Paper Menu >>
Journal Menu >>
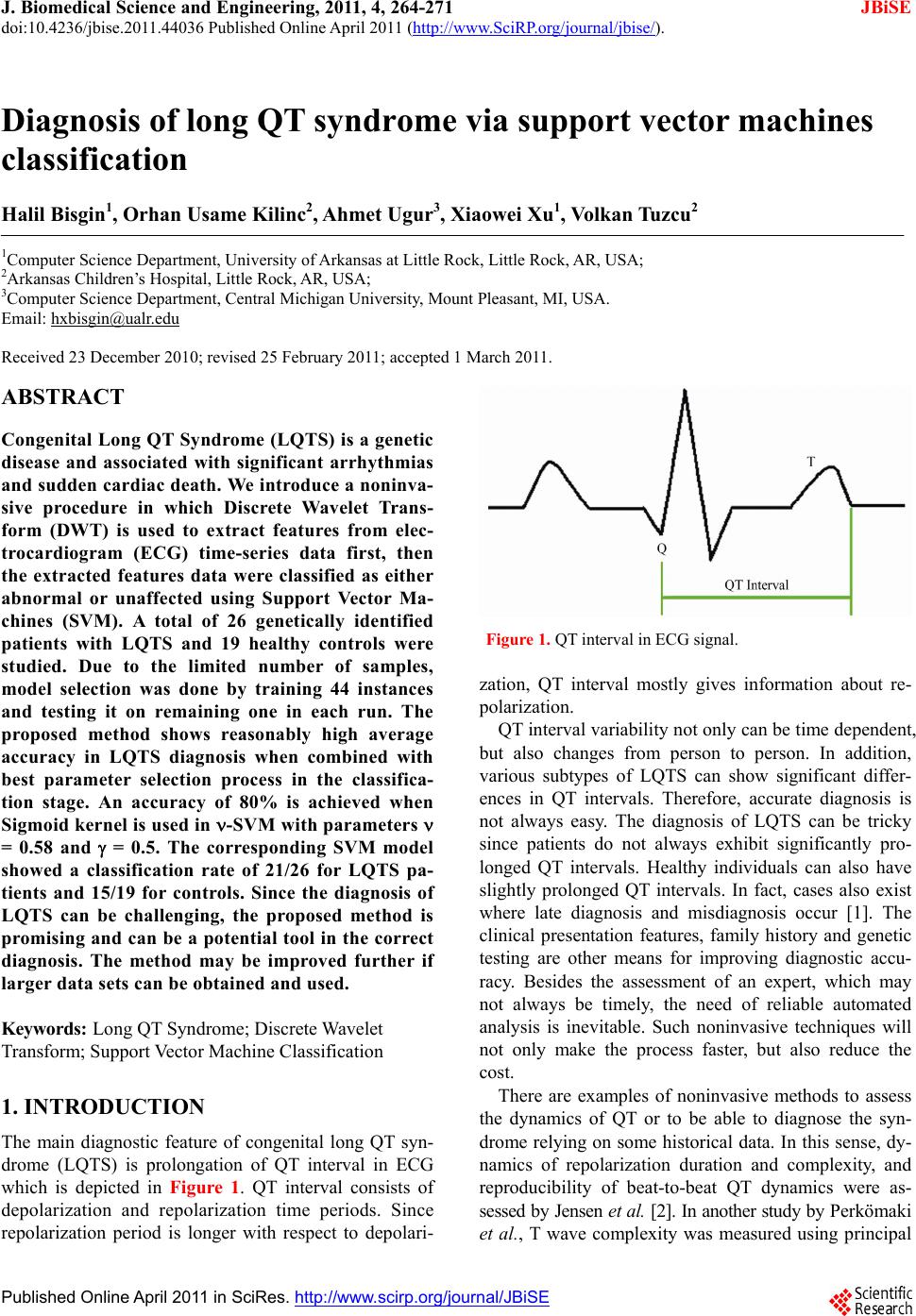 J. Biomedical Science and Engineering, 2011, 4, 264-271 JBiSE doi:10.4236/jbise.2011.44036 Published Online April 2011 (http://www.SciRP.org/journal/jbise/). Published Online April 2011 in SciRes. http://www.scirp.org/journal/JBiSE Diagnosis of long QT syndrome via support vector machines classification Halil Bisgin1, Orhan Usame Kilinc2, Ahmet Ugur3, Xiaowei Xu1, Volkan Tuzcu2 1Computer Science Department, University of Arkansas at Little Rock, Little Rock, AR, USA; 2Arkansas Children’s Hospital, Little Rock, AR, USA; 3Computer Science Department, Central Michigan University, Mount Pleasant, MI, USA. Email: hxbisgin@ualr.edu Received 23 December 2010; revised 25 February 2011; accepted 1 March 2011. ABSTRACT Congenital Long QT Syndrome (LQTS) is a genetic disease and associated with significant arrhythmias and sudden cardia c death. We introduce a noninva- sive procedure in which Discrete Wavelet Trans- form (DWT) is used to extract features from elec- trocardiogram (ECG) time-series data first, then the extracted features data were classified as either abnormal or unaffected using Support Vector Ma- chines (SVM). A total of 26 genetically identified patients with LQTS and 19 healthy controls were studied. Due to the limited number of samples, model selection was done by training 44 instances and testing it on remaining one in each run. The proposed method shows reasonably high average accuracy in LQTS diagnosis when combined with best parameter selection process in the classifica- tion stage. An accuracy of 80% is achieved when Sigmoid kernel is used in -SVM with parameters = 0.58 and = 0.5. The corresponding SVM model showed a classification rate of 21/26 for LQTS pa- tients and 15/19 for controls. Since the diagnosis of LQTS can be challenging, the proposed method is promising and can be a potential tool in the correct diagnosis. The method may be improved further if larger data sets can be obtained and used. Keywords: Long QT Syndrome; Discrete Wavelet Transform; Support Vector Machine Classification 1. INTRODUCTION The main diagnostic feature of congenital long QT syn- drome (LQTS) is prolongation of QT interval in ECG which is depicted in Figure 1. QT interval consists of depolarization and repolarization time periods. Since repolarization period is longer with respect to depolari- Figure 1. QT interval in ECG signal. zation, QT interval mostly gives information about re- polarization. QT interval variability not only can be time dependent, but also changes from person to person. In addition, various subtypes of LQTS can show significant differ- ences in QT intervals. Therefore, accurate diagnosis is not always easy. The diagnosis of LQTS can be tricky since patients do not always exhibit significantly pro- longed QT intervals. Healthy individuals can also have slightly prolonged QT intervals. In fact, cases also exist where late diagnosis and misdiagnosis occur [1]. The clinical presentation features, family history and genetic testing are other means for improving diagnostic accu- racy. Besides the assessment of an expert, which may not always be timely, the need of reliable automated analysis is inevitable. Such noninvasive techniques will not only make the process faster, but also reduce the cost. There are examples of noninvasive methods to assess the dynamics of QT or to be able to diagnose the syn- drome relying on some historical data. In this sense, dy- namics of repolarization duration and complexity, and reproducibility of beat-to-beat QT dynamics were as- sessed by Jensen et al. [2]. In another study by Perkömaki et al., T wave complexity was measured using principal  H. Bisgin et al. / J. Biomedical Science and Engineering 4 (2011) 264-271 