American Journal of Molecular Biology
Vol.2 No.2(2012), Article ID:18947,8 pages DOI:10.4236/ajmb.2012.22014
Expression of the transcription factor Xvent-2 in Xenopus laevis embryogenesis*
![]()
A. N. Bach Institute of Biochemistry, Russian Academy of Sciences, Moscow, Russia
Email: #voronina_a@mail.ru, pshennikova57@mail.ru
Received 12 March 2012; revised 26 March 2012; accepted 2 April 2012
Keywords: Whole-Mount Immunostaining; Posttranscriptional Regulation; Informosomes; Blood Differentiation
ABSTRACT
Till now the transcription factor Xvent-2 has been studied in Xenopus embryos only by the mRNA testing. We use immunochemical methods for testing of the Xvent-2 protein and gradient-centrifugation methods for estimation of activity of its mRNA. Our results show that the Xvent-2 protein is present in eggs and early embryos. The Xvent-2 mRNA is absent at any of these developmental stages. The majority of mRNA synthesized on the zygotic genome was stored in informosomes, while only its small part could be revealed in polysomes. The spatial patterning of the Xvent-2 protein at different developmental stages did not entirely agree with that of its mRNA. These data indicate that the Xvent-2 protein functioning in Xenopus embryos is regulated not only at the transcription, but at translation and posttranslation as well. We propose that the activation of translation on the masked Xvent-2 mRNA may lead to blood differentiation and cell migration.
1. INTRODUCTION
Usually an expression of a gene implies the synthesis of corresponding mRNA. There is a tacit consent that an absence or a presence of a certain mRNA suggests an absence or a presence of the protein. However there are lots of evidences that mRNA may present in a cell in inactive ribonucleoprotein complexes, called by different names, from informosomes to p-bodies [1]. As a rule the start up of protein synthesis on such messengers is regulated by the modifications of the mRNA-binding proteins and the translation factors [2]. The new-synthesized proteins also undergo some modifications, and that causes their activities. Thus the gene expression can be regulated on different levels: transcription, processing, translation and final activation of the protein molecule. The rule in molecular embryology is to see the expression of the gene by a method of in situ hybridization. It indicates the gene activity (regulation on the level of transcription) but none on availability of the protein. We compared patterning of the individual protein, the transcription factor Xvent-2, with that of its mRNA at different stages of Xenopus laevis development.
Four closely identical mRNAs (Xvent-2, Xom, Vox and Xbr) have been found in X. laevis embryos at gastrula and neurula stages [3-6]. At later stages, these mRNAs are detectable in the developing eye, tail bud, somites, bronchial arch, and about proctodeum. The mRNAs are encoded by the related genes: Xvent-2B, Xbr-1b/ Vox1, Xvent-2/Xbr-1a, Vox15, and Xom [7]. The presence of the homeodomain and its specific sequence certificate this protein to be a transcription factor of a BarH subfamily. This gene is a target of the BMP signaling pathway in Xenopus and plays an important role in the dorsoventral patterning in amphibia and fish [3-13].
The spatial patterning of the Xvent-2 mRNA in Xenopus embryos is closely studied [3-6]. The specific Ab to the Xvent-2 protein enabled us to determine spatial and temporal patterning of this protein in early embryos. We show that the Xvent-2 protein is stored in eggs when its mRNA is absent. This protein is revealed at all studied developmental stages. The Xvent-2 mRNA synthesis started and grew from midblastula on, reached maximum at neurula and then it was slowly vanishing by stages 39 - 40. The major part of the Xvent-2 mRNA is masked in informosomes and is not translated in the embryos. Spatial patterning of the Xvent-2 protein and its mRNA in Xenopus laevis embryos coincide only partially. The most coincidence is revealed at neurula. The spatial patterning permits us to propose that the activation of translation on the masked Xvent-2 mRNA may lead to blood differentiation and cell migration.
2. MATERIALS AND METHODS
2.1. Analysis of RNA
X. laevis embryos were obtained, cytoplasmic extracts prepared, and centrifugation in CsCl or sucrose density gradients were performed as described previously [14]. RNA isolation, electrophoresis, and Northern blotting followed the standard protocols [15]. Radioactive labeled probes were prepared and dot hybridization was performed as described previously [14]. For RT-PCR we used forward primer: 5’atgactaaagctttctcctctgtt 3’ and reverse primer: 5’gccttggatcctaataggccagag 3’. Amplification was performed: 10 min at 94˚C; 30 cycles of 30 s at 96˚C, 30 s at 54˚C, and 4 min at 65˚C; and 7 min at 65˚C. PCR products were analyzed by 6% PAGE.
2.2. Isolation of Protein Fraction of Embryos
The stages of the Xenopus development were determined according to the tables by [16]. The embryos were homogenized in 5 volumes of a following solution: 0.1 M Tris-HCl, pH 8.0; 0.1 M KCl; 10% glycerol; 2% b-mercaptoethanol and 1 mM PMSF. After 10 min centrifugation at 10000 g a supernatant was mixed with 1.5 volumes of 4 M ammonium sulfate and left for 2 days at 4˚C. The precipitate was collected by centrifugation at 10000 g and dissolved in Laemmli sample buffer.
2.3. Production of the Recombinant reXvent-2 Protein
Expression and isolation of reXvent-2 protein were described previously [17]. We got a construction encoding the Xvent-2 with 22 amino acids (MGHHHHHHHHHSSGHIDDDD) at the N-end that was controlled by a commercial sequence determination procedure (VGNKI, Moscow). The isolated product was subjected to a commercial MALDI (IBC RAS). The results agreed with the predicted amino acid sequence. The program MS FIT was used (http://prospector.ucsf.edu).
2.4. Production of Antiserum and Purified Antibodies (Ab)
The reXvent-2 protein was concentrated by methanol precipitation, dissolved in phosphate buffer and used to raise polyclonal antiserum in rabbits [17]. This serum was used in immunobloting. For whole-mount immunostaining anti-reXvent-2 rabbit Ab were purified and conjugated to horseradish peroxidase (IMTEK, Moscow).
