the sludge is dewatered [11]. In this context, the usage of lipolytic microorganisms in wastewater treatment could eliminate this pretreatment process [12]. Chigusa et al. [13] showed that the percentage of fat in wastewater treated with a mixed culture of nine lipase-producing yeast strains decreased by 94%.
Oily effluents can also be pretreated as an approach to conform with the CONAMA resolution, but the achievement of such pretreatment processes depends on the costs of the enzyme [14]. Many industrial processes require breakdown of solids and the prevention of fat blockage or filming in waste systems before the wastewater can be released into the sewage system. This can be accomplished 1) by degradation of organic polymers with a commercial mixture of lipase, cellulase, protease, amylase, and inorganic nutrients; or 2) by sewage treatment, cleaning of holding tanks, septic tanks, grease traps, and other systems. WW07P is produced by Environmental Oasis Ltd. and contains a range of high-performance microorganisms adapted for use in the biological treatment of wastewater containing high fat and oils. It also contains surfactants capable of liquefying heavy fat deposits, thereby assisting in their biodegradation [15].
Lipases and esterases constitute a large category of ubiquitous enzymes expressed by many organisms. Carboxylesterases (EC 3.1.1.1) have broad substrate specificity toward esters and thioesters. Esterases that hydrolyze long-chain acylglycerols (containing more than 10 carbon atoms) are termed lipases (EC 3.1.1.3) and can be considered lipolytic and esterolytic enzymes [16]. Most lipases are water-soluble enzymes that hydrolyze ester bonds of water-insoluble substrates [17]. Therefore, lipases act at the interface between a substrate phase and an aqueous phase, in which the enzyme is dissolved [18]. It is often necessary to combine two or more lipases in order to release a glycerol molecule, since all three acyl chains of a triacylglycerol molecule are rarely released by a single lipase [17].
The α-amylases (E.C.3.2.1.1) are enzymes that hydrolyze starch molecules to generate progressively smaller polymers composed of glucose units [19]. Today, a large number of microbial amylases have almost completely replaced the chemical hydrolysis of starch. The main advantage of using microorganisms for the production of amylase is the ability to bulk produce the enzyme and the easy manipulation of microbes to achieve enzymes with desired characteristics. Moreover, the stability of microbial amylases are higher than those of plant and animal [20].
Microbial proteases are used in waste treatment from various food-processing industries and household activities to solubilize proteinaceous waste and reduce the biological oxygen demand of aquatic systems [21,22]. Hydrolytic enzymes, such as lipases, amylases, and proteases, have a promising application in wastewater treatment of candy, ice cream, dairy, and meat industries. Enzymes for wastewater treatment do not require purification and thus should present a low production cost [23]. These characteristics have led to an increasing interest in enzyme production technology and the search for new microorganisms with a diverse ability to produce enzymes [24-27].
Microbial cellulases are widely used in the paper, wine, animal feed, and textile industries as well as for biofuels production, food processing, olive oil and carotenoid extraction, and waste management [28]. The wastes generated from agricultural fields and agroindustries contain a large amount of unutilized cellulose, thereby causing environmental pollution. Today, these wastes are utilized to produce valuable products, such as enzymes, sugars, biofuels, chemicals, and others [29-33].
Biosurfactants are amphiphilic molecules that possess both polar and nonpolar domains that have effective surface-active properties. There are two main types of these molecules: 1) those that reduce surface tension at the air-water interface (biosurfactants), and 2) those that reduce the interfacial tension between immiscible liquids or at the solid-liquid interface (bioemulsifiers) [34,35]. Biosurfactants usually exhibit an emulsifying capacity, but bioemulsifiers do not necessarily reduce the surface tension [34,35]. These molecules are microbial synthesized, and the different types of biosurfactants include lipopeptides synthesized by many species of Bacillus, glycolipids synthesized by Pseudomonas and Candida sp., phospholipids synthesized by Thiobacillus thiooxidans, and polysaccharidelipid complexes synthesized by Acinetobacter sp. [36-38].
The emulsification of lipids through the breakdown of lipid droplets favors the occurrence of hydrolysis, since the water-soluble lipolytic enzymes have greater surface contact with the substrate to be hydrolyzed. Natural bio-surfactants exhibit low toxicity, biodegradability, and ecological acceptability, which provide an alternative to chemically-prepared conventional surfactants. They can be produced from various substrates but are often generated from renewable resources, such as vegetable oils as well as distillery and dairy wastes [39]. They are applicable for environmental protection and management, bioremediation of soil [40], crude oil recovery, cleanup of hydrocarbon contaminated groundwater, and enhanced oil recovery [41], antimicrobial agents in healthcare [36] or in a wide variety of industrial processes involving emulsification, foaming, detergency, wetting, dispersing, or solubilization [42].
Lipase-producing microorganisms are also found in fat and oil contaminated sources. Thus, they originate from dairy, household, and biotechnology industry wastewaters. The intense competition for limited carbon sources may result in the evolution of novel genes and/or novel biochemical pathways in the specialized environment of wastewaters [43]. Therefore, in this study we isolated lipase and other hydrolytic enzyme-producing microorganisms and assessed their fat and oil degradation capabilities.
2. MATERIALS AND METHODS
2.1. Selection of Lipase-Producing Microorganisms Using Tributyrin as a Growth Substrate
Our group currently has a microbial stock composed of more than 1100 lipolytic microorganisms that were obtained randomly. Among this library, 35 strains were selected for this study that had been originally isolated from four different sewage tanks. Samples were obtained from wastewaters at 2 farm houses (A and C), a dairy industry (B), and a biotechnological industry (D). Samples or its dilutions in sterile water were spread over Spirit Blue agar (Himedia, Mumbai/India) supplemented with 3% (v/v) tributyrin emulsion (20% v/v tributyrin; 0.2% v/v tween 80). After incubation at 25˚C for 1 - 7 d, representative lipolytic colonies of each morphological type were isolated and purified in the same media. The strains were maintained in LB medium (10.0 g/l peptone, 5.0 g/l yeast extract, and 5.0 g/l sodium chloride) supplemented with 50% (v/v) fetal bovine serum and kept at −80˚C.
2.2. Determination of the Degradative Enzymatic Profile of Organic Compounds between the Lipolytic Selected Microorganisms
Lipolytic strains were inoculated in 2 ml of LB broth in a 96-well plate. After 24 h at 25˚C and 30 hz agitation, 2 µl of culture were inoculated in the follow media: 1) tributyrin-agarose [1% (w/v) agarose; 50 mM Tris-HCl pH 6,8; 1 mM CaCl2; 0.6% (v/v) tributyrin emulsion—to detect lipase]; 2) casein-agarose [1% (w/v) agarose, 1 mM CaCl2, 10% (v/v) of a casein solution in 1X PBS pH 7.4—to detect caseinase activity]; 3) corn starch-agarose [1% (w/v) agarose; 50 mM Tris-HCl pH 6.8, 1 mM CaCl2, 0.5% (w/v) corn starch—to detect amylase]; 4) carboxymethylcellulose-agarose [1% (w/v) agarose, 50 mM Tris-HCl pH 6.8, 1 mM CaCl2, 0.5% (w/v) carboxymethylcellulose—to detect cellulase modified from Akhtar et al. [44]; and 5) gelatin media [2 ml media/ assay tube: 50 mM Tris-HCl pH 6.8, 1 mM CaCl2, 10% (w/v) gelatin—to detect gelatin-specific proatease], the formulation of all those media were adapted from Vuong et al. [45]. After 24 h incubation at 25˚C, enzymatic activities were detected by assessing for the presence of a clear halo in the media tests listed above except for gelatin media (see below). For media 1, the result was obtained by direct observation. Media 2 - 4 required revelation prior to a final analysis of the results as follows: 2) 2 min incubation with 1 N HCl solution to precipitate remaining casein; 3) 2 min incubation with 2% iodine solution to colorize remaining starch; 4) 30 min incubation with Congo red [0.25% (w/v) in 0.1 M Tris-HCl, pH 8,0) followed by 5 minutes in a destaining solution (0.5 M NaCl, 0.1 M Tris-HCl, pH 8.0) [46]. Microorganisms that produced a gelatin-specific protease were capable of liquefying the gelatin medium in media 5.