Copyright © 2011 SciRes. JBiSE 265 component analysis [3]. They have shown that QT vari- ability, mean T wave complexity, and T wave complex- ity variability helped in distinguishing patients from healthy controls. Studies based on wavelet transform also helped to figureout some hidden patterns within the ECG signals. Couderc et al. pointed out how wavelet transform is a promising way of analyzing ECG signals [4]. From ECG pattern recognition to analysis of ventricular repolariza- tion, wavelet transform is considered as a technique that can highlight details of ECG signals in noninvasive electrocardiology. QRS detection via wavelet analysis by Li is a prominent example [5]. In order to analyze QT interval time frequency, Wong et al. employed DWT on a data set which consisted of exercise ECG signals [6]. They demonstrated that their QT interval automatic measurement methodology was very similar to clinical observations. Strachan et al. de- veloped a Hidden Markovian Model (HMM) based QT analysis method [7]. Specifically, they trained wave- forms from ECG signals via wavelet transform which were already annotated by the experts. They used a large data set in order to train for HMM for automated QT analysis. Besides signal processing tools, other statistical learning mechanisms may help to identify pathological cases ap- propriately. In this study, we utilize both signal process- ing method DWT and statistical learning procedure SVM. The outline of the paper is as follows. In Section 2, we describe the methods that we have use throughout the study including DWT, feature extraction with DWT, and formulation n of SVM with its variations. In Section 3, we explain data preprocessing and the grid search mechanism to find the best parameters. In Section 4, we discuss the results and present our conclusions. 2. METHODS 2.1. Discr ete Wavelet Transform Human perception or any other techniques in time do- main cannot always capture some critical details in the signal. For example, although ECG signals seem to be very similar to each other, a physician is not always able to diagnose a pathological case without using further analyses. Especially, non-stationary signals are hard to analyze even with Fourier transformation (FT) in fre- quency domain. It is because Fourier representation of a time domain signal lacks time resolution. Moreover, FT can only give global information about the frequency distribution of a signal. On the other hand, wavelet transformation is capable of transforming a signal in a multi-resolutionary pattern. Due to its multi-resolution property, it can also provide an analysis of the signal at different frequencies with different resolutions rather than a global knowledge [8]. In order to attain the characteristics of any signal in the frequency domain, the signal is expressed in terms of basis functions. Since these functions form a basis for the original signal x(t), they need to be orthogonal. FT of x(t) consists of cosine and sine functions, whereas wavelet transform is a linear combination of orthonor- mal functions, wavelets [9]. Being orthonormal requires unity in terms of length besides orthogonality. Haar wavelet is one of the well-known functions which is also adopted in the present study. The Haar wavelet can be described as: 10 12 112 1(1) 0 t tt otherwise After determining the mother wavelet, (t), its varia- tions , 1 ba tb a a are generated to form the basis. Here ,ab are said to be translation and scale parameters. Translation refers to shifting over time that enables us observe the time resolution. Resolution in terms of fre- quency is also achieved by scale parameter [9]. A dyadic transform makes it more convenient to ma- nipulate the wavelet to generate basis functions. In other words, if we take a = 2–j and b = k2–j, the following re- cursive function represents a set of functions or bases [9]. 