 International Journal of Clinical Medicine, 2013, 4, 44-50 Published Online December 2013 (http://www.scirp.org/journal/ijcm) http://dx.doi.org/10.4236/ijcm.2013.412A1008 Open Access IJCM TP53 Mutations and Chemotherapy Response to Neoadjuvant Metotrexate, Cisplatin and Adryamicin Chemotherapy in Resected Osteosarcoma Ligia Richter1,2, Marcelo Buzzi2*, Carmela Dantas-Barbosa2,3 1Faculdade LS—Coordenação do Curso de Enfermagem, Setor D Sul, Lote 5, Taguatinga Sul, DF, Brasil; 2Laboratório de Bioquimica Analítica, Patologia Molecular, Rede Sarah de Hospitais de Reabilitação, Brasília, DF, Brasil; 3Centre de Recherche en Cancérologie de Lyon, UMR INSERM 1052—CNRS 5286, Centre Léon Bérard, Cheney D, 28 Rue Laënnec, Lyon, France. Email: ligia.richter@gmail.com; *mbuzzi@sarah.br; carmeladantas@hotmail.com Received October 30th, 2013; revised November 28th, 2013; accepted December 10th, 2013 Copyright © 2013 Ligia Richter et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In accor- dance of the Creative Commons Attribution License all Copyrights © 2013 are reserved for SCIRP and the owner of the intellectual property Ligia Richter et al. All Copyright © 2013 are guarded by law and by SCIRP as a guardian. ABSTRACT Osteosarcoma is a rare and highly malignant tumor that usually affects adolescents and young adults. Despite current management protocols, up to half of patients succumb to the disease. Moreover, there is no well-characterized molecu- lar marker for diagnosis and prognosis. TP53 alterations have been associated with a poor prognosis in many cancers. The aim of this retrospective work was to find out whether TP53 functional status predicts response to neoadjuvant chemotherapy and thus may help treatment decision for osteosarcoma patients. Seventeen biopsies of osteosarcoma patients receiving primary metotrexate, cisplatin and adryamicin chemotherapy followed by surgery were analyzed. TP53 exons 5 - 9 mutations were screened. Among 17 biopsies, 4 (23.5%) displayed TP53 mutations: 3 deletions and one single-nucleotide substitution. The presence of TP53 gene mutation does not correlate with resistance to chemo- therapy according to histological Rosen grade and nevertheless is associated with patient’s age in a significant manner (p < 0.05). The presence of non-mutated TP53 is not entirely specific for a good prognosis. We found no evidence that TP53 mutations predict chemoresistance in osteosarcoma patients more over the overall survival curve, followed for more than 12 years, showing no difference between patients with tumors harboring wild type or mutated TP53 gene (p < 0.5). Further analysis to identify other genes that can influence chemotherapy response and clinical outcome in os- teosarcoma is needed to improve patient treatment. Keywords: Osteosarcoma; Chemoresistance; TP53 Gene 1. Introduction Osteosarcoma is the commonest primary bone cancer in children and young adults. Osteosarcoma presents a peak of incidence at the age of 15 - 19 years that coincides with the growth spurt. The estimated incidence rate worldwide is 4 million/year. It is highly malignant, char- acterized by a high potential to metastasize [1]. These tumors typically arise in the metaphyseal regions of