To evaluate the emulsifying capacity of the isolates, 200 µl of a culture with optical density (OD 600 nm) 0.5 were inoculated into 35 ml of LB broth supplemented with 2% soybean oil. The culture was then incubated for 35 d at 37˚C and 250 rpm agitation. When the emulsification of the oil took place in the media, it acquired a milky appearance and consistency.
2.3. Characterization of Extracellular Lipase
2.3.1. Production of Extracellular Enzymatic Extract
Cells (500 µl) were grown in LB medium for 24 h under 30 hz agitation at 25˚C and then seeded on the surface of a sterile dialysis membrane (12,000 Da) that had an equal diameter as a petri dish. The membrane was then placed on Spirit Blue agar containing 0.6% (v/v) tributyrin and incubated at 25˚C for a period of 1 - 7 d (see results section) [47]. The membranes were washed with 2 ml of buffer (10 mM Tris-HCl pH 8.4 and 40 mM NaCl). Thereafter, all experimental steps were conducted on ice. The cell suspension was centrifuged at 25,000 g for 20 min at 4˚C and filtered through a 0.22 µm filter. Then, 4 µl of enzyme extract was applied to the tributyrin-agarose (media one described previously) and incubated for 24 h at 25˚C in order to verify lipase activity.
2.3.2. Determination of Optimal ph and Temperature of Action
Optimal pH was tested using 2 ml of freshly prepared 0.3% tributyrin broth [50 mM Tris-HCl—evaluated at pH values of 4.3, 6.8, 9.8, and 12.3—1 mM CaCl2, and 1.5% (v/v) tributyrin emulsion] and 75 µl of enzymatic extract in a 1 cm cuvette. The optimal temperature was tested using 2 ml of the same broth at the optimal pH and 75 µl of enzymatic extract. Analyzed temperatures were 4˚C, 25˚C, 37˚C, and 50˚C. The reduction in OD was measured at 800 nm in a Shimadzu double beam spectrophotometer (UV-ISO-02, Kyoto, Japan) using a negative control as a standard at 0, 3, and 21 h of incubation.
2.3.3. Quantification of Lipase Activity in P-Nitrophenyl Esters
The lipase assay was performed by measuring the increase in the absorbance at 405 nm in a Thermo Plate microplate reader (TP-reader) caused by the release of p-nitrophenol after hydrolysis of p-nitrophenyl-butyrate (C4), decanoate (C10), and palmitate (C16) at 25˚C for 15 min at pH 8.0 as previously described [48], but modified by adding 10 mM CaCl2 to solution B. An enzymefree control was used as the reference. One unit of lipase (U) was defined as the amount of enzyme that releases 1 µmol p-nitrophenol per min under the assay conditions.
2.3.4. Test of Lipase Activity in the Presence of Chemical Agents and Thermal Resistance
For further characterization, thermal and chemical resistance was evaluated using p-NPB as a substrate. Solution B described above was added with one of following chemical compounds: 0.25% (v/v) H2O2, 0.1% (v/v) NaClO, 0.1% (v/v) liquid detergent, or 10 mM EDTA. For thermal resistance, extracellular enzyme extracts were incubated for 30 min at 50˚C and residual activity was also evaluated using p-NPB as a substrate.
2.4. Identification of Microorganisms
2.4.1. DNA Extraction
Genomic DNA was prepared from a loopful of cells grown in LB agar for 24 h. The cell pellet was resuspended in 250 µl of solution I (50 mM glucose, 25 mM Tris-HCl pH 8.0, and 10 mM EDTA). The cells were lysed by adding 25 µl of solution II [200 mM NaOH and 1% (w/v) SDS], and mixed for 5 min. Then, 500 µl of solution I and 2.5 µl of RNAse A (10 mg/ml) was added and incubated for 2 h at 37˚C. This methodology was adapted from alkaline lysis first described by Birnboim & Doly [49]. DNA was then purified with phenol-chloroform using a standard laboratory protocol and after precipitation, DNA was resuspended in 30 µl of TE (10 mM Tris-HCl pH 8.0 and 1 mM EDTA).
2.4.2. Ribosomal RNA Gene Amplification
Bacterial isolates were identified by sequencing rDNA. The PCR reaction was performed as previously described [50] using the primers 8F (5'-AGAGTTTGATYMTGGCTCAG-3') [51] and 907R (5'-CCGTCAATTCMTTTRAGTTT-3') [52]. Fungal identification was performed as previously described [53] by sequencing D1D2 of 26S rDNA using primers NL1 (5'-GCATATCAATAAGCGGAGGAAAAG) and NL4 (5'-GGTCCGTGTTTCAAGACGG). The sequences of PCR products were analyzed using standard protocols with a dideoxy nucleotide dye terminator (Big Dye vs. 3.1—Applied Biosystems, CA, USA) and Genetic Analyzer 3130 (Applied Biosystems, CA, USA). All 16S and 26S rRNA gene sequences were checked for quality, aligned, and analyzed with CodonCode Aligner v.3.7.1 (CodonCode Corp., Centerville, MA, USA). All sequences were compared with reference sequences in the Ribosomal Database Project (RDP) using Sequence Match and sequences in GenBank using BLASTN.
2.5. Induction of Lipase Production and Synergistic Effect between Different Strains
Pre-inoculum of five different isolates was induced with tributyrin or left untreated (group control) to evaluate if microbial metabolism is increased or not by this triglyceride (pre-induction). Then, 100 µl of pre-inoculum from each isolate were inoculated in LB broth containing 10 µl alamar blue in four different treatments groups: 1) no supplementation, 2) supplemented with 2% tributyrin, 3) supplemented with 2% soybean oil, or 4) supplemented with 2% soybean oil emulsified with sterile bacterial extract contained lipase to evaluate possible synergism between different strains. Monitoring the percentage of alamar blue reduction indicated the conditions in which the culture demonstrated higher metabolism. A calculation of standard deviation indicated the differences between the assessed values and the average. Twoway ANOVA with Bonferroni correction post test was performed using GraphPad Prism version 5.03 for Windows [54].
