Paper Menu >>
Journal Menu >>
 Advances in Bioscience and Biotechnology, 2011, 2, 52-58 ABB doi:10.4236/abb.2011.22009 Published Online April 2011 (http://www.SciRP.org/journal/abb/). Published Online April 2011 in SciRes. http://www.scirp.org/journal/ABB Polyurethane foam as substrate for fungal strains Araceli Loredo-Treviño1, Gilberto García2, Abraham Velasco-Téllez2, Raúl Rodríguez-Herrera1, Cristóbal N. Aguilar1* 1Group of Bioprocesses. School of Chemistry. University of Coahuila. Saltillo, México; 2Research and Development Division. Nemak SA, García, N.L., México; 3Biotechnology Department. School of Chemistry. University of Coahuila. Saltillo, México. Email: cristobal.aguilar@mail.uadec.mx Received 30 January 2011; revised 11 February 2011; accepted 24 February 2011. ABSTRACT Polyurethane is a versatile plastic with several indus- trial applications in the modern life, but it is consid- ered as a very recalcitrant material. Biodegradation of this plastic has been poorly explored, and most of the studies that have been published focus on bacte- rial enzymes. In this work, some fungi with the ca- pacity of growing with polyurethane foam as nutrient source were isolated from sands contaminated with this plastic and from DIA/UAdeC collection, testing their ability to grow on polyurethane as sole carbon and nitrogen sources and their enzymatic activities were determined in specific media as well as their invasion capacity on polyurethane agar plates. 22 fungal strains demonstrated their capacity of grow- ing on polyurethane. Among the enzymatic activities evaluate, the most common was the urease activity (95% of the strains).Protease, esterase and laccase activities were present in 86%, 50% and 36% respec- tively. The great ability of the isolated fungal strains to use polyurethane foam as nutrient opens an im- portant opportunity to study at detail the biodegra- dation of this plastic, with clear implications in cell biology and environmental technology. Keywords: Polyurethane Biodegradation; Fungal Enzymes; In-Plate Screening 1. INTRODUCTION Plastics are synthetic long polymer chains with resis- tance towards microbial attack. Due to their short time on Earth, nature has not been able to design new enzy- matic structures that can attack these synthetic polymers. This has brought concern about how to degrade them. There are some mechanisms like photodegradation, thermal degradation, environment erosion and biodeg- radation and biodegradation is a process in which or- ganic substances are degraded by living organisms. This could occur under different conditions depending on the environment. Plastics are potential substrates for hetero- trophic microorganisms [1] and polyurethanes are a type of plastics widely used in various industries. These plas- tics are synthesized from polyols and polyisocyantes and classified in polyesther or polyether polyurethanes de- pending on what substrates are used and the biological attack towards them is determined by the type of sub- strates used in the polymer synthesis despite its xenobi- otic origin [2]. Polyurethane (PU) biodegradation by microorganisms had become an important issue in the study of degrada- tion mechanisms and PU aging. PU biodegradation could be due to utilization of this material as carbon or nitrogen source by the microorganisms or to fortuitous biodegradation in presence of other nutrients and sub- strates [3]. Both fungi and bacteria have been isolated from the surface of buried PU and showed degradation capacity [4]. The diversity of mycromycetes and the great amount of metabolites they secrete allow them to survive in lim- ited environments [5]. Among fungi there are Gliocla- dium roseum, Aspergillus, Emericella, Fusarium, Peni- cillium, Trichoderma, Gliocladium pannorum, Nectria gliocladiodes and Penicillium ochrochloron [6]. Aureobasidium pullulans is a primary colonizer of poly- vinyl chloride [7]. The attack towards PU could be due because of the enzymes produced by microorganisms [8]. Nomura et al [9] reported that Comamonas acidovorans TB35 has two types of extracellular esterases. Meanwhile, Howard et al [10] found that the supernatant of a liquid culture of Pseudomonas chlororaphis had three different types of PU activity. A culture of Alicycliphilus sp showed es- terase activity when degrading PU [11]. The most important contributions have been made us- ing bacterial strains and the information of fungal bio- degradation of PU is scarce and old. For this reason, in  A. Loredo-Treviño et al. / Advances in Bioscience and Biotechnology 2 (2011) 52-58 Copyright © 2011 SciRes. ABB 53 this study, we report the results obtained of a study of isolation and characterization of fungal strains capable to use PU as sole carbon or nitrogen source, describing the associated enzymes to PU biodegradation. 2. MATERIALS AND METHODS 2.1. Samples and Chemicals Sand samples contaminated with PU were provided by Nemak corporation. These samples were identified as M1, M2, M3 and M4. M1 corresponded to sand mixed with soil and aluminum filings: M2 corresponded to sand that was discarded in a dumpling and contained aluminum fillings, also some plant growth was present: M3 was sand after a process that involved milling and combustion to eliminate the PU: M4, was sand contami- nated with PU before the cleaning process. All reagents were analytical grade and all analysis was performed by triplicate. PU, diisocyante and diol were provided by Nemak corporation and these PU was used for every test unless indicated otherwise. PU used in agar plates radial growth tests was Sayer Lack Mexicana, S.A. de C.V. (T-0028/A; batch: 6289513). Physicochemical charac- terization included humidity content, pH, content of phenols [12], protein [13] and content of molds and yeasts according to AOAC manual and Mexican Official Norms (NOM). All assays were made by triplicate. 2.2. Selection of Microorganisms Capable of Growing Using PU A s Nutrie nt So urce Culture media using PU as sole carbon or nitrogen source were prepared (3 g/L). Also the diisocyanate and the diol used to synthesize the PU were tested (3 g/L of each component). Composition of mineral medium used, either with the PU, the diisocyante and the diol, is de- scribed as follows. Quantity of every component is in g/L: K2HPO4, 1; KH2PO4, 0.5; MgSO4·7 H2O, 0.5; MnCl2·4 H2O, 0.001; CuCl2·2 H2O, 1.4 x 10-5; ZnCl2, 1.1 x 10-5; CoCl2·6 H2O, 2 x 10-5; Na2MoO4·2 H2O, 1.3x10-5; FeCl3·6 H2O, 7.5 x 10-5 [11]. Culture conditions were 30°C and pH 4.5. Fungal strains were isolated from the sands (coded NK strains) and Centro Internacional de Servicios Fitosanitarios (CISEF, S.A. de C.V.) provided some strains (coded CS strains) and the rest of tested fungi were from the DIA/UAdeC collection. Inocula were either spores or mycelium. Fungal growth was ob- served and the strains capable of growing in the PU me- dia were selected to continue with the research. 2.3. Fungal Growth in PU Plates As a parameter of the PU degradation capacity of the fungi, invasion of agar plates containing PU was evalu- ated. The culture media used was mineral medium and 3 g/L of PU [15]. Also 2 g/L of glucose were added to avoid stress and accomplish goo measurements. It is important to mention that no additional nitrogen source was added so the microorganism was forced to use PU as sole nitrogen source, in order for us to be able to re- late the fungal growth to the PU utilization as a nutrient source. PU used was a PU coat instead of Nemak PU because it didn’t show good properties to be used as a support for this test (data not shown). 