Open Journal of Statistics
Vol.04 No.10(2014), Article ID:51474,5 pages
10.4236/ojs.2014.410080
Estimation of Multivariate Sample Selection Models via a Parameter-Expanded Monte Carlo EM Algorithm
Phillip Li
Department of Economics, Office of the Comptroller of the Currency, Washington, DC, USA
Email: Phillip.Li@occ.treas.gov
Copyright © 2014 by Phillip Li.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 6 September 2014; revised 5 October 2014; accepted 2 November 2014
ABSTRACT
This paper develops a parameter-expanded Monte Carlo EM (PX-MCEM) algorithm to perform maximum likelihood estimation in a multivariate sample selection model. In contrast to the cur- rent methods of estimation, the proposed algorithm does not directly depend on the observed-da- ta likelihood, the evaluation of which requires intractable multivariate integrations over normal densities. Moreover, the algorithm is simple to implement and involves only quantities that are easy to simulate or have closed form expressions.
Keywords:
Multivariate Sample Selection, Heckman Correction, Incidental Truncation, Expectation Maximization

1. Introduction
Sample selection models, pioneered in [1] - [3] , are indispensable to researchers who use observational data for statistical inference. Among the many variants of these types of models, there is a growing interest in multiva- riate sample selection models. These are used to model a system of two or more seemingly unrelated equations, where the outcome variable for each equation may be non-randomly missing or censored according to its own stochastic selection variable. Applications range from modeling systems of demand equations [4] [5] to house- hold vehicle usage [6] - [8] . A common specification is to assume a correlated multivariate normal distribution underlying both the outcomes of interest and the latent variables in the system.
There are two dominant approaches in the current literature to estimate these models. One approach is to use maximum likelihood (ML) estimation. However, as noted in the literature, a major hurdle in evaluating the like- lihood is that it requires computations of multivariate integrals over normal densities, which do not generally have closed form solutions. [9] discusses the ML estimation of these models and proposes to use the popular Geweke, Hajivassiliou, and Keane (GHK) algorithm to approximate these integrals in a simulated ML frame- work. While this strategy works reasonably well, the GHK algorithm can be difficult to implement. Another popular approach is to use two-step estimation (see [10] for a survey). In general, there is a tradeoff in the statistical properties and the computational simplicity for these estimators. If efficiency and consistency are of pri- mary concern, then ML estimation should be preferred over two-step estimation.
The objective of this paper is to develop a simple ML estimation algorithm for a commonly used multivariate sample selection model. In particular, this paper develops a parameter-expanded Monte Carlo expectation maximization (PX-MCEM) algorithm that differs from [9] in a few important ways. First, the PX-MCEM algo- rithm does not use the observed-data likelihood directly, so it avoids the aforementioned integrations. Second, the proposed iterative algorithm does not require the evaluations of gradients or Hessians, which become increa- singly difficult to evaluate with more parameters and equations. Third, the algorithm is straightforward to implement. It only depends on quantities that are either easy to simulate or have closed form expressions. This last point is especially appealing when estimating the covariance matrix parameter since there are non-standard restrictions imposed onto it for identification.
This paper is organized as follows. The multivariate sample selection model (MSSM) is formulated in Section 2. Section 3 begins with a brief overview of the EM algorithm for the MSSM and continues with the develop- ment of the PX-MCEM algorithm. Methods to obtain the standard errors are discussed. Section 4 offers some concluding remarks.
2. Multivariate Sample Selection Model
The MSSM is
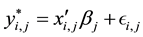 (1)
(1)
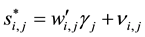 (2)
(2)
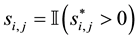 (3)
(3)
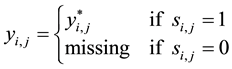 (4)
(4)
for observations , and equations
, and equations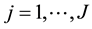 . In the previous expressions,
. In the previous expressions, 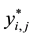 is the continuous outcome of interest for observation
is the continuous outcome of interest for observation 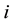 and equation
and equation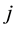 . Using similar indexing notation,
. Using similar indexing notation, 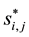 is the latent
is the latent
variable underlying the binary selection variable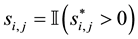 , where
, where 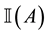 denotes an indicator function
denotes an indicator function
that equals 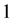 if event
if event 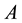 is true and 0 otherwise. Sample selection is incorporated by assuming that
is true and 0 otherwise. Sample selection is incorporated by assuming that 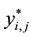 is
is
missing when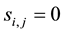 . Otherwise,
. Otherwise, 





transpose.
Furthermore, 



vectors of parameters. Define 


contain at least one exogenous covariate that does not overlap with 
trictions). The unobserved errors 





The submatrix 

The covariates and binary selection variables are always observed. Without loss of generality, assume that the outcomes for any observation 





3. Estimation
3.1. Overview of the EM Algorithm
The PX-MCEM algorithm is based on the EM algorithm of [12] . The basic idea behind the EM algorithm is to first augment 


where 
The EM algorithm then proceeds iteratively between an expectation step (E-step) and a maximization step (M-step) as follows. In iteration 

where the expectation is taken with respect to the conditional predictive distribution for the missing data,