3. RESULTS AND DISCUSSION
Our lipolytic microbial stock was selected by the presence of a halo around colonies when wastewaters were spread over Spirit Blue agar supplemented with 3% (v/v) tributyrin emulsion. Because of this triglyceride is formed by a glycerol and three four-carbon chains, esterases were selected preferably over lipases. However, because any lipase can also be classified as an esterase, some also showed lipolytic activity among the selected microorganisms.
The 16S/28S rDNA sequence analysis provides molecular identification of isolates. The microorganism collection site, identification, surfactant production [ability to emulsify 2% (v/v) soybean oil in culture medium], and enzymatic profile against agarose tributyrin, agarose casein, gelatin media, agarose corn starch, and agarose carboxymethylcellulose are shown in Table 1. Collection
Table 1. Collection site, identification, GenBank accession numbers and enzymatic profile of isolates.

(-) no enzymatic activity; (+) low enzymatic activity (colony size/halo size < 2); (++) median enzymatic activity (colony size/halo size 2 ≤ x < 5); (+++) strong enzymatic activity (colony size/halo size ≥ 5); *maximum experimental time was 36 days.
sites A and B presented a predominance of members of the Bacillaceae family and at sites C and D, we observed Enterobacteriaceae, Pseudomonas, and Bacillus spp. At site D, Pseudomonas sp. was prevalent.
The extracellular bacterial lipases are of commercial importance, as thousands of lipase units can be produced from only several liters of culture medium [55]. Bacterial lipases are mostly extracellular and are greatly influenced by nutritional and physicochemical factors, such as temperature, pH, nitrogen and carbon sources, presence of lipids, inorganic salts, agitation, and dissolved oxygen concentration [55]. Lipases are mostly inducible enzymes and are thus generally produced in the presence of a lipid source, or any other inducer, such as triacylglycerols, fatty acids, hydrolysable esters, tweens, bile salts, and glycerol [55-57]. However, their production is significantly influenced by other carbon sources, such as sugars, sugar alcohol, polysaccharides, whey, casamino acids, and other complex sources [58,59]. Therefore, 10 ml of extracellular extract were produced for lipase characterization of each of the 35 isolates. Despite the use of an inductor at this step, the lipase activity of two isolates (A9—Staphylococcus spp.- and B9—Bacillus spp.) could not be recovered, even with tributyrin induction (Table 2). In a grease trap ecosystem, the coexistence of microbial strains could supply the nutritional needs due to partial degradation of other biopolymers naturally present in wastewater. Moreover, competition for nutrients and microbial interaction could also lead to the induction of lipase expression so that it may remains active during the first few subcultures. We found that each strain required a different incubation period to express extracellular lipase (Table 2). The exact incubation period required to express lipase over the dialysis membrane was monitored by parallel incubation in spirit blue agar supplemented with tributyrin. It needs to be pointed out that the optimum growth condition was not determined for each strain separately.
Table 2. Collection site, incubation period over dialysis membrane, optimal pH, optimal temperature, and quantification in lipase units with p-NPB (C4), p-NPD (C10), and p-NPP (C16).
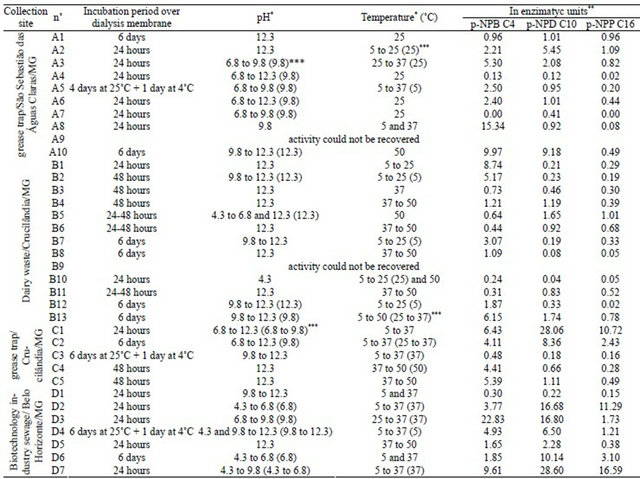
*Range of values in which the extract retains more than 70% of its activity; **One unit of lipase (U) is defined as the amount of enzyme that releases 1 µmol p-nitrophenol per min, in the assay conditions; ***Numbers between parentheses indicate the value in which the enzyme showed higher activity—it sometimes indicates a range of values.
After confirmation of lipase activity, the optimal pH and temperature of the extracts were defined (Table 2). Among the extracts, mesophilic lipases with an optimal pH in the basic range were prevalent, but psychrophiles and acidophilus were also observed. In general, bacterial lipases have optimal activity at neutral or alkaline pH [60-63]. Lipases from Bacillus species are active over a broad pH range (pH 3-12) [64], and our findings indicated that lipase extracts produced by Bacillus species often presented more than one optimal pH value. However, in contrast to the findings of the previous study, lipases secreted by ours isolates belonging to the Bacillus genus showed less activity at high temperatures, with optimal activity at mesophilic temperatures. Only lipases secreted by B. cereus were more thermotolerant, reaching optimal activity at 50˚C. Some of our lipase extracts showed thermal stability up to 50˚C and retained more than 70% of activity after thermal treatment for 30 min, including A5 (Terribacillus sp.), A10 (family Flavobacteriaceae), B3, B4 and B5 (Lysinibacillus spp.), B7, B8, and C5 (Bacillus subtilis), B10 (Mutualistic association of Bacillus sp. and Staphylococcus epidermidis), B11 (Bacillus cereus), C2, D2, D3, D5, and D7 (Pseudomonas spp.), and D1 (Bacillus megaterium) (Table 3). The thermal resistance of lipases from Bacillus and Pseudomonas has already been described [65-68].
Table 3. Percentage of lipase residual activity after various treatments.
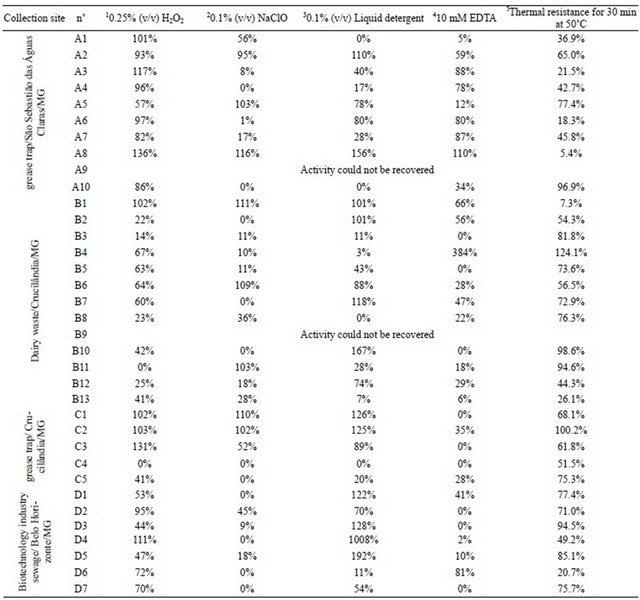
Results are shown for lipase activity after incubation in the presence of 10.25% (v/v) H2O2, 20.1% (v/v) NaClO, 30.1% (v/v) liquid detergent, 410 mM EDTA, or 5after thermal treatment for 30 min at 50˚C. Hydrolysis of p-NPB (C4) was used for all measurements. Controls with no treatment were used for comparison.