1x106 spores/mL of 10µL of mycelia suspension for non-sporulating fungi, were inoculated in the center of the plate and allowed to dry. Measurements were made every 24 hours in two axes during 96 hours. All tests were done by triplicate. 2.4. Estimation of PU Degradation and Related Enzymatic Activites To evaluate if polyurethane is better as a carbon or a nitrogen source, an experiment was performed using Trichoderma DIA-T sp and a 1:1 ratio of polyurethane and additional nutrient. Glucose was used as additional carbon source and ammonium sulfate as additional ni- trogen source. The culture was carried out in 7 mL cul- ture tubes using the mineral medium previously de- scribed. Also an experiment using PU as sole carbon and nitrogen source was performed to compare expression of protein in presence or absence of additional nutrients. The amount of spores used was 5x105 spores/mL and the agitation was oscillatory for 9.5 days. Mixing was oscil- latory and the time of culture was 9.5 days and sampling was made by sacrificing one tube every 12 hours. Analyses performed were protein [16], total phenol con- tent [12], urease activity [17] and esterase activity [18]. 2.5. Detection of Associated Enzymes To establish if the selected molds have the enzymes re- lated to PU degradation, four specific culture media were used for the detection of those enzymatic activities. YES medium to determine protease activity, Tween 80 medium for esterase activity, Christensen urea medium for urease activity and PDA-ABTS (2,2’-azino-bis (3-ethylbenzhiazoline-6-sulphhonic acid) medium [14] for laccase activity. Media composition is described as follows. Quantity of every component is given in g/L: YES medium: K2HPO4, 1; KH2PO4, 0.5; MgSO4·7 H2O, 0.5; MnCl2·4 H2O, 0.001; CuCl2·2 H2O, 1.4 x 10-5; ZnCl2, 1.1 x 10-5; CoCl2·6 H2O, 2 x 10-5; Na2MoO4·2 H2O, 1.3x10-5; FeCl3·6 H2O, 7.5 x 10-5, gelatin/peptone, 0.02. Christensen’s urea medium: Urea, 20, NaCl, 5; K2HPO4, 2; glucose, 1; phenol red, 0.12. The urea is sterilized by filtration and then added to the rest of the medium. Tween 80 medium: peptone, 10; NaCl, 5; CaCl2, 0.1; Tween 80, 10 mL [11]. PDA-ABTS medium: Potato-dextrose agar 49; ABTS, 0.1M. To carry out the experiments, 5 mL were disposed in tubes and inocu-  A. Loredo-Treviño et al. / Advances in Bioscience and Biotechnology 2 (2011) 52-58 Copyright © 2011 SciRes. ABB 54 lated with the fungal strains and incubated 8 days at 30 °C, except for PDA-ABTS medium were the incubation was carried out in plate. Fungal growth was observed in YES medium, and this indicated a positive protease ac- tivity; urease activity was determined by the turn in color from yellow to pink; esterase activity was evi- denced by the apparition of a white precipitate in the medium and laccase activity was evidenced by the appa- rition of a blue-green halo round the fungal colony. 3. RESULTS AND DIS CUSSION From 32 fungal strains, 22 grew using PU as nutrient source. The other 10 grew using either the diol or the diisocyanate. These 22 strains were isolated from the sand samples, the group collection and provided by CISEF. Among the selected genera of fungi were As- pergillus, Trichoderma, Paecelomyces, Penicillium, Al- ternaria, and Fusarium. These results are similar to re- sults reported by Lagauskas et al [5]. They isolated a great amount of fungal species from several types of plastics including PU and species of Trich oderma, Peni- cillium, Paecellomyces and Alternaria. Cosgrove et al [4] also isolated and identified some Penicillia and Alter- naria among others from buried PU coupons and 3 also from buried samples of PU, isolated species of Nectria, Penicillium and Geomyces. These fungal genera were isolated from plasticized polyvinyl chloride (PVC) [7]. The recurrent presence of these mycromycetes in soil buried plastics may be due to the great amount of exoenzymes that fungi secrete and these enzymes allow the fungi to degrade