Denote the maximal values as


For the MSSM, 



of complete data, 



with 

normal with mean 


Equation (10) is a degenerate density since conditioning on 



3.2. PX-MCEM Algorithm
The standard EM algorithm using (7) and (8) is difficult to implement for the MSSM as the E-step and M-step are intractable. The PX-MCEM algorithm addresses this issue by modifying the E-step in two ways and leads to an M-step that can be evaluated with closed form quantities. Stated succinctly, the PX-MCEM algorithm is as follows.
1. Initialize



At iteration
2. Draw 


3. PX-MC E-step: Estimate 

4. PX-MC M-step: Maximize 
obtain the maximizing parameters


5. Reduction step: Apply reduction functions to





6. Repeat Steps 2 through 5 until convergence. The converged values are the ML estimates


Each step is described in more detail in the subsequent sections.
3.2.1. PX-MC E-Step
Following [13] , the first modification is to expand the parameter space of the complete-data likelihood function from 



with


are defined analogously to 


Second, instead of computing 






where

for








Similarly, for the latent variables,

for
















The Gibbs sampler recursively samples from the full conditional distributions in (14) and (15) in the usual way. After a sufficient burn-in period, the last 
3.2.2. PX-MC M-Step and Reduction Step
By recognizing that (11) is proportional to the log-likelihood function of a seemingly unrelated regression model with 


and

where 






and (17) recursively until convergence. Denote the converged values as


In the reduction step, set





maining 


3.3. Standard Errors
The observed information matrix is

where



estimated by taking additional draws from the Gibbs sampler and constructing their Monte Carlo analogs. The standard errors are the square roots of the diagonals of the inverse estimated quantity in (18).
4. Concluding Remarks
A new and simple ML estimation algorithm is developed for multivariate sample selection models. Roughly speaking, the implementation of this algorithm only involves iteratively drawing sets of missing data from well- known distributions and using IGLS on the complete data, both of which are inexpensive to perform. By using parameter expansion and Monte Carlo methods, the algorithm only depends on quantities with closed form expressions, even when estimating the covariance matrix parameter with correlation restrictions. This algorithm is readily extendable to other types of selection models, including extensions to various types of outcome and selection variables with an underlying normal structure, and modifications to time-series or panel data.
Acknowledgements
I would like to thank the referee, Alicia Lloro, Andrew Chang, Jonathan Cook, and Sibel Sirakaya for their helpful comments.
References
- Heckman, J. (1974) Shadow Prices, Market Wages, and Labor Supply. Econometrica, 42, 679-694. http://dx.doi.org/10.2307/1913937
- Heckman, J. (1976) The Common Structure of Statistical Models of Truncation, Sample Selection and Limited Dependent Variables and a Simple Estimator for Such Models. Annals of Economic and Social Measurement, 5, 475-492.
- Heckman, J. (1979) Sample Selection Bias as a Specification Error. Econometrica, 47, 153-161. http://dx.doi.org/10.2307/1912352
- Su, S.J. and Yen, S.T. (2000) A Censored System of Cigarette and Alcohol Consumption. Applied Economics, 32, 729-737. http://dx.doi.org/10.1080/000368400322354
- Yen, S.T., Kan, K. and Su, S.J. (2002) Household Demand for Fats and Oils: Two-Step Estimation of a Censored Demand System. Applied Economics, 34, 1799-1806. http://dx.doi.org/10.1080/00036840210125008
- Hao, A.F. (2008) A Discrete-Continuous Model of Households’ Vehicle Choice and Usage, with an Application to the Effects of Residential Density. Transportation Research Part B: Methodological, 42, 736-758. http://dx.doi.org/10.1016/j.trb.2008.01.004
- Li, P. (2011) Estimation of Sample Selection Models with Two Selection Mechanisms. Computational Statistics & Data Analysis, 55, 1099-1108. http://dx.doi.org/10.1016/j.csda.2010.09.006
- Li, P. and Rahman, M.A. (2011) Bayesian Analysis of Multivariate Sample Selection Models Using Gaussian Copulas. Advances in Econometrics, 27, 269-288. http://dx.doi.org/10.1108/S0731-9053(2011)000027A013
- Yen, S.T. (2005) A Multivariate Sample-Selection Model: Estimating Cigarette and Alcohol Demands with Zero Observations. American Journal of Agricultural Economics, 87, 453-466. http://dx.doi.org/10.1111/j.1467-8276.2005.00734.x
- Tauchmann, H. (2010) Consistency of Heckman-Type Two-Step Estimators for the Multivariate Sample-Selection Model. Applied Economics, 42, 3895-3902. http://dx.doi.org/10.1080/00036840802360179
- Puhani, P.A. (2000) The Heckman Correction for Sample Selection and Its Critique. Journal of Economic Surveys, 14, 53-68. http://dx.doi.org/10.1111/1467-6419.00104
- Dempster, A.P., Laird, N.M. and Rubin, D.B. (1977) Maximum Likelihood from Incomplete Data via the EM Algorithm. Journal of the Royal Statistical Society, 39, 1-38. http://dx.doi.org/10.2307/2984875
- Liu, C., Rubin, D.B. and Wu, Y.N. (1998) Parameter Expansion to Accelerate EM: The PX-EM Algorithm. Biometrika, 85, 755-770. http://dx.doi.org/10.1093/biomet/85.4.755