Bacterial lipases generally have optimal activity in the temperature range of 30˚C - 60˚C; however some reports have shown that bacterial lipases exist with optimal activity at both low and high temperature ranges [60,61,63, 69]. Lipases quantification using pNP-butyrate (C4), decanoate (C10), and palmitate (C16) was done in a fixed condition for all enzymes, at 25˚C and pH 8.0 (Table 2). The majority of lipase extracts were active on short chain esters, which was expected since the selection was made with tributyrin. In addition, these findings are consistent with the fact that all lipases are also esterases and harbor esterolytic activity. However, isolates A2, A10, B4, B5, B6, C1, C2, D2, D3, D4, D5, D6, and D7 showed a greater affinity for long chain esters, therefore characterizing a “true” lipase extract, since a “true” lipase hydrolyses esters with more than 10 carbon atoms (Table 2).
There are three categories of microbial lipases: nonspecific, regiospecific, and fatty acid-specific [55]. Nonspecific lipases act randomly on triacylglyceride molecules, which results in the complete breakdown tofatty acid and glycerol. Regiospecific lipases are 1, 3-specific lipases that hydrolyze only primary ester bonds, which is observed in lipases produced by some Bacillus species. The third group, fatty acid-specific lipases, comprise those with a pronounced fatty acid preference [55]. Despite showing activity on tributyrin-agarose, the isolated A7 (Bacillus pumillus) was inactive when evaluated for hydrolysis of pNP-ester with 4 and 16 carbons and exhibited poor activity for pNP-ester with 10 carbon atoms. This can be explained by the fact that some lipases have affinity for triacylglycerols, exhibiting no or little activity against monoand diglycerides [17,70]. Another possibility is that A7 isolate was originally capable of producing more than one type of esterase. However, over the course of the passages in vitro, the strain began to only express the esterase that hydrolyzes the ester of 10 carbon atoms. This behavior has been observed in some species in vitro. In addition, it is possible that our system was not sensitive enough to detect the reduced expression after several passages in culture media.
Some biochemical similarities can be observed between microorganisms with the same identification. Lysinibacillus spp. isolates showed many similarities, including the expression of active lipases within 48 h of incubation, having an optimal pH that was alkaline and an optimal temperature of 37˚C of higher, and the absence of gelatinase, caseinase, or amylase activity. Twenty strains belonged to the Bacillaceae family, of which 15 belonged to the Bacillus genus. All strains identified as B. megaterium were gelatin-specific protease producers and were unable to produce cellulase. They expressed extracellular lipase after 24 h of incubation over a dialysis membrane and their lipases had optimal activity at a pH in the alkaline range. In addition, these strains are mesophilic/psicrophilic and are more active against esters with only 4 carbon atoms. Among the five strains identified as B. subtilis, all were amylase and cellulase producers. Five were also capable of producing gelatinspecific proteases, but only 4 strains were able to produce casein-specific proteases as well. All five B. subtilis lipases had better activity at an alkaline pH and against 4 carbon esters. Among the five strains identified as Pseudomonas spp. all were amylase and cellulase producers, and 4 were still capable of producing gelatinand caseinspecific proteases All Pseudomonas lipases presented considerable activity at 37˚C and against 10 carbon atom esters, indicating the presence of at least one type of lipase (Tables 1 and 2).
For preliminary assessment of the potential use of lipases in sewage treatment, the thermal and chemical resistance of the enzymes was evaluated using p-NPB as a substrate (Table 3). An abundance of cleaning products and other chemical compounds are released daily from grease traps. None of the extracts maintained more than 70% residual activity in all analyzed conditions. Isolate A2 (unclassified Saccharomycetales) exhibited the best results for all conditions combined and maintained more than 50% of residual activity in the presence of H2O2, NaClO, liquid detergent, EDTA, and thermal treatment. Other extracts that showed good results included B6 (from Bacillus cereus) and B12 (from Acetobacter pasteurianus). They showed residual activity in greater than 20% of all of the conditions. In addition to the A2 extract, a combination of extracts could also be potentially advantageous for the development of a biological method for cleaning O&G from grease traps and sewage treatment plant equipment. The strong inhibition caused by 0.1% (v/v) detergent could be due to inactivation of the enzyme as a result of a disruption of its tertiary structure. When dishes, clothes or floor are washed, high concentrations of detergents and soaps are released into the sewer over a short period of time. Therefore, the choice of a stable enzyme is an important aspect for sewage treatment. Therefore, we analyzed the ability of isolates to produce multiple degradative enzymes and found that isolates B2, B8, C4, C5, D1, D2, and D7 presented a wide range of ability in utilizing biopolymers commonly present in wastewater (Table 3). These isolates all produced proteases and at least one other hydrolytic enzyme besides lipase. Among the 35 strains, more than 70% were able to produce at least two enzymes or more. This multiple approach allows for the use of a small number of isolates in sewage treatment, since each one individually has the ability to secrete more than one enzyme at a time. This can also reduce the requirement for nutritional supplementation of the system. Among the extracellular lipases produced from the 35 strains, approximately 45% were resistant to 0.25% (v/v) H2O2, approximately 20% were resistant to 0.1% (v/v) of NaClO or 10 mM EDTA, and more than 50% were resistant to 0.1% (v/v) liquid detergent (Table 3).
Bioremediation techniques in situ include the introduction of different strains of live microorganisms to wastewater at various stages of its treatment. Almost all known methods for sludge treatment introduce microbial strains in the log phase of growth. These microbes are in active phase of multiplication, however their action requires time to degrade the substrats [71]. Estera et al. [72] previously developed a method for reducing the time for degradation, which includes first providing an enzyme mixture capable of digesting natural polymeric materials, and only the adding at least one species of fermenting bacteria to the system that is able to ferment the resulting suspension. Dash et al. [71] described a composition for the treatment of wastewater to remove pollutants that was comprised of a synergistic composition of microbes, enzymes, and cofactors/nutrients. The microbes in the composition were selected Pseudomonas aeruginosa, Pseudomonas fluorescens, Pseudomonas putida, Pseudomonas desmolyticum, Coriolus versicolour, Lactobacillus sp., Bacillus subtilis, Bacillus cereus, Staphylococcus sp., and Phanerochaete chrysosporium, alone or in combination. The enzymes produced include proteases, lipases, amylases, glucose oxidases, and others. This composition exhibits synergy and effectively removes pollutants from the wastewater. Enzymes act by dissociating the molecules to simpler forms, and microbes utilize these intermediates in their metabolism, which results in the complete degradation of the pollutants in the wastewater. Microbes will grow faster due to the increasing availability of intermediates and therefore will produce more enzymes that can further degrade the pollutants. Thus, enzymes and microbes are interdependent and work together to facilitate faster degradation of the pollutant molecules [71].
Diverse microorganisms are able to hydrolyze different types of oil. However, the biodegradation process can be lengthy due to the low water solubility of oil [73]. In natural or induced conditions, many microorganisms are able to produce emulsifying agents, which minimizes the time required for biodegradation of O&G by enhancing hydrophobic substrate bioavailability [74]. Cell-bound esterase synthesis has been recently reported in association with the generation of surface active substances, indicating the coupled function of emulsification with lipolytic activity [75,76]. Biosurfactants increase the uptake of microorganisms when grown on insoluble substrates and also increase the efficiency of bioremediation [77].