plastics and probably use them as nutrient source [5]. Also, these reports used standardized soil or soil from template regions. Food Research De- partment and NK strains were isolated from a Mexican semidesertic region of extreme environmental conditions (where temperatures and solar radiation are elevated and water content very low). It was important to know if the selected strains had the enzymatic activities related to PU biodegradation so the enzymatic characterization was performed. These results are shown in Table 2. All the fungi grew on the media, but not every mold showed all the enzymatic activities. In table 1 it can be observed that the majority of the fungi presented urease activity (95.45%). The second more common activity was protease activity (86.36%), then esterase activity (50%) and the less common activity detected was lac- case activity (36.26%). These enzymes, as stated previ- ously, are related to PU degradation, as reported by Bar- rat, et al [6]. In recent years the research about PU bio- degradation has focused in bacteria and their enzymes. The major part of the enzymes reported as PU degrading enzymes showed esterase activity (19; 20; 10; 11; 21) and some showed also protease activity (20; 10) but none of these reports mention urease activity. The diver- sity of PUs due to the different components used for their synthesis could be the reason of the difference be- tween the enzymes produced by the microorganisms. In this work the PU used was a polyether PU (information provided by personal of the automotive company. The main components used to synthesize the PU are 4,4'-Methylenebis(phenyl isocyanate) and phenol for- maldehyde) and the fungi capable of using it as nutrient source must be able to produce enzymes that hydrolyze the plastic and since there are not ester bonds in the molecule, the fungi that were isolated must have the ca- pacity to secrete enzymes other than esterases to attack the PU molecule and use it degradation products as nu- trient source. In this experiment we determined the presence of this type of enzymes since they are the main group reported responsible of polyurethane degradation. The invasive capacity of the fungi was evaluated on agar plates with mineral medium and PU. Results are shown in Figure 1. It can be seen that the fungi have different growth rate but Trichoderma DIA-T and PSS showed the fastest growth velocity (0.7917 mm/h and 0.7813 mm/h respectively). Invasive capacity of fungi has been used to indicate Table 1. Enzymatic characterization of the fungi capable of using polyurethane as nutrient source. Fungi esterase protease urease laccase Fungi esterase protease urease laccase DIA-T - + + + CS3 + + + FP310 + + + - CS4 - - + - PSH + + + - CS6 - - + - EH2 + + + + CS7 - - + - EH3T1 + + + + NK2 - + + - PSS - + - - NK4 + + + - NH4 + + + + NK6 + + - GS + + + - NK8 + + + + GH1 - + + + NK10 - + + + CS1 - + + - NK12 - + + - CS2 - + + + NK23 - + + - 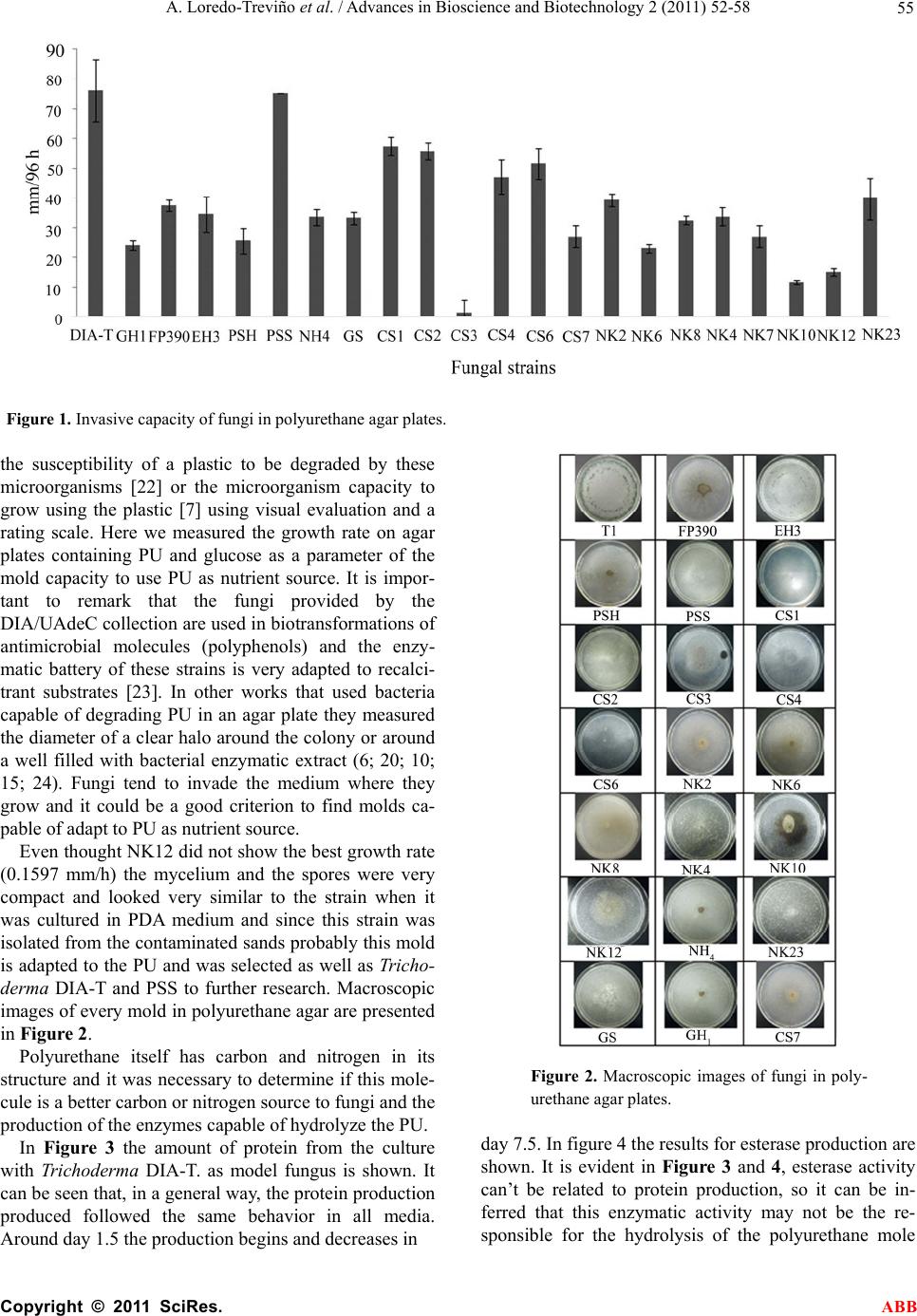 A. Loredo-Treviño et al. / Advances in Bioscience and Biotechnology 2 (2011) 52-58 Copyright © 2011 SciRes. ABB 55 Figure 1. Invasive capacity of fungi in polyurethane agar plates. the susceptibility of a plastic to be degraded by these microorganisms [22] or the microorganism capacity to grow using the plastic [7] using visual evaluation and a rating scale. Here we measured the growth rate on agar plates containing PU and glucose as a parameter of the mold capacity to use PU as nutrient source. It is impor- tant to remark that the fungi provided by the DIA/UAdeC collection are used in biotransformations of antimicrobial molecules (polyphenols) and the enzy- matic battery of these strains is very adapted to recalci- trant substrates [23]. In other works that used bacteria capable of degrading PU in an agar plate they measured the diameter of a clear