long bones. A preference for the distal femur, proximal tibia and proximal humerus is observed [2]. Almost all osteo- sarcomas are high grade and have a poor prognosis. Pa- tients diagnosed with osteosarcoma usually present a large tumor and numerous lung micrometastases which markedly decrease the potential for cure [3]. The use of multiple chemotherapy agents pre- and postoperatively has significantly improved the outcome in the last 30 years, leading to a 5-year disease-free survival for pa- tients with localized tumors in the range of 20% to 60% range [4]. Despite these improvements, approximately 30% of patients with localized disease and 60% of pa- tients with pulmonary metastases succumb to the illness. Osteosarcoma is a complex neoplasia. Cytogenetic analysis has revealed multiple chromosomal rearrange- ments with a high degree of aneuploidy, gene amplifica- *Corresponding author.  TP53 Mutations and Chemotherapy Response to Neoadjuvant Metotrexate, Cisplatin and Adryamicin Chemotherapy in Resected Osteosarcoma 45 tion, and multiple unbalanced chromosomal rearrange- ments without a typical translocation, as it can be seen in other sarcomas [5]. Osteosarcoma markers are required to further characterize this complex, multifactorial neoplasia and improve diagnosis and prognosis. The TP53 gene was defined as the tumor suppressor gene due to its ability to suppress the malignant growth of transformed cells as well as tumors [6]. The TP53 gene locates in chromosome 17 (17p13), a region fre- quently lost in tumors (LOH) and extensive TP53 muta- tion searches revealed that over 50% of human tumors carry mutations in this gene [7]. The majority of the TP53 mutations are missense mutations occurring in the highly conserved DNA binding domain of TP53 (exons 5 - 8), plus exon 9 [8]. According to IARC TP53 mutation database, exons 5, 7 and 8 encompass 30, 26 and 24% of all TP53 mutations respectively. Exon 10 harbors only 1.1% of reported TP53 mutations [9]. Of Li-Fraumeni family members, who present an increased risk for de- velopment of cancers, including osteosarcoma, it was observed a correlation between tumor type, family struc- ture and TP53 mutation type [10]. TP53 is a transcription factor which responds to di- verse cellular stresses to regulate target genes that induce cell cycle arrest, apoptosis, senescence, DNA repair, or changes in metabolism [11]. TP53 also fulfills functions during development in normal tissues [12] and in respon- se to inflammation [13]. The p53 protein is expressed at lower levels in normal cells compared to transformed cells where higher levels of p53 was observed, suggest- ing that p53 can contribute to transformation and mali- gnancy. Many attempts to establish TP53 mutations as a tumor marker have been done and the results are often contradictory. It is difficult to compare all the studies, be- cause different methodologies have been employed. Im- munohistochemistry does not allow the precise identifi- cation of the mutation. DNA sequencing revealed that in human cancers, >1800 distinct TP53 missense mutations have been identified. In breast cancer, it was demonstra- ted that TP53 mutation is a marker of biologically ag- gressive disease [14] and