As shown by Gautam et al. [78], Saharam et al. [79], and Pattanathu et al. [80], several species belonging to the genera Pseudomonas are capable of producing different classes of biosurfactants. In our study, we found that various isolates belonging to those genera and Bacilaceae class (A3, A4, A5, A6, A7, B3, B4, B5, B10, C2, C4, C5, D2, and D5) were able to emulsify 2% soybean oil, although we did not identify the class of biosurfactant produced (Table 1).
Acetobacter pasteurianus is an acetogenic bacterial species normally associated with wine production and spoilage [81]. It produces acetic acid due to the incomplete oxidation of a carbon source into CO2 [81]. In our study, isolates B12 and B13 were associated with wastewater and lipase production. These enzymes preferentially hydrolyzed triglycerides with esters of 4 carbon atoms and were active in the basic pH range. Despite these similarities, they showed different chemical and thermal resistance, indicating that they are most likely different enzymes. In addition, these two strains were not able to produce any other type of hydrolytic enzyme or biosurfactant among those evaluated.
According to the literature, emulsification of lipids would favor its hydrolysis, since the water-soluble lipolytic enzymes have greater surface contact with the substrate to be hydrolyzed due to the breakdown of lipid droplets. To evaluate the behavior of some of our isolates, five strains with different emulsification and hydrolysis profiles were subjected to metabolic quantification by reduction of alamar blue, in four different conditions: 1) LB broth culture media, 2) LB broth with 2% tributyrin (triglyceride of 4 carbons), 3) LB with 2% soybean oil, and (iv) LB with 2% lipid emulsion (Figure 1). The initial curve of alamar blue reduction indicated the best volume of pre-inoculum and the optimal period of incubation in the presence of the reagent for each strain. The graphs in Figure 1 show that the presence of an emulsified lipid in the culture medium did not increased the metabolic rates of any of the microorganisms, and rather the metabolic rate was reduced. However, previous induction of lipase production by the addition of tributyrin to pre-inoculum media proved effective in raising the metabolism in all experimental conditions for the A3 and B1 strains. The only situation in which the induction was not efficient was for strain B13, wherein the pre-inoculum that was not induced was more metabolically efficient in all experimental conditions. However, because lipase production may be influenced by the carbon source used in the induction process, the absence of lipase production for strain B13 in the presence of tributyrin is justifiable, since no other inducer was evaluated. The metabolism of strains B1 and B12 was markedly increased in presence of tributyrin compared to the presence of soybean oil, and these findings were in agreement with the hydrolysis of p-nitrophenol esters. Strains B12 and B13 were also more efficient at hydrolyzing
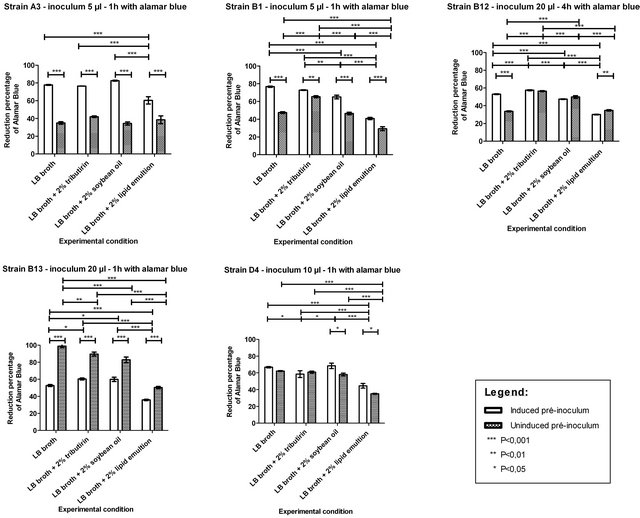
Figure 1. Metabolic status of five different strains when induced or non-induced pre-inocula were challenged against a simple trigliceride, a complex mixture of triglicerides, or an oily emulsion of a complex mixture of triglicerides. Quantification of metabolism was performed by determining the percent reduction of 10% (v/v) alamar blue reagent.
p-nitrophenol butyrate and more metabolically active in LB media containing 2% tributyrin. Similar results were observed for strain D4, which was more efficient in hydrolyzing p-nitrophenol decanoate and more metabolically active in LB media containing 2% soybean oil (Figure 1).
Cellulose is the most common organic polymer. It is the most prevalent material in waste from agriculture and the most abundant renewable biopolymer on Earth [82]. A promising strategy for utilization of this energetic renewable source is microorganism-mediated hydrolysis of discarded lignocellulose, followed by fermentation of the resulting compound, which produces the desired metabolites or biofuel [82]. Among our selected lipolytic microorganisms, 34.3% presented also hydrolytic activity against CM cellulose. Parmar et al. [83] showed that a mixture of hydrolytic enzymes, such as cellulases, proteases, and lipases, in equal proportion by weight, reduced total suspended solids (TSS) by 30% - 50% and improved sedimentation of solids in sludge. An increase in solid reduction was observed with increasing enzyme concentration. That reduction occurred due to the hydrolysis of residual polymers, proving that enzymatic synergism can effectively reduce the organic matter in industrial wastewater pretreatment plants.
4. CONCLUSION
Cultures isolated from the vast diversity of microorganisms provide a major source of biological material for industrial biocatalysts and other environmental applications. Lipases and esterases obtained in this study presented different resistances and affinities. Enzymes were characterized with the aim of identifying suitable candidates for use in wastewater treatment. The good stability of isolated lipases in the presence of chemical agents, thermal stability, wide range of pH activity and tolerance, and affinity for different lengths of ester chains indicates that some of these enzymes may be good candidates for the hydrolysis of organic compounds and polymers present in the wastewater of diverse industries. As bacterial enzymes are highly robust, being active over a wide range of pH and temperature and possessing a diverse range of substrate specificity, they could easily be used in pretreatment sludge processes, since they possess adequate resistance of some chemical elements and can be produced at a low cost. The absence of purification requirements contributes to the cost reduction of using these enzymes for sewage treatment. In addition, it is possible that a combination of two or more enzymes may facilitate the process of complete hydrolysis of triglycerides, proteins, and lignocellulose that normally occurs in the wastes of industrial processes. However, careful selection of the strains to be used in sewage treatment is essential, because it may be possible to use fewer strains to achieve the same purpose, since several strains showed the capability of producing two or more enzymes. This will ensure that the hydrolysis of all compounds commonly discarded in wastewater will be sufficiently achieved. In order to give a more real reflection of degradation capabilities of those microorganisms, further studies will use a simulation condition of common sewage as culture to test the degradation capabilities, and also focus on optimizing hydrolysis conditions with the aim of using combined enzyme/microbial strategies for improving industrial wastewater treatment processes.
5. ACKNOWLEDGEMENTS
We thank CNPQ for financial support, projects number: 580311/ 2008-2, 560912/2010-2, 551113/2011-1, 300721/2012-9.