2.5. Immunoblotting
Proteins were electrophoresed in SDS-PAAG, transferred onto nitrocellulose membrane and stained with 0.1% Ponceaus S to control the transfer. Then the membrane was washed in water and treated consequently with blocking solution (5% milk), rabbit anti-reXvent-2 serum (1:1000) and goat anti-rabbit Ab conjugated to horseradish peroxidase p-SAR Iss (IMTEK, Moscow) (1:20000) as it was proposed by Promega. The peroxidase substrate TMB (tetramethilbenzydine) (“BioTestSystems” Moscow) served for staining.
2.6. Whole-Mount Immunostaining of Embryos
Embryos were fixed according to Klymkowsky lab manual [18] in 20% DMSO: 80% methanol (Dent’s fixative). After 2 h gentle shaking at room temperature the embryos were transferred to ice cold acetone for10 min and to 100% methanol where they could be stored at –20˚C indefinitely. For staining, embryos were washed twice with Tris-buffered saline (TBS) for five minutes each time and if necessary bleached in 5% formamide, 0.5X SSC, 1% H2O2 for a night. They were then incubated overnight at 4˚C in blocking solution: 20% normal rabbit serum, 2% DMSO diluted into TBS and after that for a day in antiXvent-2 rabbit Ab conjugated to horseradish peroxidase diluted 1:500 into the blocking solution. The embryos were then washed in TBS once for 15 min, once for 30 min, once for 1 h, once for 2 h. Bound peroxidase-conjugated Ab were visualized using the peroxidase substrate TMB for 1 to 2 h (dark blue staining). The reaction was stopped by changing the substrate for TBS.
2.7. Control Whole-Mount Immunostainings
In the absence of Ab we found no staining of embryos with TMB. We used some controls to avoid any nonspecific Ab staining. Control 1: immunostaining with conjugate of anti-human-immunoglobulins rabbit Ab with peroxidase. Control 2: immunostaining with the conjugate of rabbit antiXvent-2 Ab with peroxidase depleted with the reXvent-2 protein. 2ml of the working solution of the conjugated anti-Xvent-2 Ab were sequentially incubated with three pieces of nitrocellulose membrane 3 × 3 cm carrying 30 - 60 mcg of the reXvent-2 protein each. The third membrane after appropriate washing could hardly be stained with peroxidase substrate and thus we considered these anti-reXvent-2 Ab to be depleted with the reXvent-2 protein.
3. RESULTS AND DISCUSSION
3.1. Detection of the Xvent-2 Protein and mRNA in the Embryos at Different Developmental Stages
The anti-Xvent-2 immune rabbit serum was characterized in our earlier paper [17]. Control immunoblotting experiments showed that the serum reacted with a major band in the extract of E coli cells transformed with the plasmid containing the Xvent-2 cDNA insert and did not stain any protein in the extract of E. coli cells transformed with the empty vector.
The PAGE and immunoblotting of protein fractions obtained from X. laevis embryos at different developmental stages from eggs to tadpoles are shown in Figure 1. The immune serum revealed a single protein band in eggs and embryos of all developmental stages examined (Figure 1(a)). The same staining of the membrane treated with nonimmune rabbit serum with secondary Ab did not reveal any bands (not shown). The electrophoretic mo-
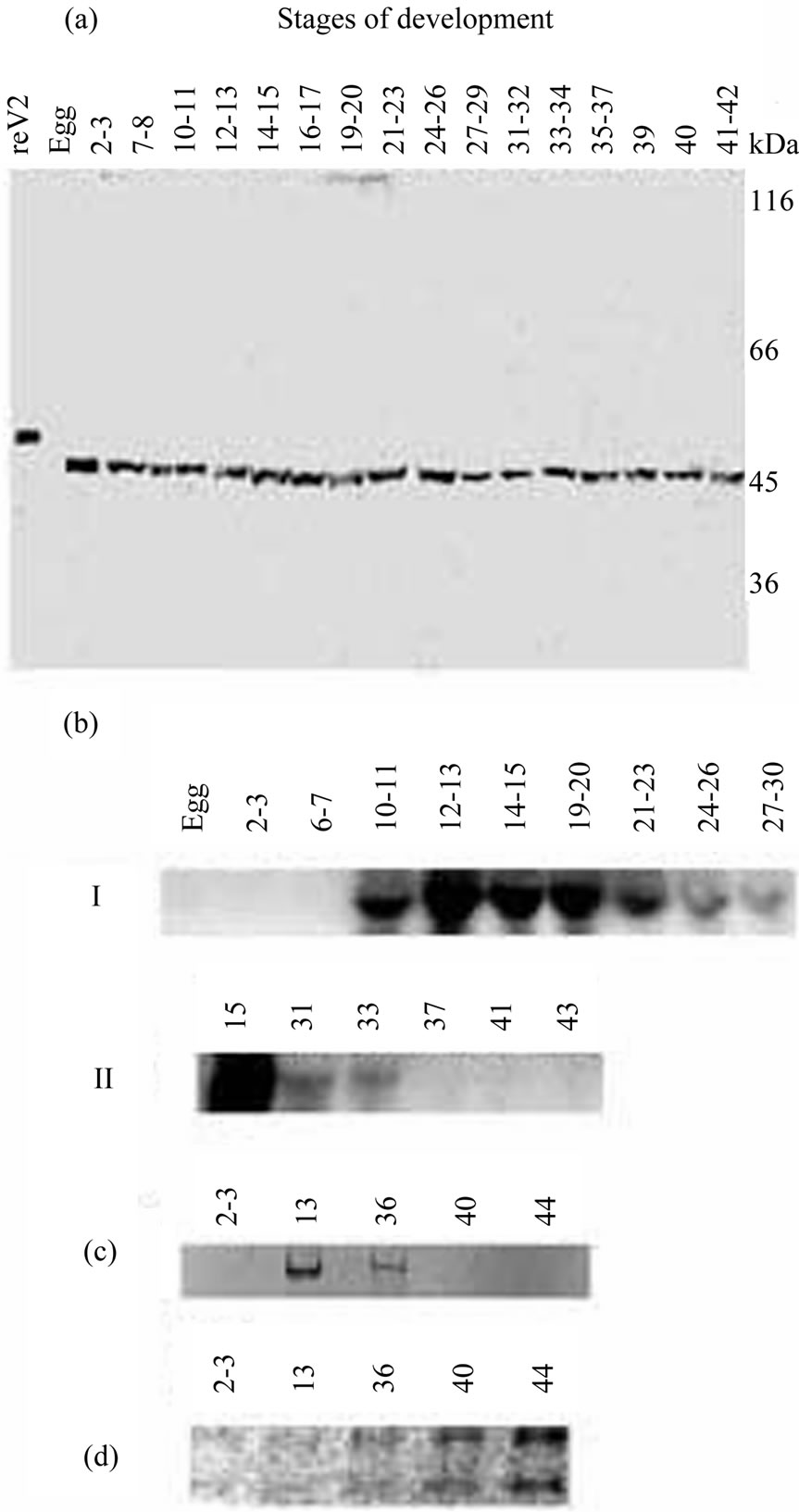
Figure 1. Immunoblotting of the Xvent-2 protein (a) and Northern blot analysis of total RNA with Xvent-2 probe (b) from Xenopus laevis embryos at different developmental stages. The exposure to X-ray film 20 h (I), and 2 weeks (II). (c) 6% PAGE of the RT-PCR products, EtdBr staining; (d) 1% agarose electrophoresis of the same samples of total RNA, EtdBr staining.