2 ,22 jj jk ttk (2) Hence any signal x(t) can be expressed by the series 2 , , 22 jj jk jk x ta tk (3) Eq.3 can be simplified as ,, , jk jk jk x ta t (4) Both Eqs. 3 and 4 imply that aj,k are the coefficients of base functions j,k (t) as we discussed earlier. Since we have an orthonormal basis, it’s clear that any coefficient aj,k can be derived from the inner product, < j,k (t), x(t) >. The two-dimensional set of coefficients, aj,k, are said to be DWT of x(t) [9]. In the light of the definition of DWT, it’s worth to state that equation 4 can be also written as ,, , , jk jk jk x ttxtt (5) Referring to the wavelet, shifting over the time and variation of the scale contribute to time-scale representa- tion of a signal. In discrete case, the above analysis is  H. Bisgin et al. / J. Biomedical Science and Engineering 4 (2011) 264-271 Copyright © 2011 SciRes. JBiSE 266 achieved in a way that the signal is passed through com- bined high-pass and low-pass filters incorporated with subsampling [8]. If a signal is filtered by changing cut- off frequencies in a cascaded filter model, a frequency resolution (scale in wavelet terminology) is obtained. Subsampling is applied to attain the resolution over time. That is to remove some samples from the signal. If a signal is subsampled by 2, it is implied that every other sample remains in the sequence. The implementation of the idea above is done by a digital filtering approach where the signal is filtered at different cut-off frequencies at different scales. The process is summarized as filter bank approach in the literature [10]. At each level of the procedure, low-pass and high-pass filters are employed to the signals which are subsampled by 2. Output of the highpass filtered part of the signal is processed based on the same principle. In other words, the output becomes an input for the next filter pair where cut-off is halved and subsampling oc- curs again. This process continues until no subsampling is possible [10]. If we let x[n], g[n] and h[n] be the signal, high pass filter, and low pass filter respectively, the following convolutions in Eqs.6 and 7 give us different representa- tions from x[n]. Figure 2 shows the cascading filter model for this iterative procedure. 2 high n yk xngkn (6) 2 lown yk xnhkn (7) At the end of the iterative process, a series of coeffi- cients, which come from the output of the low-pass fil- ters at every level, is obtained. Due to the subsampling, the number of coefficients decreases in log2 scale at each stage. The concatenation all of the coefficients becomes the DWT presentation of the signal, x[n] and yields the same signal length. 2.2. Feature Extraction via DWT Wu et al. show that DWT is more capable of capturing the underlying shape of the original signal than Discrete Fourier Transform (DFT) and DWT has less error of distance estimates on the transformed domains than DFT in its nature [11,12]. Therefore, in this study DWT is preferred to extract features. However, coefficients ob- tained at every level and the energy that those coeffi- cients preserve can be considered as representing com- ponents of the original signal. Mörchen compares dif- ferent manipulations of those coefficients [13]. In this study, we have adopted the proposed method by Wu. We have selected k largest coefficients since they have the largest amount of the energy, which means that these are the most representative. Instead of using just Figure 2. Cascading filter design. coefficients, energy contained by coefficients is taken into account in an aggregated pattern. As Mörchen sug- gests, ordered coefficients and their energies are accu- mulated in steps where we have 2 length signal [13]. Emerging partial sums are considered as fea- tures of the input signal. Hence, in the classification process, instead of the entire signal, we are able to do our experiments with a very small representative data set. This will not only decrease computational burden, but also discard irrelevant information embedded in the sig- nal [13]. 