a response to therapy and survi- val [15]. The correlation between p53 protein functional status and a response to neoadjuvant chemotherapy was demonstrated in head and neck squamous cell carcinoma (HNSCC) in the oral cavity. The results indicate that the p53 loss of function may predict a significant low rate of complete remission and suboptimal response to cisplatin- based neoadjuvant chemotherapy in patients with oral cavity SCC [15]. The purpose of this paper is to investi- gate whether TP53 mutational status predicts a response to neoadjuvant chemotherapy and thus may help treat- ment decision in osteosarcoma patients. 2. Material and Methods 2.1. Patient’s Selection Patients with a confirmed diagnosis of osteosarcoma, at any age of both genders that had the biopsies effectuated in our center (Sarah Network of Hospitals, Brasília, Bra- zil), without previous treatment of chemotherapy or ra- diotherapy, were eligible for the study. Both fresh tissue and paraffin embedded biopsies were considered. To be included patients should be submitted to the treatment comporting neoadjuvant chemotherapy, followed by sur- gery and adjuvant chemotherapy according to the proto- col established in May 1996. Of the 33 eligible patients, 15 were excluded due to insufficient DNA quantity and one patient left our institution before the chemotherapy treatment. The remaining 17 patients were analyzed in the present work. This study was previously approved by the Sarah Hospital Ethics Committee. After neoadjuvant chemotherapy, all patients were ra- diologically re-evaluated and surgery of primary tumor was performed. The type of surgery (amputation or limb salvage) was chosen according to the location and exten- sion of the tumor. To evaluate chemotherapy response, pathologists re- viewed histological material to determine the primary tumor response according to a method previously descri- bed [16]. Responses were defined as “good” (>95% tu- mor necrosis) or “poor” (<95% tumor necrosis). 2.2. TP53 Gene Amplification Genomic DNA was extracted from 50 mg of frozen tis- sue or from 3 to 6 10 µM sections of formalin-fixed, par- affin-embedded blocks using the QIAamp DNA Mini Kit (Qiagen). PCR analysis of exons 5 to 9 was performed using the primers shown in the Table 1. TP53 exons were amplified during 30 cycles of 94˚C for 45 s, 65˚C for 45 s and 72˚C for 1 min in a 25 l volume reaction containing from 50 to 200 ng of genomic DNA, 1.5 mM Table 1. TP53 primers sequence. Primer Sequence 5’ 3’ Amplicon Size in bp E5-53-Actcttcctgcagtactcccctgc E5-53-Bgccccagctgctcaccatcgcta 211 E6-53-Agattgctcttaggtctggcccctc E6-53-Bggccactgacaaccacccttaacc 185 E7-53-Agtgttctctcctaggttggctctg E7-53-Bcaagtggctcctgacctggagtc 139 E8-53-Aacctgatttccttactgcctctggc E8-53-Bgtcctgcttgcttacctcgcttagt 200 E9-53-Agcctctttcctaggactgcccaac E9-53-Bcgcaagacttagtacctgaagggtg 102 Open Access IJCM  TP53 Mutations and Chemotherapy Response to Neoadjuvant Metotrexate, Cisplatin and Adryamicin Chemotherapy in Resected Osteosarcoma 46 of MgCl2, 0.2 mM dNTP’s, 50 M of each primer and 0.3 U of Taq DNA Polymerase Platinum (Life Techno- logies). 