bility of X. laevis Xvent-2 was slightly higher than that of the recombinant protein reXvent-2 owing to the lack of a polyhistidine tag and corresponded to a protein of about 45 kDa. The calculated molecular mass for the Xvent-2 is 36.5 kDa, but some proteins, for example transcription factor Ybox1 [19], demonstrate the less mobility than theoretically counted one. The amount of the Xvent-2 was approximately the same at all the developmental stages and slightly higher in the eggs. This was quite unexpected, because the Xvent-2 mRNA synthesis did not start until the middle blastula [3-6]. To verify this, the RNA was examined by Northern blotting and RTPCR (Figures 1(b), (c)). The results confirmed that the Xvent-2 mRNA was undetectable at cleavage and early blastula stages. Then, its amount increases, reaches maximum at the stages 12 - 15, and decreases in further development. At the stages 39 - 40 neither Northern blotting nor RT-PCR could detect Xvent-2 mRNA. It is not connected with worse RNA extraction for the amount of rRNA is increasing in these samples (Figure 1(d)). The Xvent-2 protein content in embryos does not correlate with the accumulation and degradation of the Xvent-2 mRNA. The discrepancy in the detection of the mRNA and protein suggests a strict regulation of the amount of Xvent-2 protein at transcriptional, translational and protein-stability levels.
3.2. Spatial Patterning of the Xvent-2 Protein in Embryos at Cleavage and Blastula Stages
Intensive nonspecific staining of embryos was revealed in our preliminary experiments where the standard procedure with primary and secondary Ab was performed. That is why in the sequel we used the affine-isolated rabbit anti-Xvent2 Ab conjugated to the horseradish peroxidase. A normal rabbit serum diluted 1:5 in TBS served as a blocking solution and as a solution for anti-Xvent-2 Ab to exclude any nonspecificity.
The results of the whole-mount immunostaining of embryos at the stages of 16 - 32 blastomers and blastula are presented in Figure 2. The Xvent-2 is revealed in the cytoplasm apical area of animal blastomers at cleavage stages (Figures 2(a)-(c)). At midblastula the Xvent-2 is seen in the animal hemisphere (Figures 2(d), (e)). The higher magnification shows the possible nuclear localization of the Xvent-2 protein at stage 8 (Figure 2(e*)). It seems that the transcription factor Xvent-2 migrates from cytoplasm to nucleus just before the transcription start at zygote genome. As it was shown earlier [20] there was revealed specific proteinase activity against the Xom/Xvent2 protein in embryos at stage 7. At the stages 9 - 10 this activity decreased dramatically and then it grew at gastrulation. One can suppose this proteinase destroyed the cytoplasmic Xvent-2 which had not entered the nucleus.
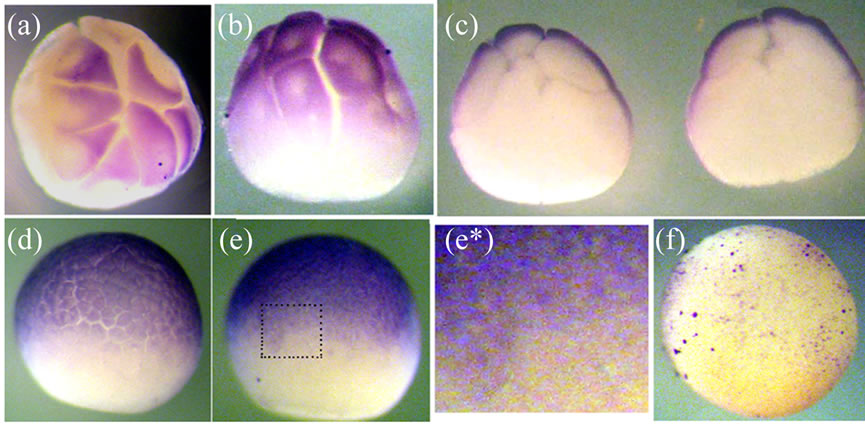
Figure 2. The whole-mount immunostaining of the Xvent-2 protein in the embryos at cleavage and blastula stages. (a) Stage 5, animal view; (b) Stage 6, lateral view. Note that the nuclear areas are excluded from the staining; (c) Stage 6, internal view of two halves; (d) Stage 7; (e) Stage 8, lateral views, up—animal; (e*) The close-up view of the framed area in (e); The nuclear localization of the Xvent-2 is seen; (f) Control immunostaining by conjugate of rabbit anti-human-γ-globulin Ab with peroxidase; stage 8, lateral view.