2.3. Support Vector Machine Classification Linear models such as linear regression, logistic regres- sion as well as perceptron algorithm for linear classifica- tion are the most prominent ones that can be used for classification when all the attributes are numeric. How- ever, their ability to draw boundaries between two classes is restricted to some point. Specifically, for many practical problems, they become too simple in handling a complicated data set. On the other hand, SVM over- comes such problems using also linear models, but in a higher dimensional space. It behaves as if it handles a linear case; therefore, its capability of putting non-linear boundaries is possible [14]. If we assume a two-class case, we are supposed to find a hyperplane which divides our data set into two parts accordingly. Many times, it is possible to find more than one boundary to partition the set. The question is to find the best division. This is simply an optimization problem. In other words, one has to find most appropri- ate border via maximizing distances from each set to the separating hyperplane. Actually, the goal of employing SVM is to determine the separating boundary with the maximum margin [15]. Let 1 ,,1,1 n p ii iii Dxyx y where xi is a p-dimensional vector and yi’s are the class labels. If we define a hyperplane H for set D, we have: T T 11 11 i i bify bify wx wx (8) Then, our aim is to maximize the distance2 w, which is also depicted in the Figur e 3.  H. Bisgin et al. / J. Biomedical Science and Engineering 4 (2011) 264-271 Copyright © 2011 SciRes. JBiSE 267 Figure 3. Margin to be maximized. In order to maximize 2 w, it is obvious that the denominator needs to be minimized. One approach to solve this problem is to define T 1 2 www which exactly fits to the quadratic programming approach [15]. More formally, our primal form is as follows: T T 1 min 2 such that 1 ii yb www wx (9) where w and b are design variables. If we rewrite our model in a dual form where we have Lagrange multiplier i for each constraint in the primal form, we obtain the following model: T 1 max 2 such that 0, 0 iijijij iii yy y xx (10) Then we derive that iii y wx and T kk bywx for any k x such that 0 k . As a matter of fact, xk values which correspond to 0 k are called support vectors [15]. Those data points are the ones which con- stitute the classification function fx. Namely, if we suppose that x is a test point and we are supposed to de- termine a label for it, we simply plug it into the follow- ing: T iii f yb xxx (11) In a non-separable linear case, we have a regulariza- tion parameter C in the presence of slack variables i which helps to manage the misclassification and the noise. Slightly modified optimization problem turns out to be in primal form like: T T 1 min 2 such that 1and 0 i iii i C yb www wx (12) That implies the dual form is going to be T 1 max 2 such that 0, 0 iijijij iii yy Cy xx (13) where slack variables do not exist anymore. Since C is a regularization parameter, it prevents from any over fit- ting problem. We will refer to this kind of procedure as C-Support Vector Machine classification (C-SVM). The trade off between minimizing the training error and the maximizing the margin is managed by the constant C [15]. Since there is no way of determining it a priori, n -Support Vector Machine classification (n-SVM) was introduced by Schölkopf and Smola [16]. Optimization procedure was changed in a way that