halo around the colony or around a well filled with bacterial enzymatic extract (6; 20; 10; 15; 24). Fungi tend to invade the medium where they grow and it could be a good criterion to find molds ca- pable of adapt to PU as nutrient source. Even thought NK12 did not show the best growth rate (0.1597 mm/h) the mycelium and the spores were very compact and looked very similar to the strain when it was cultured in PDA medium and since this strain was isolated from the contaminated sands probably this mold is adapted to the PU and was selected as well as Tricho- derma DIA-T and PSS to further research. Macroscopic images of every mold in polyurethane agar are presented in Figure 2. Polyurethane itself has carbon and nitrogen in its structure and it was necessary to determine if this mole- cule is a better carbon or nitrogen source to fungi and the production of the enzymes capable of hydrolyze the PU. In Figure 3 the amount of protein from the culture with Trichoderma DIA-T. as model fungus is shown. It can be seen that, in a general way, the protein production produced followed the same behavior in all media. Around day 1.5 the production begins and decreases in Figure 2. Macroscopic images of fungi in poly- urethane agar plates. day 7.5. In figure 4 the results for esterase production are shown. It is evident in Figure 3 and 4, esterase activity can’t be related to protein production, so it can be in- ferred that this enzymatic activity may not be the re- sponsible for the hydrolysis of the polyurethane mole  A. Loredo-Treviño et al. / Advances in Bioscience and Biotechnology 2 (2011) 52-58 Copyright © 2011 SciRes. ABB 56 Figure 3 Extracellular production of Trichoderma DIA-T under dif- ferent culture conditions. Polyurethane as sole carbon and nitrogen source (); ammonium sulfate as additional nitrogen source (); glucose as additional carbon source (). Figure 4. Esterase activity of Trichoderma DIA-T. under different cultura conditions. Polyurethane as sole carbon and nitrogen source (); ammonium sulfate as additional nitrogen source (); glucose as additional carbon source (). Figure 5. Release of phenolic compounds to the cultura media by Trichoderma DIA-T under different cultura conditions. Polyurethane as sole carbon and nitrogen source (); ammonium sulfate as addi- tional nitrogen source (); glucose as additional carbon source (). cule and/or its modification. In 2005, Santerre et al, re- ported that cholesterol esterase could degrade ether type polyurethanes although there was no presence of ester bonds in the molecule [25] so, it is possible for a fungal esterase to hydrolyze this type of PU in later stages of the culture process. For urease activity (Figure 5), tendency of the fungi is to produce this enzyme in the first stages of the culture and this, related with phenolic compounds released to the culture media (Figure 5, lines). The enzyme could modify the polyurethane structure so, from day 5 the phenolic compounds begin to increase. The measure- ment of phenolic compounds released to the culture me- dia could be a parameter to estimate the polyurethane degradation due to the phenolic molecules contained in its estructure. Maximum U/L of urease activity reported in this in- vestigation (4985 U/L) is higher than reported for bacte- ria by other authors like Shigeno-Akutsu et al [26] that report a maximum of 3300 U but for esterase activity. It’s important to highlight that the majority of the articles adjudge to esterases the hydrolysis of the polyurethanes that were tested. In 2009, Ibrahim et al, reported degra- dation of an ester type polyurethane by Alternaria sp. and found that the strain produced protease, esterase and urease enzymes when tested in specific media, but when they look for the enzymes in the polyurethane culture filtrate they did not found any urease activity [27]. The present work would be the first report to our knowledge to attribute the polyurethane degradation to an urease enzyme. 