3.3. Most of the Xvent-2 mRNA Is Not Translated
The expression of many genes in embryo development of eukaryotes is regulated at the translational level by variety of mechanisms [2]. The individual templates differ in specific localization within the embryo, activity time, and mechanisms of masking and translational activation. In our previous works we showed that some zygotic mRNAs starting to be expressed immediately at midblastula transition came to polysomes only at middle gastrula [14]. A translational regulation has been demonstrated for several Xenopus genes involved in the BMP signaling pathway [21]. The stored mRNAs arrived to polysomes at different periods: translation on the Smad1 and ALK2 mRNAs started in oocyte maturation, on the BMP-4 and XSTK9 mRNAs—soon after fertilization and only in early gastrula—on the ALK3 mRNA. Specific mRNAs encoding for the proteins participating in dorsoventral differentiation (Xwnt-11, Xnot-2, Xbra, and Xgsc) demonstrated individual dynamics of activation and inactivation during early embryogenesis [22]. Here we show the distribution of Xvent-2 mRNAs between polysomes and informosomes in X. laevis embryos at the gastrula (Figure 3). It is seen that most of the Xvent-2 mRNA sedimented in sucrose gradient at post ribosome zone (Figure 3(a), II), only few—at polysome zone (Figure 3(a), I). It was confirmed with Northern blotting (Figure 3(b)). The buoyant density in CsCl of Xvent-2 mRNP from zone I is 1.54 g/cm3 as that of polysomes (Figure 3(c)) and the buoyant density in CsCl of Xvent-2 mRNP from zone II (free mRNP) is 1.4 g/cm3 like that of informosomes. The Northern blotting of the RNA from the CsCl gradient confirmed the existence of the most of the Xvent-2 mRNA in inactive form (Figure 3(d)). The most part of Xvent-2 mRNA was detected in informo-
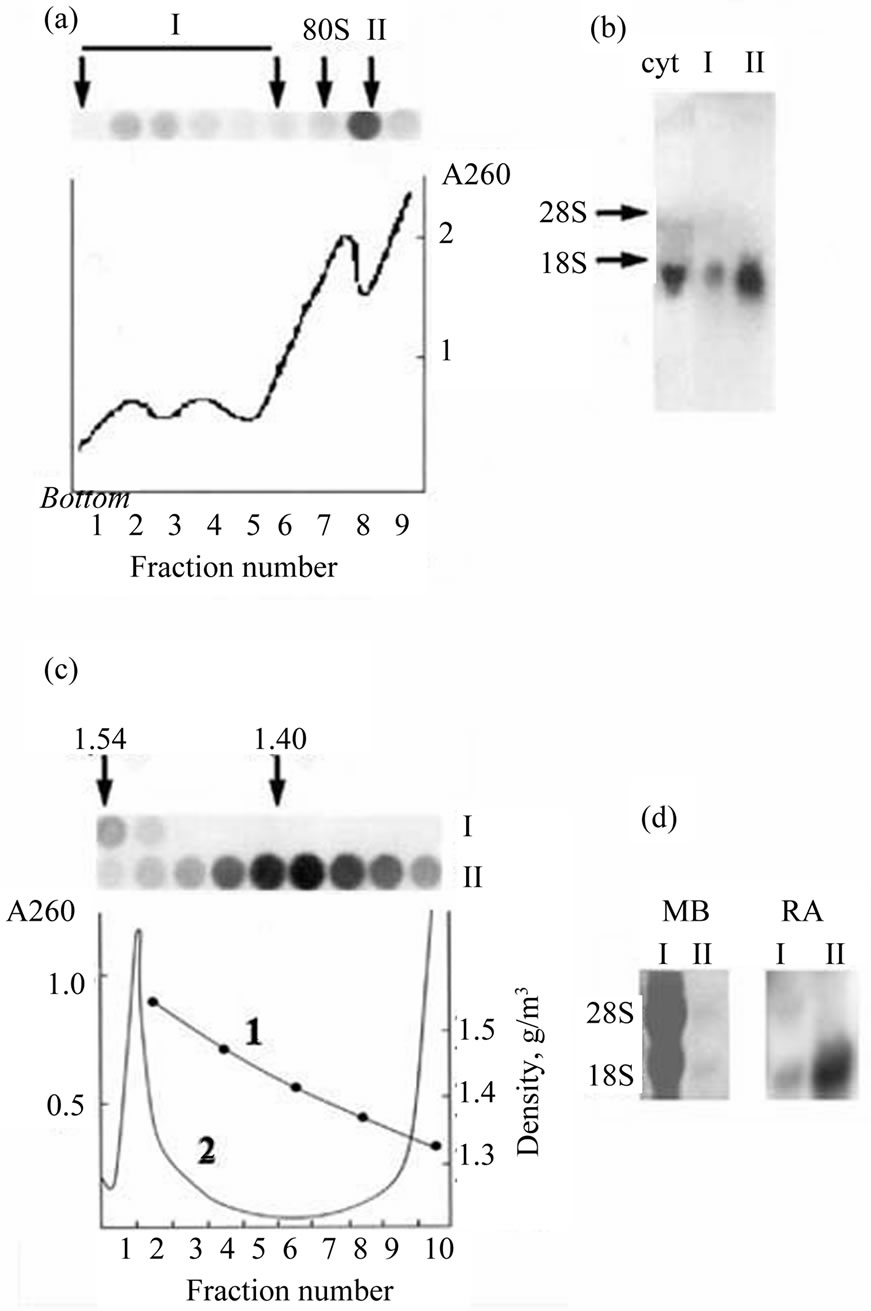
Figure 3. Sedimentation in the sucrose gradient (a) and banding in the CsCl density gradient (c) of Xvent-2 mRNPs from the embryos at gastrula. (a) The line A260 demonstrates distribution of ribosomes and polysomes. Dot-hybridizations of the fractions from the sucrose gradient with P32-Xvent-2 probe are seen on the top. Polysomes (I) and free mRNPs (II) were collected; (b) Northern-blot of RNA from I and II fractions of the sucrose gradient or RNA from the cytoplasm extract of embryos (cyt). Hybridizations with P32-Xvent-2 probe; (c) Polysomes (I), free mRNPs (II) or the cytoplasm extract were centrifuged in CsCl density gradients. The line 1—density. The line 2 (A260) demonstrates buoyant density of ribosomes and polysomes (1.54 g/cm3) in the cytoplasm. Dot-hybridizations with P32-Xvent-2 probe of the fractions from I and II CsCl gradients are seen on the top; (d) RNA was isolated from the CsCl gradient, I—from polysomes (fractions 1 - 2) and II—from informosomes (fractions 4 - 7) and analyzed with Northern-blot. On the left—the staining of the membrane with methylene blue (MB), on the right—hybridizations with P32-Xvent-2 probe. RA-radioactivity.
somes at all the developmental stages examined (from 9 to 17.5), and only its minor fraction was associated with polysomes [23]. From our data it follows that the most of the Xvent-2 mRNA detected by in situ hybridization does not participate in the protein synthesis and thus, the in situ hybridization cannot reproduce the protein pattern.