parameter C was removed while a new parameter, n was introduced. This modification aims to control not only the number of margin errors by n, but also the number of support vec- tors. The new formulation in terms of dual problem is as follows: T 11 1 max 2 such that 1 0, 0, mm ijiji j ij iiii yy y m xx (14) In the case of non-separable data set, kernel functions : xx are used to map the data points into a higher dimensional space where the set is not non-sepa- rable anymore. The inner product T ij xxin the optimiza- tion problem above is replaced by a kernel function T , ij i K xxx . We applied kernel trick with radial basis and sigmoid functions, which are expressed below respectively [15]. 2 T 0 ,e (, )tanh ij ij ij ij K K a xx xx xx xx (15) 2.4. Model Validation During training period it is possible to capture all the details of the training set. However, obtained model may not necessarily perform well on a testing set. As a matter of fact, if the model tries to handle every data point in- cluding the noise, overfitting may be a potential problem. T1 ibwx T0 ibwx T1 ibwx  H. Bisgin et al. / J. Biomedical Science and Engineering 4 (2011) 264-271 Copyright © 2011 SciRes. JBiSE 268 Therefore, a classifier should be validated while training is done to avoid the overfitting, which can cause a high error rate on the testing data [17]. While SVM classifiers learn, we aim to build a model incorporated with a validation scheme not to fail on a testing set. Although limited data set can be thought as a constraint to generate a very suitable model for classifi- cation, it is still statistically possible to manipulate such small data sets. In order to reduce the error rate in this kind of problems, k-fold cross-validation is a method that can be applied. Samples are split into k disjoint sub- sets and procedure is run k times. In each run, one of the folds is removed and classifier learns from k-1 folds. Removed set is used to test the model [14]. Figure 4 illustrates a 4-fold cross-validation example where red rectangles represent held sets to be tested and blue ones are used for training. Usually k is taken 10 to generate a classification model, but if there is a very small amount of instances leave-one-out cross-validation (LOOCV) is preferred since it can handle the computational cost. LOOCV is simply done in a way that each instance is held once and training is done based on remaining data points. Trained model is tested on the held instance. This procedure is repeated for every instance so that we can utilize the data maximally and have a higher chance to have an accurate classifier. Another motivation for using LOOCV is that it is deterministic [14]. Let n be the sample size of any data set. Then, the pseudocode of LOOCV can be summarized like below. 1: res ult = 0 {initialize variable which sums the ac- curacies from each run from 1 to n} 2: for i = 1 to n do 3: model = train(D-ri) {remove the ith instance from the data set and train based on remaining por- tion} 4: re sul t = res ult + test(model, ri) {test the model on the unseen ith instance and return the accuracy rate} 5: end for 6: accuracy = re sult/n {take the average of summed accuracies} 3. EXPERIMENTAL DESIGN AND DATA PREPROCESSING Our data set was obtained from a three-year study in Arkansas Children’s Hospital. It was collected from 45 children in which 19 of them were clinically and geneti- cally identified as LQTS patient. Beat-to-beat QT inter- vals were measured via 24 hour Holter monitoring by using the Delmar-Reynolds holter analysis system. Figure 4. k-fold cross validation scheme example. Since the method of recording data is to measure the time for a QT interval, patients did not have the same data length. It forced us to consider the minimum signal length as a