4. CONCLUSIONS From two collections and from sand contaminated with PU, 22 fungal strains were capable of growing using PU as carbon source, and all of these strains showed at least one of the enzymatic activities related with PU biodegradation being the protease activity the most common one. Two fungal strains showed the fastest growth on PU plates. Three strains were selected for further research on degrading PU based on their invasive capacity, their morphology on PU plates and their enzy- matic activities and one of them was used to evaluate the PU degradation in culture media using different addi- tional nutrients. Probably, urease is the main enzyme responsible of the fungal attack towards PU as the re- sults from different culture media using Trichoderma DIA-T showed. 5. ACKNOWLEDGEMENTS The authors thank Nemak corporation for materials and samples pro- vided; CISEF for fungal strains and Consejo Nacional de Ciencia y Tecnología (CONACYT) for the student grant (258623/221603).  A. Loredo-Treviño et al. / Advances in Bioscience and Biotechnology 2 (2011) 52-58 Copyright © 2011 SciRes. ABB 57 REFERENCES [1] A. Shah, F. Hasan, A. Hameed and S. Ahmed, “Biologi- cal degradation of plastics: A comprehensive review”, Biotechnology Advances, 2008, Vol. 26, pp.246-265. [2] R. E. Vega, T. Main and G. T. Howard, “Cloning and expression in Escherichia coli of a polyure- thane-degrading enzyme from Pseudomonas fluorescens” International Biodeterioration and Biodegradation, 1999, Vol. 43, pp. 49-55. [3] M. Urgun-Demirtas, D. Sing and K, Pagilla, “Laboratory investigation of biodegradability of a polyurethane foam under anaerobic conditions”, Polymer Degradation and Stability, 2007, Vol. 92, pp. 599-1610. [4] L. Cosgrove, P.L. McGeehan, G. D. Robson and P.S. Handley “Fungal Communities Associated with Degra- dation of Polyester Polyurethane in Soil”, Applied and Environmental Microbiology, 2007, Vol. 73, pp. 5817-5824. [5] Lagauskas, L. Levinskaitė and D. Pečiulytė, “Micromy- cetes as deterioration agents of polymeric materials”, In- ternational Biodeterioration and Biodegradation, 2009, Vol. 52, pp. 233-242. [6] S. R. Barrat, A. R. Ennos, M, Greenhalgh, G.D. Robson and P.S. Handley, "Fungi are the predominant mi- cro-organisms responsible for degradation of soil-buried polyester polyurethane over a range of soil water holding capacities”, Journal of Applied Microbiology, 2003, Vol. 95, pp. 78-85. [7] J. S. Webb, M. Nixon, I. M. Eastwood, M. Greenhalgh, G. D. Robson and P. S. Handley, “Fungal colonization and Biodeterioration of Plasticized Polyvinyl Chloride”, Ap- plied and Environmental Microbiology, 2000, Vol. 66, pp. 3194-3200. [8] Ruiz, T. Main, N. P. Hilliard, and G. T. Howard, “Purifi- cation and characterization of two polyurethanases en- zymes from Pseudomonas chlororaphis”, International Biodeterioration and Biodegradation, 1999, Vol. 43, pp. 43-47. [9] N. Nomura, Y. Shigeno-Akutsu, T. Nakajima-Kambe and T. Nakahara, “Cloning and Sequence Analysis of a Poly- urethane Esterase of Comamonas acidovorans TB-35”, Journal of Fermentation and Bioengineering, 1998, Vol. 86, pp. 339-345. [10] G. T. Howard and N. P. Hilliard, “Use of Coomasie blue-polyurethane interaction in screening of polyure- thanase proteins and polyurethanolytic bacteria”, Interna- tional Biodeterioration and Biodegradation, 1999, Vol. 43, pp. 23-30. [11] Oceguera-Cervantes, A. Carrillo-García, N. López, S. Bolaños-Núñez, M. J. Cruz-Gómez, C. Wacher and H. Loza-Tavera, “Characterization of the Polyurethanolytic Activity of Two Alicycliphilus sp. Strains Able To De- grade Polyurethane and N-Methylpyrrolidone”, Applied and Environmental Microbiology, 2007, pp. 6214-6223. [12] H. P. S. Makkar, M. Blümmel, N. K. Borowy