3.4. Spatial Patterning of the Xvent-2 Protein in Embryos at Gastrula, Neurula and Tadpoles
Figure 4 shows the Xvent-2 protein immunostaining of the embryos at the gastrula stages. It is seen that at early gastrula the Xvent-2 protein is located in the animal hemisphere with a shift towards dorsal side of blastopore. At the late gastrula (st. 11.5 - 12) the Xvent-2 protein spreads all over the embryo excepting blastopore (Figures 4(e), (f)). From the literature [3-6] it is clear that the Xvent-2 mRNA is detected at the gastrula in the same aria except for a region above the dorsal blastopore lip (prospective the neural plate and the notochord) (Figure 4(i)). To the termination of gastrulation Xvent-2 mRNA disappear on each side (Figure 4(j)). According to the data shown (Figure 3) Xvent-2 mRNA is practically not translated at gastrula. Thus with immunostaining we probably see to a greater extent the Xvent-2 protein which had been stored in eggs.
An animal pole-derived ectoderm plays a critical role in blood cells formation [24]. Expression of BMP-4 not only in ventral mesoderm but in the overlying ectoderm is important for primitive red blood cells induction. As it was shown earlier, the Xvent-2 have activated the BMP-4 and Xvent-1 genes involved in ventral mesoderm formation in embryo development of fish and amphibians [7-12]. We propose that just the Xvent-2 protein (stored in the animal hemisphere of an egg and being found in the ectoderm due to cell movements at gastrulation) activates the synthesis of the BMP-4 and as a result the red blood cells formation in mesoderm.
At the neurula and early tailbud stages (Figure 5)
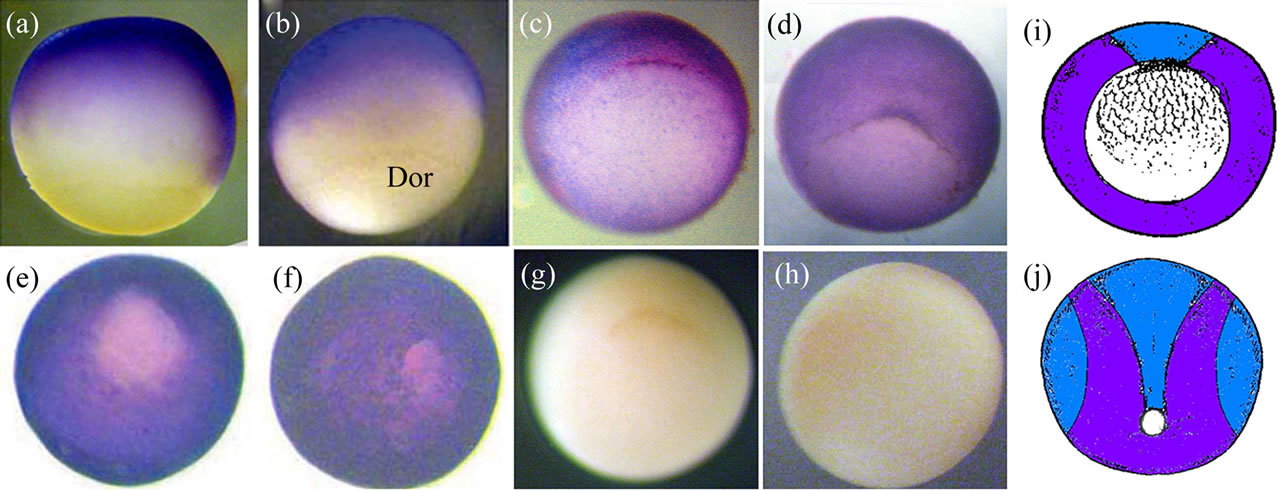
Figure 4. The whole-mount immunostaining of the Xvent-2 protein in the embryos at gastrula. Stage10.25: lateral view, dorsal on the right (a), dorsal view (b). Vegetal views of stages 10.25 (c), 10.5 (d), 11.5 (e), 12 (f). Dor—dorsal. (g), (h) Control immunostaining by the conjugate depleted of the reXvent-2 protein; stage 10.25; (g)—vegetal view, (h)—animal view. (i), (j) Diagrams of areas occupied by the expression domains of the protein (blue) and coincidence of mRNA and protein (violet) within (i)—the early gastrula (st. 10.25), vegetal view and (j)—the late gastrula/early neurula (st. 12 - 13), posterior-dorsal view.