baseline. Therefore, the first 16 384 data points for each patient were analyzed because of the computational restrictions of DWT. That corresponds to approximately 4-hour data and takes a power of 2 num- bers of points, which is more appropriate for DWT. Since beat-to-beat QT intervals are obtained over time, the x-axis is no more like in a conventional time series. Due to the variability of QT intervals, instead of equally spaced time intervals, QT interval data are ordered over time in order to constitute QT time-series data. As a re- sult, x values represent order of QT intervals and y val- ues represent QT interval measurements in msec. Figure 5 shows five samples from QT measurements. Recall that DWT with the cascading filter gives coef- ficients at each level from 0 to 1 where the number of data points is 2. Therefore, the proposed DWT pro- cedure implemented in MATLAB [13] successfully ex- tracted 15 attributes from 16 384 time points. As a result, every instance was represented in a lower dimensional vector to proceed into classification step. The specified length of the data was labeled with respect to their diag- nosis. We assigned +1 for the ones with LQTS and -1 for healthy children. In addition, every feature column has been normalized in order to avoid the attribute domi- nancy. We have performed our experiments to find out the most suitable SVM model for our data. LIBSVM soft- ware package has been used for all SVM experiments [18]. As noted earlier, the parameters of SVM were tuned in order to obtain the classification. For this reason, grid-search optimization was used for the parameters C, n and g equations 12, 14, 15 respectively. The potential values for searching the optimum classification were selected around default values of LIBSVM. These were [0.01,0.8] for nwith Dn = 0.01, g = 1/i where 1, ,15i and [1,50] for C with DC = 1. Since kernel functions may also vary throughout the experiments, we considered both radial basis function and sigmoid func- tion. In this way, we expand the domain for the grid-search. 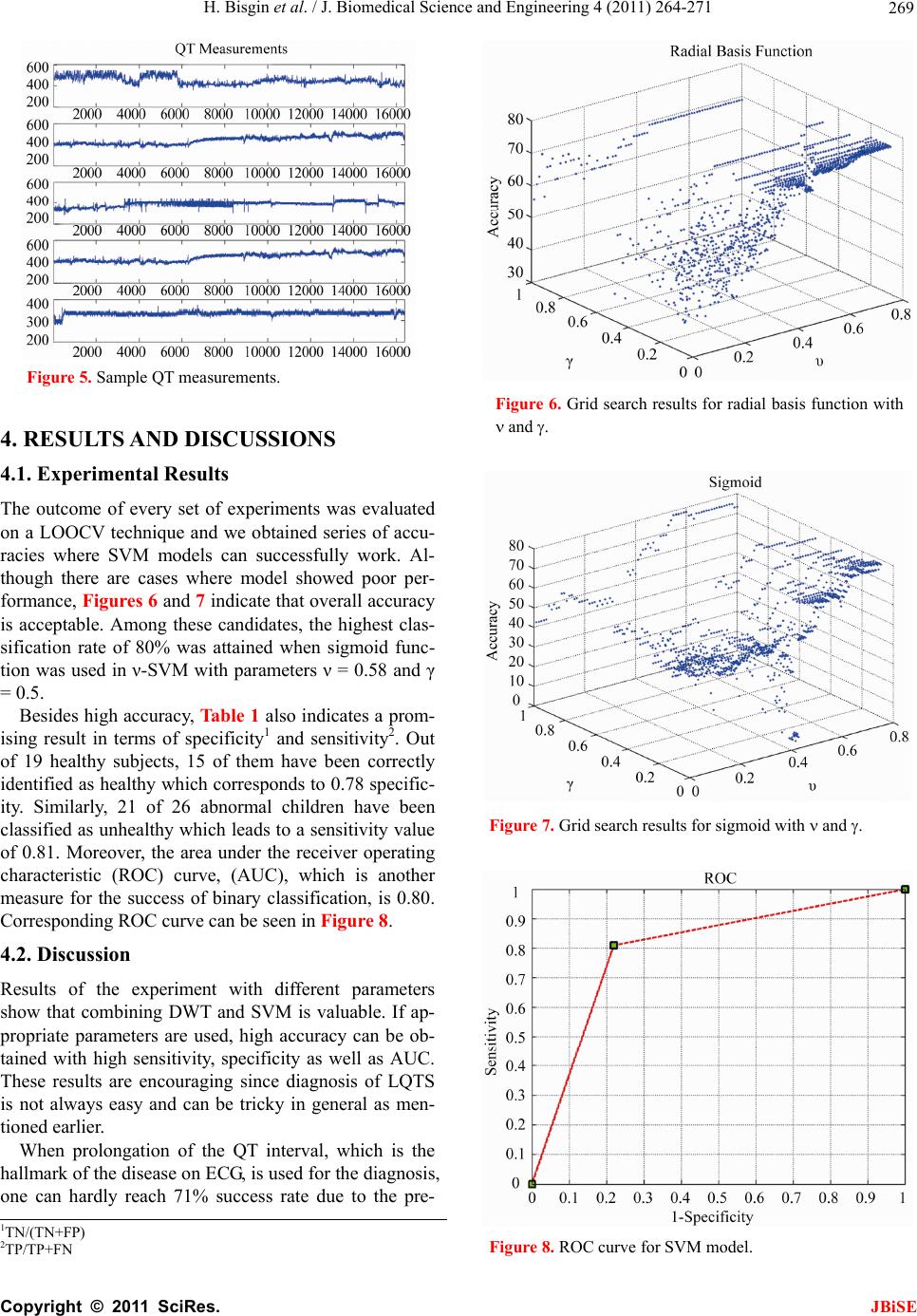 H. Bisgin et al. / J. Biomedical Science and Engineering 4 (2011) 264-271 Copyright © 2011 SciRes. JBiSE 269 Figure 5. Sample QT measurements. 4. RESULTS AND DISCUSSIONS 4.1. Experimental Results The outcome of every set of experiments was evaluated on a LOOCV technique and we obtained series of accu- racies where SVM models can successfully work. Al- though there are cases where model showed poor per- formance, Figures 6 and 7 indicate that overall accuracy is acceptable. Among these candidates, the highest clas- sification rate of 80% was attained when sigmoid func- tion was used in ν-SVM with parameters ν = 0.58 and γ = 0.5. Besides high accuracy, Table 1 also indicates a prom- ising result in terms of specificity1 and sensitivity2. Out of 19 healthy subjects, 15 of them have been correctly identified as healthy which corresponds to 0.78 specific- ity. Similarly, 21 of 26 abnormal children have been classified as unhealthy which leads to a sensitivity value of 0.81. Moreover, the area under the receiver operating characteristic (ROC) curve, (AUC), which is another measure for the success of binary classification, is 0.80. Corresponding ROC curve can be seen in Figure 8. 4.2. Discussion Results of the experiment with different parameters show that combining DWT and SVM is valuable. If ap- propriate parameters are used, high accuracy can be ob- tained with high sensitivity, specificity as well as AUC. These results are encouraging since diagnosis of LQTS is not always easy and can be tricky in general as men- tioned earlier. When prolongation of the QT interval, which is the hallmark of the disease on ECG, is used for the diagnosis, one can hardly reach 71% success rate due to the pre- Figure 6. Grid search results for radial basis function with and Figure 7. Grid search results for sigmoid with and Figure 8. ROC curve for SVM model. 