2.3. Single Strand Conformation Polymorphism (SSCP) and Sequencing Analysis The single strand conformation polymorphism (SSCP) analysis was performed using the PhastSystem automat- ed electrophoresis apparatus (GE Healthcare/Amersham Pharmacia) and 12.5% and 20% polyacrilamid gels. The run settings were adapted from Kurvinen and coworkers (1995) [17]. Pre-run: 400 V at 10 mA, 2.5 W, 100 Avh; gel loading: 400 V, 1 mA, 2.5 W, 2 Avh, separation: 400 V, 10 mA, 2.5 W, 400 Avh*. All runs were performed at 15˚C. *Avh (accumulated volts per hour) the run time varies according to the DNA size. Exon 5: 300 Avh, Exon 6: 350 Avh, Exon 7: 350 Avh, Exon 8: 300, Avh, Exon 9: 200 Avh. After electrophoresis the gels were silver stained with the Plus-One DNA Silver Staining kit (GE Healthcare/ Amersham Pharmacia) following manufactures’ indica- tions. Using a vacuum gel drying system the gels were dry with Whatman paper. Samples with mutations were identified by the presence of an abnormal electrophoretic migration pattern in comparison with a control carrying wild-type (wt) TP53 and were further excised from the gel, for PCR reamplification. The excised band was placed in a 1.5 mL tube, following the addition of 100 µl of water the material were frozen, macerated and heated at 100˚C for 10 minutes. Following a 10,000 g centrifu- gation for 10 minutes the supernatant were transferred to a new tube and 5 µL of it used for PCR reamplification in only 20 PCR cycles. After checking in an agarose gel for the presence of a PCR product, the material was cloned into a pGEM-T Easy vector (Promega). Plasmid purification was performed using the Kit Flexi Prep (GE Healthcare) according to instructions. The sequencing reactions were prepared using the universal primer (Promega) and the Big Dye Terminator kit (Applied Biosystems), following manufacturers’ in- structions. Sequencing reactions were resolved in the ABI310 automatic sequencer (Applied Biosystems) Se- quence alignments and translations were made with BioEdit software. 2.4. Statistical Analysis The association between TP53 gene mutations and the response to chemotherapy was calculated using the Fi- sher’s two-tailed exact test. Students’ test was applied to compare ages. The criterion for statistical significance was = 0.05. Kaplan and Meier survival curves and sta- tistical analyses were conducted with GraphPad Prism software (GraphPad Software). 3. Results Seventeen patients aged 10 - 75 years (median 17 years) entered the study. Ten patients were male (59%) and 7 female (41%). Sixteen patients had an osteoblastic tumor and only one patient had a chondroblastic subtype. We selected only biopsies of primary tumors before treatment aiming to correlate the TP53 status to the neoadjuvant treatment response. The identified somatic mutations associated with hotspot region from the tumor suppressor gene TP53 are presented in Table 2. Among the 17 biopsies, 4 (23.5%) displayed TP53 mutations. In total, 6 mutations were identified in 4 osteosarcoma tu- mor samples: The patient 2 presented deletion of exon 9, Ex9 c.924_936del102, patient 14 presented a silent mu- tation at position E x 7 c.697 C > T and a deletion at po- sition E x 7 c.732del1 corresponding to codon 244 which engender a frameshit. The patient 25 also harbors a silent mutation at position E x 7 