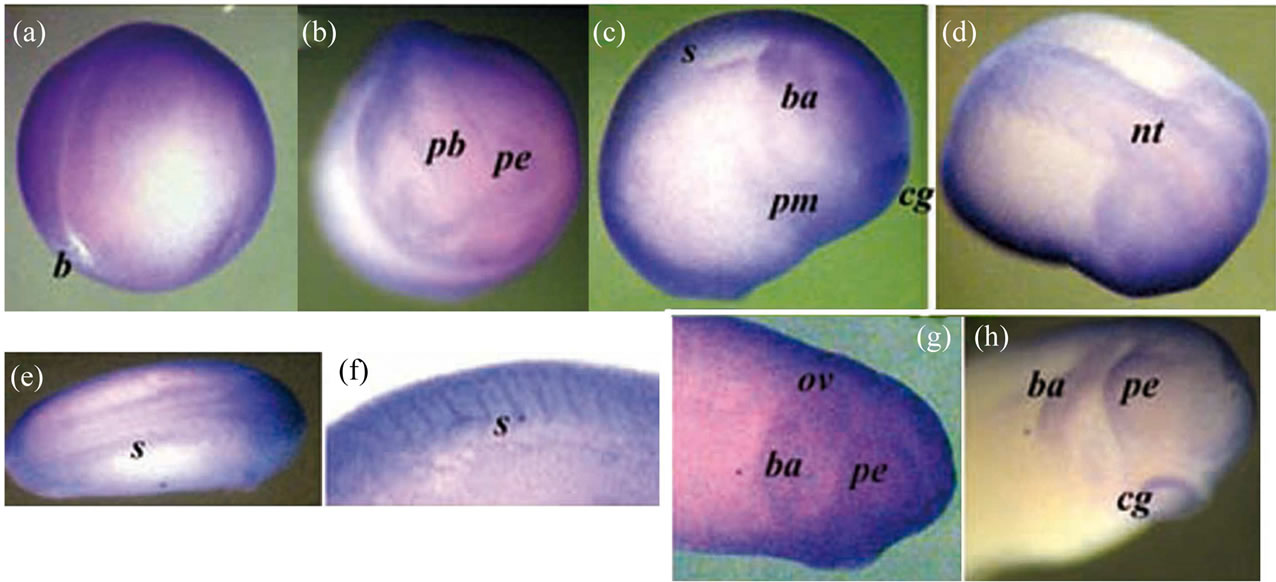
Figure 5. The whole-mount immunostaining of the Xvent-2 protein in the embryos at neurula and tailbud stages. The late neurula, st. 18 - 19: posterior-lateral view (a) and anterior view (b). The early tailbud, st. 20 - 22: lateral view (c) and dorsal view (d). (e) Tailbud st. 23 - 25, dorsal view, the head at right. (f) Somites at stage 27, lateral view. (g) The head, stage 26, lateral view. (h) The head, stage 27, lateral view. b—blastopore, pb—presumptive brain, nt—nerve tube, pe —presumptive eye, cg—cement gland, s—somites, ba—bronchial arch, ov—otic vesicle, pm— presumptive myeloid blood cells.
immunostaining reveals Xvent-2 along the edge of the neural tube, the presumptive brain and eyes, in the bronchial arc, in the otic vesicle, in the somites and the cement gland. The staining of the cement gland may be an artifact, as an occasional nonspecific staining of it was found earlier [18]. Presumptive myeloid blood cells were concentrated at these stages in the middle of ventral area of embryo [25]. We saw the Xvent-2 protein staining in this area too (Figure 5(c)).
Results of other authors on the in situ hybridization at the neurula and early tailbud stages revealed that the Xvent-2 mRNA patterns were generally overlapped with those of the protein. The mRNA was found along the edge of the neural tube [3,5,6,9] along the edge of the presumptive forebrain [13], in the presumptive eyes, bronchial arch, tail bud [3-6] somites [3] and in the areas of the mieloid ventral blood island [4,5,25]. The Xvent-2 mRNA was absent in the cement gland [3-6].
At the stages 30-35 the Xvent-2 mRNA was seen in three areas: in the tail tip, the dorsal parts of eyecups and in the bronchial arch [3,5,6,9].
Our study revealed the Xvent-2 protein at stage 35 in both anterior and posterior red blood island [24,26], in the lymphatic caudal dorsal vessel, in the tail fin and around the eyes, the bronchial arches and the proctodeum (Figure 6). Thus coincidence of mRNA and protein patterning at these stages is only partial (Figure 6(c)). The immunostaining of albino tadpoles at the stage 45 revealed stained granules chains along the lymphatic caudal dorsal vessel, near the proctodeum and in the head (Figure 7). In addition there were stained numerous filaments winding the eyes and ramifying throughout the head. These filaments might be blood or lymphatic vessels. The chains observed are rather like lymphatic vessels.
It was shown earlier [23] that the Xvent-2 mRNA from cut tail at tailbud stages was present in informosomes and so it was not translated. The presence of the Xvent-2
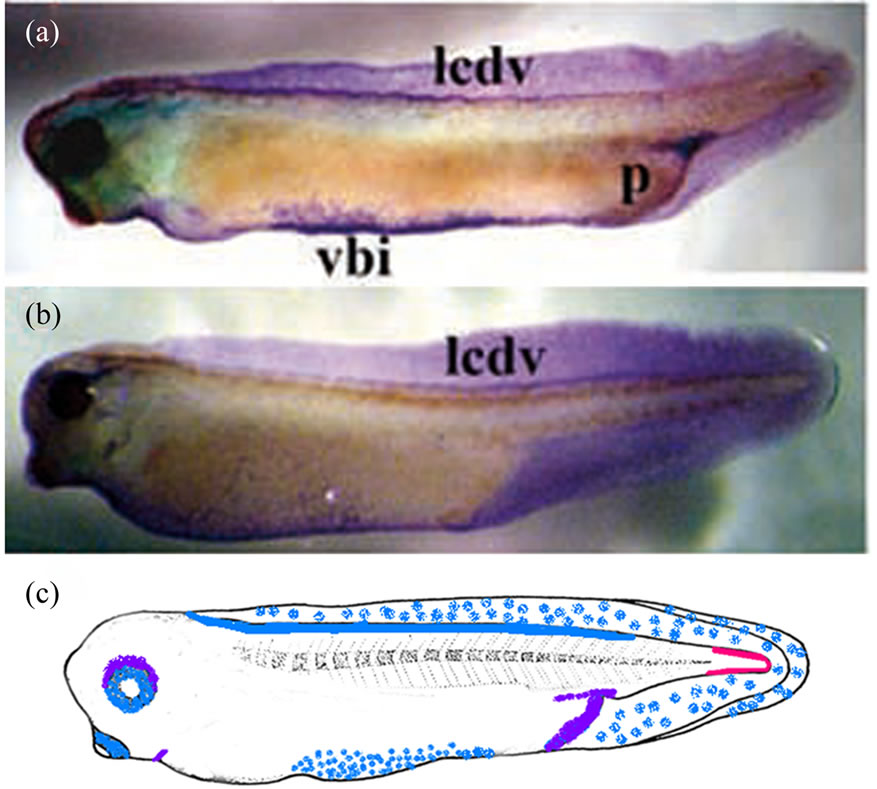
Figure 6. The whole-mount immunostaining of the Xvent-2 protein in the embryos at stages 35 (a) and 37 (b). (c) Diagrams of areas occupied by the expression domains of Xvent2 mRNA (red), the protein (blue) and coincidence of mRNA and protein (violet) within a tadpole at stage 35. vbi—ventral blood island, lcdv—lymphatic caudal dorsal vessel, p—proctodeum.