1TN/(TN+FP) 2TP/TP+FN 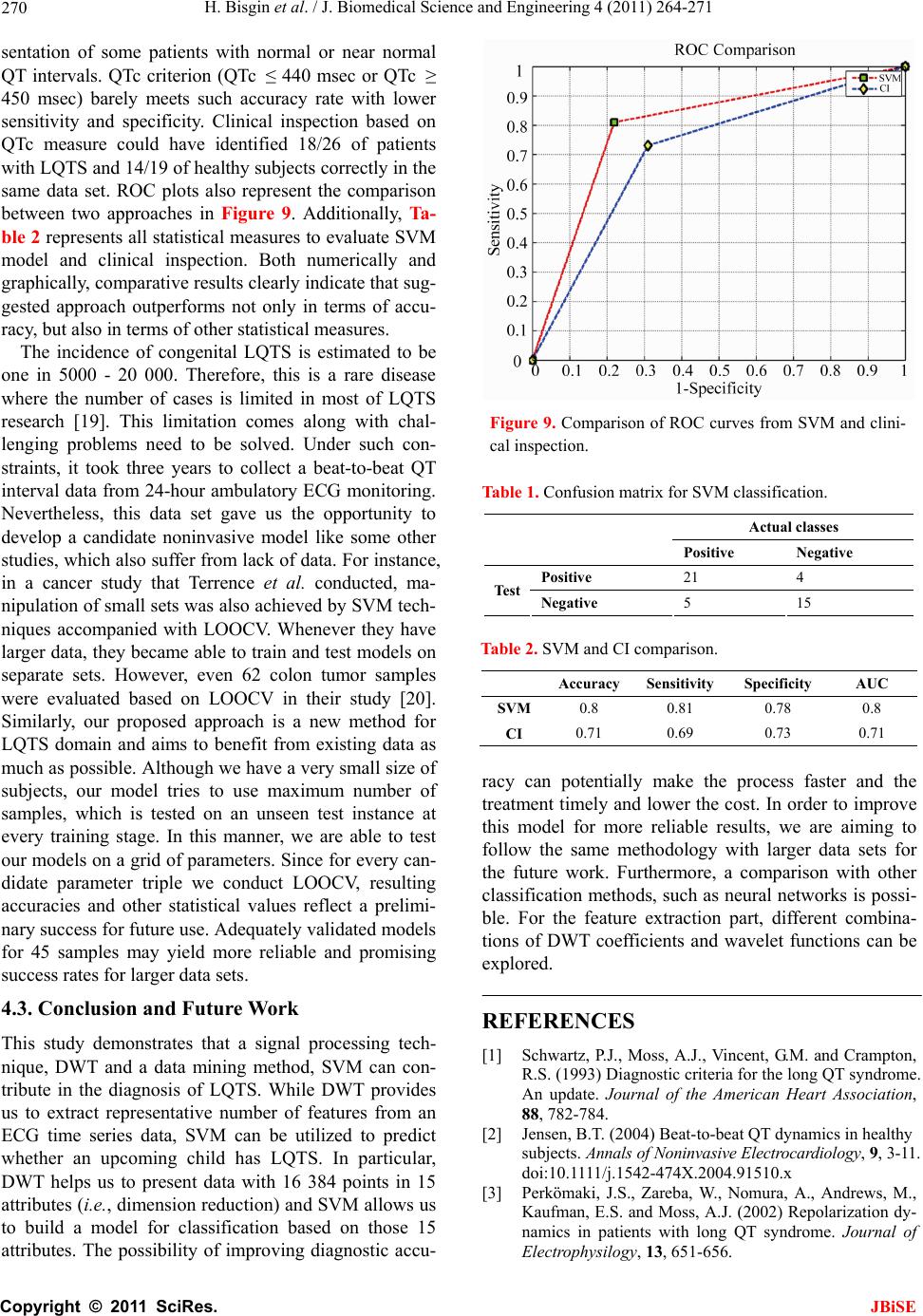 H. Bisgin et al. / J. Biomedical Science and Engineering 4 (2011) 264-271 Copyright © 2011 SciRes. JBiSE 270 sentation of some patients with normal or near normal QT intervals. QTc criterion (QTc ≤ 440 msec or QTc ≥ 450 msec) barely meets such accuracy rate with lower sensitivity and specificity. Clinical inspection based on QTc measure could have identified 18/26 of patients with LQTS and 14/19 of healthy subjects correctly in the same data set. ROC plots also represent the comparison between two approaches in Figure 9. Additionally, Ta- ble 2 represents all statistical measures to evaluate SVM model and clinical inspection. Both numerically and graphically, comparative results clearly indicate that sug- gested approach outperforms not only in terms of accu- racy, but also in terms of other statistical measures. The incidence of congenital LQTS is estimated to be one in 5000 - 20 000. Therefore, this is a rare disease where the number of cases is limited in most of LQTS research [19]. This limitation comes along with chal- lenging problems need to be solved. Under such con- straints, it took three years to collect a beat-to-beat QT interval data from 24-hour ambulatory ECG monitoring. Nevertheless, this data set gave us the opportunity to develop a candidate noninvasive model like some other studies, which also suffer from lack of data. For instance, in a cancer study that Terrence et al. conducted, ma- nipulation of small sets was also achieved by SVM tech- niques accompanied with LOOCV. Whenever they have larger data, they became able to train and test models on separate sets. However, even 62 colon tumor samples were evaluated based on LOOCV in their study [20]. Similarly, our proposed approach is a new method for LQTS domain and aims to benefit from existing data as much as possible. Although we have a very small size of subjects, our model tries to use maximum number of samples, which is tested on an unseen test instance at every training stage. In this manner, we are able to test our models on a grid of parameters. Since for every can- didate parameter triple we conduct LOOCV, resulting accuracies and other statistical values reflect a prelimi- nary success for future use. Adequately validated models for 45 samples may yield more reliable and promising success rates for larger data sets. 4.3. Conclusion and Future Work This study demonstrates that a signal processing tech- nique, DWT and a data mining method, SVM can con- tribute in the diagnosis of LQTS. While DWT provides us to extract representative number of features from an ECG time series data, SVM can be utilized to predict whether an upcoming child has LQTS. In particular, DWT helps us to present data with 16 384 points in 15 attributes (i.e., dimension reduction) and SVM allows us to build a model for classification based on those 15 attributes. The possibility of improving diagnostic accu- Figure 9. Comparison of ROC curves from SVM and clini- cal inspection. Table 1. Confusion matrix for SVM classification. Actual classes Positive Negative Positive 21 4 Test Negative 5 15 Table 2. SVM and CI comparison. Accuracy Sensitivity Specificity AUC SVM 0.8 0.81 0.78 0.8 CI 0.71 0.69 0.73 0.71 racy can potentially make the process faster and the treatment timely and lower the cost. In order to improve this model for more reliable results, we are aiming to follow the same methodology with larger data sets for the future work. Furthermore, a comparison with other classification methods, such as neural networks is possi- ble. For the feature