REFERENCES
- Becker, P., Koster, D., Popov, M.N., Markossian, S., Antranikian, G. and Markl, H. (1999) The biodegradation of olive oil and treatment of lipid-rich wool wastewater under aerobic thermophilic condition. Water Research, 33, 653-660. doi:10.1016/S0043-1354(98)00253-X
- Stoll, U. and Gupta, H. (1997) Management strategies for oil and grease residues. Waste Management & Research, 15, 23-32. doi:10.1006/wmre.1996.0062
- Cadoret, A., Conrad, A. and Block, J.C. (2002) Availability of low and high molecular weight substrates to extracellular enzymes in whole and dispersed activated sludges. Enzyme and Microbial Technology, 31, 179-186. doi:10.1016/S0141-0229(02)00097-2
- Sheng, G.P. and Yu, H.Q. (2006) Characterization of extracellular polymeric substances of aerobic and anaerobic sludge using three-dimensional excitation and emission matrix fluorescence spectroscopy. Water Research, 40, 1233-1239. doi:10.1016/j.watres.2006.01.023
- Wagner, M., Loy, A., Nogueira, R., Purkhold, U., Lee, N. and Daims, H. (2002) Microbial community composition and function in wastewater treatment plants. Antonie van Leeuwenhoek, 81, 665-680. doi:10.1023/A:1020586312170
- Perle, M., Kimchie, S. and Shelef, G. (1995) Some biochemical aspects of the anaerobic degradation of dairy wastewater. Water Research, 29, 1549-1554. doi:10.1016/0043-1354(94)00248-6
- Lefebvre, X. Paul, E. and Mauret, M. (1998) Kinetic characterization of saponified domestic lipid residues aerobic biodegradation. Water Research, 32, 3031-3038. doi:10.1016/S0043-1354(98)00053-0
- Vidal, G., Carvalho, A., Méndez, R. and Lema, J.M. (2000) Influence of the content in fats and proteins on the anaerobic biodegradability of dairy wastewaters. Bioresource Technology, 74, 231-239. doi:10.1016/S0960-8524(00)00015-8
- Masse, L., Kennedy, K.J. and Chou, S. (2001) Testing of alkaline and enzymatic hydrolysis pretreatments for fat particles in slaughterhouse wastewater. Bioresource Technology, 77, 145-155. doi:10.1016/S0960-8524(00)00146-2
- CONAMA 357—Conselho Nacional do Meio Ambiente. (2005) Legislação Ambiental Federal, resolution 357. www.mma.gov.br
- Cheryan, M. and Rajagopalan, N. (1998) Membrane processing of oil streams. Wastewater treatment and waste reduction. Journal of Membrane Science, 151, 13-28. doi:10.1016/S0376-7388(98)00190-2
- Bhumibhamon, O., Koprasertsak, A. and Funthong, S. (2002) Biotreatment of high fat and oil wastewater by lipase producing microorganisms. Kasetsart Journal Natural Science, 36, 261-267.
- Chigusa, S., Hasegawa, T., Yamamoto, N. and Watanabe Y. (1996) Treatment of wastewater form oil manufacturing plant by yeast. Water Science and Technology, 34, 51-58. doi:10.1016/S0273-1223(96)00820-7
- Alberton, D., Mitchell, D.A., Cordova, J., Peralta-Zamora, P. and Krieger, N. (2010) Production and application of R. microsporus lipases. Food Technology and Biotechnology, 48, 28-35.
- Environmental Oasis Ltd. (2012) WW07P—Grease removal and food processing. http://www.oasisenviro.co.uk/ww07pproductinfo.html
- Wong, H. and Schotz, M.C. (2002) The lipase gene family. Journal of Lipid Research, 43, 993-999. doi:10.1194/jlr.R200007-JLR200
- Gilham, D. and Lehner, R. (2005) Techniques to measure lipase and esterase activity in vitro. Methods, 36, 139-147. doi:10.1016/j.ymeth.2004.11.003
- Bussamara, R., Fuentefria, A.M., Oliveira, E.S., Broetto, L., Simcikova, M., Valente, P., Schrank, A. and Vainstein, M.H. (2010) Isolation of a lipase-secreting yeast for enzyme production in a pilot-plant scale batch fermentation. Bioresource Technology, 101, 268-275. doi:10.1016/j.biortech.2008.10.063
- Windish, W.W. and Mhatre, N.S. (1965) Microbial amylases. In: Wu, W., Ed., Advances in Applied Microbiology, Elsevier Inc., Houston, 273-304.
- Tanyildizi, M.S., Ozer, D. and Elibol, M. (2005) Optimization of -amylase production by Bacillus sp. using response surface methodology. Process Biochemistry, 40, 2291-2296. doi:10.1016/j.procbio.2004.06.018
- Gupta, R., Beg, Q.K. and Lorenz, P. (2002) Bacterial alkaline proteases: Molecular approaches and industrial applications. Applied Microbiology and Biotechnology, 59, 15-32. doi:10.1007/s00253-002-0975-y
- Ichida, J.M., Krizova, L., LeFevre, C.A., Keener, H.M., Elwell, D.L. and Burtt Jr., E.H. (2001) Bacterial inoculum enhances keratin degradation and biofilm formation in poultry compost. Journal of Microbiological Methods, 47, 199-208. doi:10.1016/S0167-7012(01)00302-5
- Rigo, E., Rigoni, R.E., Lodea, P., Oliveira, D., Freire, D.M.G. and Luccio, M. (2008) Application of different lipases as pretreatment in anaerobic treatment of wastewater. Environmental Engineering Science, 25, 1243-1248. doi:10.1089/ees.2007.0197
- Hu, W.C., Thayanithy, K. and Forster, C.F. (2002) A kinetic study of the anaerobic digestion of ice-cream wastewater. Process Biochemistry, 37, 965-971. doi:10.1016/S0032-9592(01)00310-7
- Mongkolthanaruk, W. and Dharmsthiti, S. (2002) Biodegradation of lipid-rich wastewater by a mixed bacterial consortium. International Biodeterioration & Biodegradation, 50, 101-105. doi:10.1016/S0964-8305(02)00057-4
- Cavalcanti, E.A.C., Gutarra, M.L.E., Freire, D.M.G., Castilho, L.R. and Sant’Anna Jr., G.L. (2005) Lipase production by solid-state fermentation in fixed-bed bioreactors. Brazilian Archives of Biology and Technology, 48, 79-84. doi:10.1590/S1516-89132005000400010
- Leal, M.C.C.R., Freire, D.M.G., Cammarota, M.C. and Sant’Anna Jr., G.L. (2006) Effect of enzymatic hydrolysis on anaerobic treatment of dairy wastewater. Process Biochemistry, 41, 1173-1178. doi:10.1016/j.procbio.2005.12.014
- Kuhad, R.C., Rishi, G. and Singh, A. (2011) Microbial cellulases and their industrial applications. Enzyme Research, 2011, Article ID: 280696. doi:10.4061/2011/280696
- Kuhad, R.C., Gupta, R. and Khasa, Y.P. (2010) Bioethanol production from lignocellulosic biomass: An overview. In: Lal B, Ed., Wealth from Waste, Teri Press, New Delhi, 53-106.