c.703 C > A single-nucleotide substitution and a serine→ cysteine substitution at codon 241 (E x 7 c.722 C > G S241C). For these patients, the mutations can lead to a loss of function of p53 protein. The patient 30 presented a deletion in intron 7 at position In7 c14115del1. The patients included in this study were treated with the protocol established in the Sarah Hospital in May 1996 that consisted of a neoadjuvant chemotherapy, sur- gery and adjuvant chemotherapy. The drugs concentra- tions were estimated after a detailed clinical evaluation of patients. Parameters such as body surface, age, even- tual diseases, clinical and laboratorial conditions were taken in account before the beginning of treatment and between each cycle. The goal of neoadjuvant chemo- therapy is to shrink the cancer. The osteosarcoma neoad- juvant chemotherapy is composed of a high-dose of Me- totrexate (12 g/m2 of body surface area), cisplatin at 120 mg/ m2 of body surface area and Adriamycin at 75 mg/m2 of body surface area and the schedule was as follow: first and second weeks: Metotrexate, third week: Cisplatin and Adriamycin, 4th and 5th weeks: rest, during 3 cycles. The four mutated samples presented the tumor necro- sis rate under 95% (Table 3) that lead us to conclude that none of those samples presented a good response to the chemotherapy. Nevertheless, 10 other samples also pre- sented a tumor necrosis rate under 95%, indicating a bad response to the chemotherapy. Only 3 samples had a good response (>95%). However there is no correlation between TP53 mutation and response to chemotherapy, p = 1.2 (p > 0.05), that means the difference is not signifi- cant for this population. The response to the neoadjuvant chemotherapy is summarized in Table 2. There is a sig- Open Access IJCM 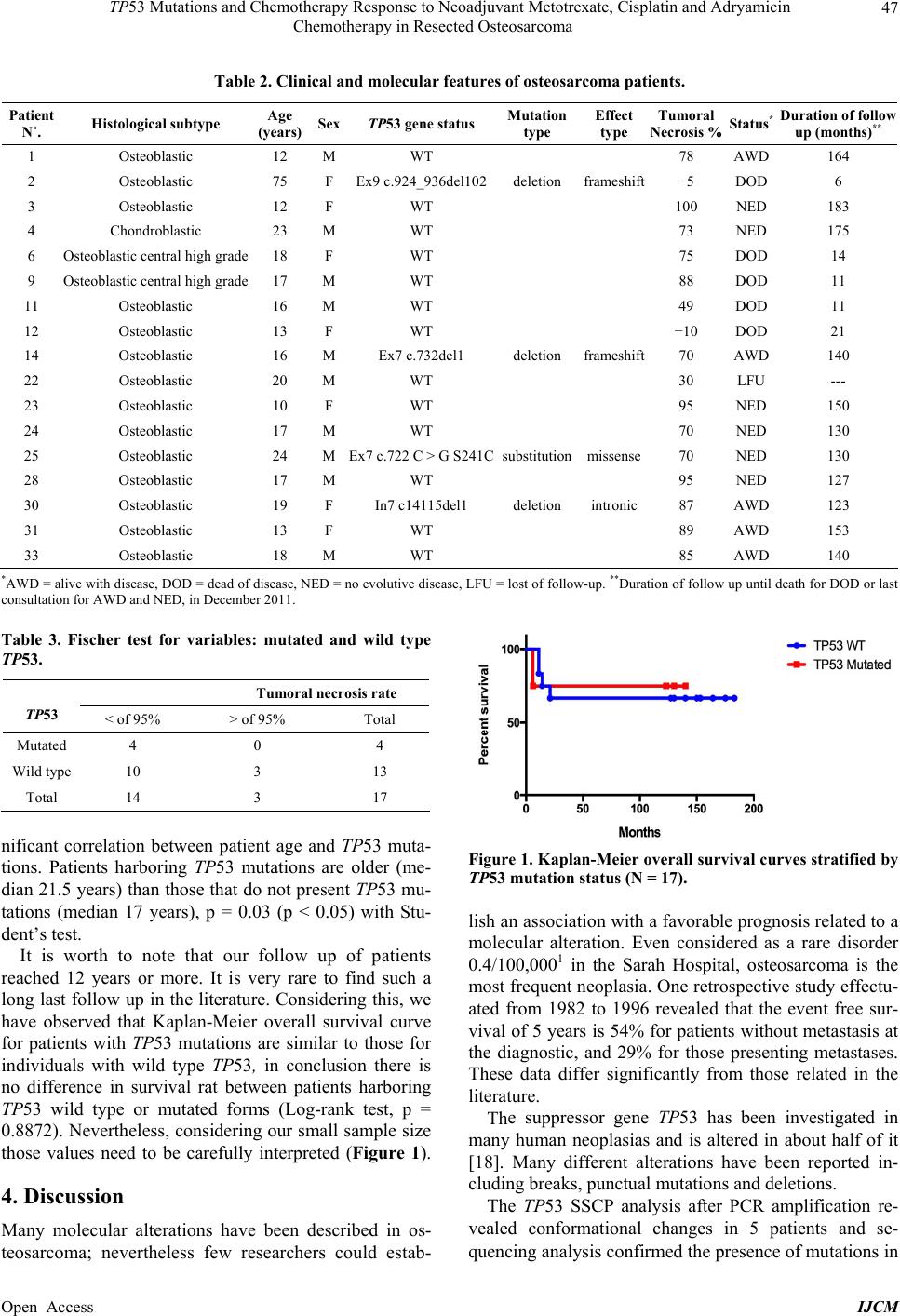 TP53 Mutations and Chemotherapy Response to Neoadjuvant Metotrexate, Cisplatin and Adryamicin Chemotherapy in Resected Osteosarcoma Open Access IJCM 47 Table 2. Clinical and molecular features of osteosarcoma patients. Patient N˚. Histological subtype Age (years) Sex TP53 gene status Mutation type Effect type Tumoral Necrosis % Status* Duration of follow up (months)** 1 Osteoblastic 12 M WT 78 AWD 164 2 Osteoblastic 75 F Ex9 c.924_936del102deletion frameshift−5 DOD 6 3 Osteoblastic 12 F WT 100 NED 183 4 Chondroblastic 23 M WT 73 NED 175 6 Osteoblastic central high grade 18 F WT 75 DOD 14 9 Osteoblastic central high grade 17 M WT 88 DOD 11 11 Osteoblastic 16 M WT 49 DOD 11 12 Osteoblastic 13 F WT −10 DOD 21 14 Osteoblastic 16 M Ex7 c.732del1 deletion frameshift70 AWD 140 22 Osteoblastic 20 M WT 30 LFU --- 23 Osteoblastic 10 F WT 95 NED 150 24 Osteoblastic 17 M WT 70 NED 130 25 Osteoblastic 24 M Ex7 c.722 C > G S241Csubstitutionmissense70 NED 130 28 Osteoblastic 17 M WT 95 NED 127 30 Osteoblastic 19 F In7 c14115del1 deletion intronic87 AWD 123 31 Osteoblastic 13 F WT 89 AWD 153 33 Osteoblastic 18 M WT 85 AWD 140 *AWD = alive with disease, DOD = dead of disease, NED = no evolutive disease, LFU = lost of follow-up. **Duration of follow up until death for DOD or last consultation for AWD and NED, in December 2011. Table 3. Fischer test for variables: mutated and wild type TP53. Tumoral