extraction part, different combina- tions of DWT coefficients and wavelet functions can be explored. REFERENCES [1] Schwartz, P.J., Moss, A.J., Vincent, G.M. and Crampton, R.S. (1993) Diagnostic criteria for the long QT syndrome. An update. Journal of the American Heart Association, 88, 782-784. [2] Jensen, B.T. (2004) Beat-to-beat QT dynamics in healthy subjects. Annals of Noninvasive Electrocardiology, 9, 3-11. doi:10.1111/j.1542-474X.2004.91510.x [3] Perkömaki, J.S., Zareba, W., Nomura, A., Andrews, M., Kaufman, E.S. and Moss, A.J. (2002) Repolarization dy- namics in patients with long QT syndrome. Journal of Electrophysilogy, 13, 651-656.  H. Bisgin et al. / J. Biomedical Science and Engineering 4 (2011) 264-271 Copyright © 2011 SciRes. JBiSE 271 [4] Couderc, J.P. and Zareba, W. (1998) Contribution of the wavelet analysis to the noninvasive electrocardiology. Annals of Noninvasive Electrocardiology, 3, 54-62. doi:10.1111/j.1542-474X.1998.tb00030.x [5] Li, Y. and Zhang, C. (1993) QRS detection by wavelet transform. Proceedings of Conference of IEEE Engi- neering in Medicine and Biology, 15, 330-331. [6] Wong, S.N., Mora, F., Passariello, G. and Almeida, D. (1998) QT interval time frequency analysis using Haar wavelet. Computers in Cardiology, 25, 405-408. [7] Strachan, I.G.D., Hughes, N.P., Poonawala, M.H., Wason, J.W. and Tarassenko, L. (2009) Automated QT analysis that learns from cardiologist annotations. Annals of Non- invasive Electrocardiology, Supplement 1, 9-21. doi:10.1111/j.1542-474X.2008.00259.x [8] Polikar, R. (2011) The wavelet tutorial. http://users.rowan.edu/~polikar/wavelets/wttutorial.html [9] Burrus, C.S., Gopinath, R.A. and Guo, H. (1998) Intro- duction to Wavelets and Wavelet Transforms: A Primer. Prentice Hall, Inc., New Jersey, USA. [10] Mallat, S.G. (1989) A theory for multiresolution signal decomposition: The wavelet representation. IEEE Trans- actions on Pattern Analysis and Machine Intelligence, 11, 674-693. doi:10.1109/34.192463 [11] Chan, K. and Fu, A.W. (1999) Efficient time series matching by wavelets. In: ICDE, 12-133. [12] Wu, Y., Agrawal, D. and Abbadi, A.E. (2000) A com- parison of DFT and DWT based similarity search in time-series databases. Proceedings of the 9th Interna- tional Conference on Information and Knowledge Man- agement, McLean, November 2009, 488-495. [13] Mörchen, F. (2003) Time series feature extraction for data mining using DWT and DFT. Data Bionics, Philipps- University Marburg, Germany, 1-31. [14] Witten, I.H. and Frank, E. (2005) Data mining practical machine learning tools and techniques. Morgan Kaufman, San Francisco, USA. [15] Schölkopf, B. and Smola, A.J. (2002) Learning with Kernels: Support vector machines, regularization, opti- mization, and beyond. MIT Press, Cambridge. [16] Schölkopf, B., Smola, A.J., Williamson, R.C. and Bartlett, P.L. (2000) New support vector algorithms. Neural Compu- tation, 12, 1207-1245. [17] Alpaydin, E. (2004) Introduction to Machine Learning (Adaptive Computation and Machine Learning). The MIT Press, Cambridge, USA. [18] Chang, C.C. and Lin, C.J. (2001) LIBSVM: A library for support vector machines. Software available at http://www.csie.ntu.edu.tw/~cjlin/libsvm [19] Schwartz, P.J., Stramba-Badiale, M., Crotti, L., Pedrazz- ini, M., Besana, A., Bosi, G., Gabbarini, F., Goulene, K., Insolia, R., Mannarino, S., Mosca, F., Nespoli, L., Rimini, A., Rosati, E., Salice, P. and Spazzolini, C. (2009) Preva- lence of the congenital long-QT syndrome. Circulation, 120, 1761-1767. doi:10.1161/CIRCULATIONAHA.109.863209. [20] Terrence, S.F., Nello, C., Nigel, D., David, W., Schum- mer, M. and Haussler, D. (2000) Support vector machine classification and validation of cancer tissue samples us- ing microarray expression data. Bioinformati cs, 16, 906-914. |