- Karmakar, M. and Ray, R.R. (2011) Current trends in research and application of microbial cellulases. Research Journal of Microbiology, 6, 41-53. doi:10.3923/jm.2011.41.53
- Gupta, R., Mehta, G., Khasa, Y.P. and Kuhad, R.C. (2011) Fungal delignification of lignocellulosic biomass improves the saccharification of cellulosics. Biodegradation, 22, 797-804. doi:10.1007/s10532-010-9404-6
- Gupta, R., Khasa, Y.P. and Kuhad, R.C. (2011) Evaluation of pretreatment methods in improving the enzymatic saccharification of cellulosic materials. Carbohydrate Polymers, 84, 1103-1109. doi:10.1016/j.carbpol.2010.12.074
- Gupta, R., Sharma, K.K. and Kuhad, R.C. (2009) Separate hydrolysis and fermentation (SHF) of Prosopis juliflora, a woody substrate, for the production of cellulosic ethanol by Saccharomyces cerevisiae and Pichia stipitisNCIM 3498. Bioresource Technology, 100, 1214-1220. doi:10.1016/j.biortech.2008.08.033
- Batista, R.M., Rufino, R.D., Luna, J.M., Souza, J.E.G. and Sarubbo, L.A. (2010) Effect of medium components on the production of a biosurfactant from Candida tropicalis, applied to the removal of hydrophobic contaminants in soil. Water Environment Research, 82, 418-425. doi:10.2175/106143009X12487095237279
- Luna, J.M., Rufino, R.D., Campos-Takaki, G.M. and Sarubbo, L.A. (2012) Properties of the biosurfactant produced by Candida sphaerica cultivated in low-cost substrates. Chemical Engineering Transactions, 27, 67-72. doi:10.3303/CET1227012
- Moussa, T.A.A., Ahmed, A.M. and Abdelhamid, S.M.S. (2006) Optimization of cultural conditions for biosurfactant production from Nocardia amarae. Journal of Applied Sciences Research, 11, 844-850.
- Nitschke, M. and Pastore, G.M. (2006) Production and properties of a surfactant obtained from Bacillus subtilis grown on cassava wastewater. Bioresource Technology, 97, 336-341. doi:10.1016/j.biortech.2005.02.044
- Anyanwu, C.U. (2010) Surface activity of extracellular products of a Pseudomonas aeruginosa isolated from petroleum contaminated soil. International Journal of Environmental Sciences, 1, 225-235.
- Abouseoud, M., Maachi, R. and Amrane, A. (2007) Biosurfactant Production from olive oil by Pseudomonas fluorescens. In: Méndez-Vilas, A., Ed., Communicating Current Research and Educational Topics and Trends in Applied Microbiology, Formatex, Badajoz, 340-347.
- Van Dyke, M.I., Lee, H. and Trevors, J.T. (1991) Application of microbial surfactants. Biotechnology Advances, 9, 241-252. doi:10.1016/0734-9750(91)90006-H
- Ron, E.Z. and Rosenberg, E. (2001) A review of natural roles of biosurfactants. Environmental Microbiology, 3, 229-236. doi:10.1046/j.1462-2920.2001.00190.x
- Desai, J.D. and Banat, I.M. (1997) Microbial production of biosurfactants and their commercial potential. Microbiology and Molecular Biology Reviews, 61, 47-64.
- Bramucci, M., Kane, H., Chen, M. and Nagarajan, V. (2003) Bacterial diversity in an industrial wastewater bioreactor. Applied Microbiology and Biotechnology, 62, 594-600. doi:10.1007/s00253-003-172-x
- Akhtar, N., Ghauri, M.A., Iqbal, A., Anwar, M.A. and Akhtar, K. (2008) Biodiversity and phylogenetic analysis of culturable bacteria indigenous to Khewra Salt Mine of Pakistan and their industrial importance. Brazilian Journal of Microbiology, 39, 143-150. doi:10.1590/S1517-83822008000100029
- Vuong, C., Gotz, F. and Otto, M. (2000) Construction and characterization of an agr deletion mutant of Staphylococcus epidermidis. Infection and Immunity, 68, 1048- 1053. doi:10.1128/IAI.68.3.1048-1053.2000
- Ruegger, M.J.S. and Tauk-Tornisielo, S.M. (2004) Atividade da celulase de fungos isolados do solo da Estação Ecológica de Juréia-Itatins, São Paulo, Brasil. Brazilian Journal of Botany, 27, 205-211. doi:10.1590/S0100-84042004000200001
- Christen, G.L. and Marshall, R.T. (1984) Selected properties of lipase and protease of Pseudomonas fluorescens 27 produced in four media. Journal of Dairy Science, 67, 1680-1687. doi:10.3168/jds.S0022-0302(84)81492-7
- Fakhreddine, L., Kademi, A., Ait-Abdelkader, N. and Baratti, J.C. (1998) Microbial growth and lipolytic activities of moderate thermophilic bacterial strains. Biotechnology Letters, 20, 879-883. doi:10.1023/A:1005371727699
- Birnboim, H.C. and Doly, J. (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Research, 7, 1513-1523. doi:10.1093/nar/7.6.1513
- Pontes, D.S., Pinheiro, F.A., Lima-Bittencourt, C.I., Guedes, R.L,, Cursino, L., Barbosa, F., Santos, F.R., ChartoneSouza, E. and Nascimento, A.M. (2009) Multiple antimicrobial resistance of gram-negative bacteria from natural oligotrophic lakes under distinct anthropogenic influence in a tropical region. Microbial Ecology, 58, 762-772. doi:10.1007/s00248-009-9539-3
- Felske, A., Rheims, H., Wolterink, A., Stackebrandt, E. and Akkermans, A.D.L. (1997) Ribosome analysis reveals prominent activity of an uncultured member of the class Actinobacteria in grassland soils. Microbiology, 143, 2983-2989. doi:10.1099/00221287-143-9-2983
- Lane, D.J. (1991) 16S/23S rDNA sequencing. In: Stackebrandt, E. and Goodfellow, M., Eds., Nucleic Acid Techniques in Bacterial Systematics, John Wiley &Sons, New York, 115-148.
- Lachance, M.A., Bowles, J.M., Starmer, W.T. and Barker, J.S.F. (1999) Kodamaea kakaduensis and Candida tolerans, two new ascomycetous yeast species from Australian Hibiscus flowers. Canadian journal of microbiology, 45, 172-177.