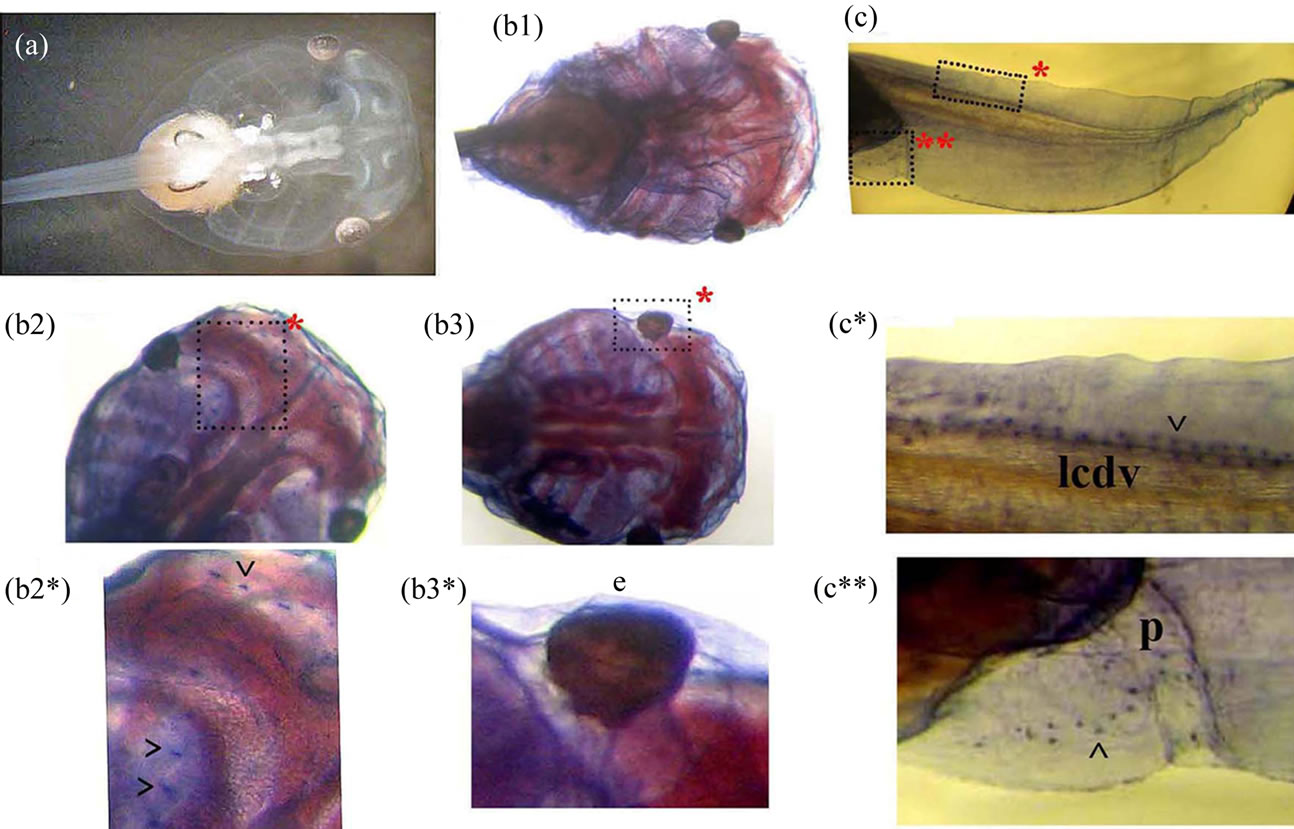
Figure 7. The whole-mount immunostaining of the Xvent-2 protein in the tadpole at stage 45. (a) The unstained head of albino tadpole. (b1, b2, b3) Stained heads at different definitions in depth. (c) Stained posterior part of tadpole. (b2*, b3*, c*, c**) The framed areas close-up. ^—granules, e—eye, lcdv—lymphatic caudal dorsal vessel, p—proctodeum.
mRNA and lack of the protein in a tail tip confirm it. We propose, that as soon as a cell (under any signal) starts to synthesize the Xvent-2 protein, it moves towards a developing lymphatic or blood vessel. That is why Xvent-2 mRNA had not been detected either in globins-containing ventral blood island or in lymphatic or blood vessels. We propose that a possible function of the Xvent-2 protein may be not only the dorsoventral axis formation in the early embryo, but also participating in the blood and lymph development. That is why the cells which synthesize the Xvent-2 protein can be revealed along the edge of some presumptive organs at neurula. They may participate in creation of the blood system of these organs.
Our results corroborate our hypothesis on the role of activation of some mRNAs in cell transition from a competence to the determination at an embryogenesis [2].
4. CONCLUSION
A pattern of mRNA cannot sometimes give a perfect reflection of a corresponding protein pattern because of the existence of inactive mRNAs or long-living proteins. It also may be a case when after activation of some protein synthesis, cells migrate and the protein appears at another place. Thus one must investigate the pattern of gene expression not only with in situ hybridization but with immunostaining of the protein too. For transcription factor Xvent-2 we showed, that it was stored in eggs and in animal blastomers when its mRNA was absent. The major of the Xvent-2 mRNA synthesized after midblastula transition was not translated in the embryos. The spatial patterning permits us to propose that the activation of translation on the masked Xvent-2 mRNA may lead to the blood differentiation and cell migration.
REFERENCES
- Voronina, A.S. and Pshennikova, E.S. (2010) mRNPs: From informosomes to stress granules. Molecular Biology, 44, 520-528. doi:10.1134/S0026893310040035
- Voronina, A.S. (2002) Translational regulation in early development of eukaryotes. Molecular Biology, 36, 956- 969. doi:10.1023/A:1021669506664
- Onichtchouk, D., Gawantka, V., Dosch, R., Delius, H., Hirschfeld, K., Blumenstock, C. and Niehrs, C. (1996) The Xvent-2 homeobox gene is part of the BMP-4 signalling parthway controlling dorsoventral patterning of Xenopus mesoderm. Development, 122, 3045-3053.