and K. Becker, “Gravimetric determination of tannins and their correlations with chemical and protein precipitation methods” Journal of the Science of Food and Agricul- ture,1993, Vol. 61, pp. 161-165. [13] H. Lowry, N. J. Rosebrough, A. Farr and J. Randall, “Protein measurements with the Folin-phenol reagents”, Journal of Biological Chemistry, 1951, Vol. 193, pp. 265-275. [14] P. Chairattanamanokorn, T. Imai, R. Kondo, M. Ukita and P. Prasertsan, “Screening thermotolerant white-rot fungi for decolorization of wastewaters”, Applied Biochemistry and Biotechnolgy, 2006, Vol. 3, pp. 195-204. [15] G. T. Howard, J. Vicknair and R. I. Mackie, “Sensitive plate assay for screening and detection of bacterial poly- urethanase activity”, Letters in Applied Microbiology, 2001, Vol. 32, pp. 211-214. [16] M. M. Bradford, “A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding” Analytical Bio- chemistry, 1952, Vol. pp. 248–254. [17] P. Witte and N. Medina-Escobar, “In-gel detection of urease with nitroblue tetrazolium and quantification of the enzyme from different crop plants using the indo- phenol reaction. Analytical Biochemistry, 2001, Vol. 290, pp. 102–107. [18] Bastida, P. Sabuquillo, P. Armisen, R. Fernan- dez-Lafuente, J. Huguet and J. N. Guisán, ”A single step purification, immobilization, and hyperactivation of li- pase via interfacial adsorption on strong hydrophobic supports” Biotechnology and bioengineering. 1998, Vol. 58, pp. 486-493. [19] Y. Akutsu, T. Nakajima-Kambe, N. Nomura and T. Na- kahara, “Purification and Properties of a Polyester Poly- urethane-Degrading Enzyme from Comamonas acido- vorans TB-35” Applied and Environmental Microbiology, 1998, Vol.64, pp. 62-67. [20] G. T. Howard and R. C. Blake, “Growth of Pseudomonas fluorescens on a polyester-polyurethane and the purifica- tion and characterization of a polyurethanase-protease enzyme”, International Biodeterioration and Biodegrada- tion, 1998, Vol. 42, pp. 213-220. [21] L. Rowe and G. T. Howard, “Growth of Bacillus subtilis on polyurethane and the purification and characterization of a polyurethanase-lipase enzyme” International Biode- terioration and Biodegradation, 2002, Vol. 50, pp. 33-40. [22] R. Ghazali, L. C. Mei, N. Z. K. Shaari, N. Z. K., M. Yu- sof and S. Ahmad, “Preliminary study on microbial deg- radation of flexible polyurethane foams-physico-mechanical and weight changes during fungal deterioration” Journal of Oil Palm Research, 2005, Vol. 17, pp. 103-109. [23] M. Cruz-Hernández, J. C. Contreras-Esquivel, F. Lara, R. Rodríguez and C. N. Aguilar, “Isolation and evaluation of tannin-degrading fungal strains from the mexican desert”, Verlag der Zeitschrift für Naturforschung, Tübingen, 2005, Vol. 60, pp. 843-848. [24] G. T. Howard, B. Crother and J. Vicknair, “Cloning, nu- cleotide sequencing and characterization of a polyure- thanase gene (pueB) from Pseudomonas chlororaphis” International Biodeterioration and Biodegradation, 2001, Vol. 47, pp. 141-149. [25] J. P. Santerre, K. Woodhouse, G. Laroche and R. S. Labow, “Understanding the biodegradation of polyure- thanes: From classical implants to tissue engineering materials”, Biomaterials, 2005, Vol. 26, pp. 7457-7470.  A. Loredo-Treviño et al. / Advances in Bioscience and Biotechnology 2 (2011) 52-58 Copyright © 2011 SciRes. ABB 58 [26] Y. Shigeno-Akutsu, T. Nakajima-Kambe, N. Nomura and T. Nakahara, “Purification and properties of cul- ture-broth-secreted esterase from the polyurethane de- grader Comamonas acidovorans TB-35”, Journal of Bio- science and Bioengineering, 1999, Vol. 88, pp. 484-487. [27] N. I. Ibrahim, A. Maraqa, K. M. Hameed, I. M. Saadoun, H. M. Maswadeh and T. Nakajima-Kambe, “Polyes- ter-polyurethane biodegradation by Alternaria solani, isolated from Northern Jordan”, Advances in Environ- mental Biology, 2009, Vol. 2, pp. 162-170. |