- GraphPad Software (2009) Prism 5 for Windows: Version 5.03. www.graphpad.com
- Gupta, R., Gupta, N. and Rathi, P. (2004) Bacterial lipases: An overview of production, purification and biochemical properties. Applied Microbiology and Biotechnology, 64, 763-781. doi:10.1007/s00253-004-1568-8
- Sharma, R., Chisti, Y. and Banerjee, U.C. (2001) Production, purification, characterization and applications of lipases. Biotechnology Advances, 19, 627-662. doi:10.1016/S0734-9750(01)00086-6
- Rathi, P., Saxena, R.K. and Gupta, R. (2001) A novel alkaline lipase from Burkholderia cepacia for detergent formulation. Process Biochemistry, 37, 187-192. doi:10.1016/S0032-9592(01)00200-X
- Ghanem, E.H., Al-Sayeed, H.A. and Salch, K.M. (2000) An alkalophilic thermostable lipase produced by a new isolate of Bacillus alcalophilus. World Journal of Microbiology and Biotechnology, 16, 459-464. doi:10.1023/A:1008947620734
- Rashid, N., Shimada, Y., Ezaki, S., Atomi, H. and Imanaka, T. (2001) Low-temperature lipase from psychrotrophic Pseudomonas sp. strain KB700A. Applied and Environmental Microbiology, 67, 4064-4069. doi:10.1128/AEM.67.9.4064-4069.2001
- [61] Dharmsthiti, S. and Luchai, S. (1999) Production, purification and characterization of thermophilic lipase from Bacillus sp. THL027. FEMS Microbiology Letters, 179, 241-246. doi:10.1111/j.1574-6968.1999.tb08734.x
- [62] Lee, O.-W., Koh, Y.-S., Kim, K.-J., Kim, B.-C., Choi, H.- J., Kim, D.-S., Suhartono, M.T. and Pyun, Y.-R. (1999) Isolationa and characterization of a thermophilic lipase from Bacillus thermoleovorans ID-1. FEMS Microbiology Letters, 179, 393-400. doi:10.1111/j.1574-6968.1999.tb08754.x
- [63] Kanwar, L. and Goswami, P. (2002) Isolation of a Pseudomonas lipase produced in pure hydrocarbon substrate and its applications in the synthesis of isoamyl acetate using membrane-immobilized lipase. Enzyme and Microbial Technology, 31, 727-735. doi:10.1016/S0141-0229(02)00191-6
- [64] Sunna, A., Hunter, L., Hutton, C.A. and Bergquist, P.L. (2002) Biochemical characterization of a recombinant thermoalkalophilic lipase and assessment of its substrate enantioselectivity. Enzyme and Microbial Technology, 31, 472-476. doi:10.1016/S0141-0229(02)00133-3
- [65] Bradoo, S., Saxena, R.K. and Gupta, R. (1999) Two acidothermotolerant lipases from new variants of Bacillus spp. World Journal of Microbiology and Biotechnology, 15, 87-91. doi:10.1023/A:1008835015132
- [66] Hassan F, Shah AA, and Abul-Hameed A. (2006) Influence of culture conditions on lipase production by Bacillus sp. FH5. Annals of Microbiology, 56, 247-252. doi:10.1007/BF03175013
- [67] Bora, L. and Kalita, M.C. (2007). Production and optimization of thermostable lipase from a thermophilic Bacillus sp. LBN 4. The Internet Journal of Microbiology 4. http://www.ispub.com/journal/the-internet-journal-of-microbiology/volume-4-number-1/production-and-optimization-of-thermostable-lipase-from-a-thermophilic-bacillus-sp-lbn-4.html#sthash.qRHstqSc.dpbs
- [68] Zhang, J.W. and Zeng, R.Y. (2008) Molecular cloning and expression of a cold-adapted lipase gene from an Antarctic deep sea psychrotrophic bacterium Pseudomonas sp. 7323. Marine Biotechnology, 10, 612-621. doi:10.1007/s10126-008-9099-4
- [69] Kumar, S., Kikon, K., Upadhyay, A., Kanwar, S.S. and Gupta, R. (2005) Production, purification, and characterization of lipase from thermophilic and alkaliphilic Bacillus coagulans BTS-3. Protein Expression and Purification, 41, 38-44. doi:10.1016/j.pep.2004.12.010
- [70] Litthauer, D., Ginster, A. and Skein, E.V.E. (2002) Pseudomonas luteola lipase: a new member of the 320-residue Pseudomonas lipase family. Enzyme and Microbial Technology, 30, 209-215. doi:10.1016/S0141-0229(01)00469-0
- [71] Xu, X. (2000) Production of specific-structured triacylglycerols by lipase-catalyzed reactions: a review. European Journal of Lipid Science and Technology, 102, 287- 303. doi:10.1002/(SICI)1438-9312(200004)102:4<287::AID-EJLT287>3.0.CO;2-Q
- [72] Dash, S.S., Subramani, R. and Kompala, D.S. (2011). A method for rapid treatment of wastewater and a composition thereof. World Intellectual Property Organization (WIPO), Geneva.
- [73] Estera, S.D., Lund, S., Olof, N. and Helsingborg, S. (2006). Method for digestion of sludge in water purification. US Patent No. 20060086659, PCT No. PCT/SE03/ 01436.
- [74] Snape, I., Ferguson, S., Harvey, P.M. and Riddle M. (2006) Investigation of evaporation and biodegradation of fuel spills in Antarctica: II extent of natural attenuation at Casey station. Chemosphere, 63, 89-98. doi:10.1016/j.chemosphere.2005.07.040
- [75] Banat, I.M., Makkar, R.S. and Cameotra, S.S. (2000) Potential commercial applications of microbial surfactants. Applied Microbiology and Biotechnology, 53, 495-508. doi:10.1007/s002530051648
- [76] Bach, H., Bedichevsky, Y. and Gutnick, D. (2003) An exocellular protein from the oil-degrading microbe Acinetobacter venetianums RAG-1 enhances the emulsifying activity of the polymeric bioemulsifier emulsan. Applied and Environmental Microbiology, 69, 2608-2615. doi:10.1128/AEM.69.5.2608-2615.2003
- [77] Mathur, C., Prakash, R., Ali, A., Kaur, J., Cameotra, S.S. and Prakash, N.T. (2010) Emulsification and hydrolysis of oil by Syncephalastrum racemosum. Defence Science Journal, 60, 251-254.
- [78] Saimmai, A., Rukadee, O., Onlamool, T., Sobhon, V. and Maneerat, S. (2012) Isolation and functional characterization of a biosurfactant produced by a new and promising strain of Oleomonas sagaranensis AT18. World Journal of Microbiology Biotechnology, 28, 2973-2986. doi:10.1007/s11274-012-1108-0
- [79] Gautam, K.K. and Tyagi, V.K. (2006) Microbial surfactants: a review. Journal of Oleo Science, 55, 155-166. doi:10.5650/jos.55.155
- [80] Saharan, B.S., Rahu, R.K. and Sharma, D. (2011) A review on biosurfactants: Fermentation, current developments and perspectives. Genetic Engineering and Biotechnology Journal, 2011, GEBJ-29.
- [81] Pattanathu, K.S.M., Rahman, K.S.M. and Gakpe, E. (2008) Production, characterisation and applications of biosurfactants—Review. Biotechnology, 7, 360-370. doi:10.3923/biotech.2008.360.370
- [82] Prieto, C., Jara, C., Mas, A. and Romero, J. (2007) Application of molecular methods for analysing the distribution and diversity of acetic acid bacteria in Chilean vineyards. International Journal of Food Microbiology, 115, 348-355.
- [83] Sukumaran, R.K., Singhania, R.R. and Pandey, A. (2005) Microbial cellulases—Production, applications and challenges. Journal of Scientific & Industrial Research, 64, 832-844.
- [84] Parmar, N., Singh, A. and Ward, O.P. (2001) Enzyme treatment to reduce solids and improve settling of sewage sludge. Journal of Industrial Microbiology and Biotechnology, 26, 383-386. doi:10.1038/sj.jim.7000150