- Schmidt, J.E., von Dassow, G. and Kimelman, D. (1996) Regulation of dorsal-ventral patterning: The ventralizing effects of the novel Xenopus homeobox gene Vox. Development, 122, 1711-1721.
- Ladher, R., Mohun, N.J., Smith, J.C. and Snape, A.M. (1996) Xom: A Xenopus homeobox gene that mediates the early effects of BMP-4. Development, 122, 2385- 2394.
- Papalopulu, N. and Kintner, C. (1996) A Xenopus gene, Xbr-1, defines a novel class of homeobox genes and is expressed in the dorsal ciliary margin of the eye. Developmental Biology, 174, 104-114. doi:10.1006/dbio.1996.0055
- Rastegar, S., Friedle, H., Frommer, G. and Knochel, W. (1999) Transcriptional regulation of Xvent homeobox genes. Mechanisms of Development, 81, 139-149. doi:10.1016/S0925-4773(98)00239-1
- Onichtchouk, D., Glinka, A. and Niehrs, C. (1998) Requirement for Xvent-1 and Xvent-2 gene function in dorsoventral patterning of Xenopus mesoderm. Development, 125, 1447-1456.
- Melby, A.E., Clements, W.K. and Kimelman, D. (1999) Regulation of dorsal gene expression in Xenopus by the ventralizing homeodomain gene Vox. Developmental Biology, 211, 293-305. doi:10.1006/dbio.1999.9296
- Trindade, M., Tada, M. and Smith, J.C. (1999) DNAbinding specificity and embryological function of Xom (Xvent-2). Developmental Biology, 216, 442-456. doi:10.1006/dbio.1999.9507
- Melby, A.E., Beach, C., Mullins, M. and Kimelman, D. (2000) Patterning the early Zebrafish by the opposing actions of bozozok and vox/vent. Developmental Biology, 224, 275-285. doi:10.1006/dbio.2000.9780
- Schuler-Metz, A., Knochel, S., Kaufmann, E. and Knochel, W. (2000) The homeodomain transcription factor Xvent-2 mediates autocatalytic regulation of BMP-4 expression in Xenopus embryos. The Journal of Biological Chemistry, 275, 34365-34374. doi:10.1074/jbc.M003915200
- Martynova, N., Eroshkin, F., Ermakova, G., Bayramov, A., Gray, J., Grainger, R. and Zaraisky, A. (2004) Patterning the forebrain: FoxA4a/Pintallavis and Xvent-2 determine the posterior limit of Xanf1 expression in the neural plate. Development, 131, 2329-2338. doi:10.1242/dev.01133
- Voronina, A.S. and Potekhina, E.S. (1999) Translational regulation of synthesis of proteins responsible for dorsoventral differentiation of Xenopus laevis embryos. Russian Journal of Developmental Biology, 30, 83-90.
- Maniatis, T., Fritsch, E.F. and Sambrook, J. (1982) Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor.
- Nieuwkoop, P.D. and Faber, J. (1956) Normal table of Xenopus laevis (daudin): A systematical and chronologica survey of the development from the fertilized egg till the end of metamorphosis. North-Holland, Amsterdam.
- Pshennikova, E.S. and Voronina, A.S. (2008) Detection of the Xvent-2 transcription factor in early development of Xenopus laevis. Molecular Biology, 42, 901-905. doi:10.1134/S0026893308060101
- Dent, J.A., Polson, A.G. and Klymkowsky, M.W. (1989) A whole-mount immunocytochemical analysis of the expression of the intermediate filament protein vimentin in Xenopus. Development, 105, 61-74.
- Evdokimova, V.M., Wei, C.L., Sitikov, A.S., Simonenko, P.N., Lazarev, O.A., Vasilenko, K.S., Ustinov, V.A., Hershey, J.W. and Ovchinnikov, L.P. (1995) The major protein of messenger ribonucleoprotein particles in somatic cells is a member of the Y-box binding transcription factor family. The Journal of Biological Chemistry, 270, 3186-3192. doi:10.1074/jbc.270.7.3186
- Zhu, Z. and Kirschner, M. (2002) Regulated proteolysis of Xom mediates dorsoventral pattern formation during early Xenopus development. Developmental Cell, 3, 557- 568. doi:10.1016/S1534-5807(02)00270-8
- Fritz, B.R. and Sheets, M.D. (2001) Regulation of the mRNAs encoding proteins of the BMP signaling pathway during the maternal stages of Xenopus development. Developmental Biology, 236, 230-243. doi:10.1006/dbio.2001.0324
- Voronina, A.S. and Pshennikova, E.S. (2006) Activity of specific mRNAs in early development of Xenopus and Rana embryos. Journal of Biological Sciences, 6, 115- 120. doi:10.3923/jbs.2006.115.120
- Voronina, A.S., Pshennikova, E.S. and Shatilov, D.V. (2003) Distribution of the Xvent-2 mRNA between informosomes and polysomes in early frog development. Molecular Biology, 37, 429-435. doi:10.1023/A:1024295528924
- Kikkawa, M., Yamazaki, M., Izutsu, Y. and Maéno, M. (2001) Two step induction of primitive erythrocytes in Xenopus laevis embryos: Signals from the vegetal endoderm and the overlying ectoderm. The International Journal of Developmental Biology, 45, 387-396.
- Ciau-Uitz, A., Liu, F. and Patient, R. (2010) Genetic control of hematopoietic development in Xenopus and zebrafish. The International Journal of Developmental Biology, 54, 1139-1149. doi:10.1387/ijdb.093055ac
- Iraha, F., Saito, Y., Yoshida, K., Kawakami, M., Izutsu, Y., Daar, I.O. and Maéno, M. (2002) Common and distinct signals specify the distribution of blood and vascular cell lineages in Xenopus laevis embryos. Development, Growth and Differentiation, 44, 395-407. doi:10.1046/j.1440-169X.2002.00653.x
NOTES
*This study was supported by the Russian Foundation for Basic Research N 09-04-00276.
#Corresponding author.