necrosis rate TP53 < of 95% > of 95% Total Mutated 4 0 4 Wild type 10 3 13 Total 14 3 17 nificant correlation between patient age and TP53 muta- tions. Patients harboring TP53 mutations are older (me- dian 21.5 years) than those that do not present TP53 mu- tations (median 17 years), p = 0.03 (p < 0.05) with Stu- dent’s test. Figure 1. Kaplan-Meier overall survival curves stratified by TP53 mutation status (N = 17). lish an association with a favorable prognosis related to a molecular alteration. Even considered as a rare disorder 0.4/100,0001 in the Sarah Hospital, osteosarcoma is the most frequent neoplasia. One retrospective study effectu- ated from 1982 to 1996 revealed that the event free sur- vival of 5 years is 54% for patients without metastasis at the diagnostic, and 29% for those presenting metastases. These data differ significantly from those related in the literature. It is worth to note that our follow up of patients reached 12 years or more. It is very rare to find such a long last follow up in the literature. Considering this, we have observed that Kaplan-Meier overall survival curve for patients with TP53 mutations are similar to those for individuals with wild type TP53, in conclusion there is no difference in survival rat between patients harboring TP53 wild type or mutated forms (Log-rank test, p = 0.8872). Nevertheless, considering our small sample size those values need to be carefully interpreted (Figure 1). The suppressor gene TP53 has been investigated in many human neoplasias and is altered in about half of it [18]. Many different alterations have been reported in- cluding breaks, punctual mutations and deletions. 4. Discussion The TP53 SSCP analysis after PCR amplification re- vealed conformational changes in 5 patients and se- quencing analysis confirmed the presence of mutations in Many molecular alterations have been described in os- teosarcoma; nevertheless few researchers could estab-  TP53 Mutations and Chemotherapy Response to Neoadjuvant Metotrexate, Cisplatin and Adryamicin Chemotherapy in Resected Osteosarcoma 48 4 out 5. The SSCP is an efficient method for screening genetic variations, including single mutations. The sensi- bility allows detecting between 80% to 90% of the muta- tions in 200 - 400 bp length DNA fragments [19]. It is an affordable method for institutions that do not dispose of a high throughput sequencer. Our analysis of TP53 mutations extended from exons 5 to 9, a region that encompasses about 90% of the muta- tions [8]. We found mutations in 23.5% of our patients. In osteosarcoma, there are no consensual data concerning the TP53 mutations rate and the response to chemother- apy. In a study published in 1998, Goto and coworkers (1998) observed TP53 mutations in 41% of the samples, and among them, 15% were good responders while 64% of patients without TP53 mutations presented a good response, with a significant correlation (p < 0.05). They considered as good responders who presented tumoral necrosis rate >90% [20]. This criterion is different from those suggested by Rosen and coworkers (1993) and adopted by us in this study, in which the good responders present >95% of tumor necrosis rate [21]. Radig and coworkers (1996) analyzed 40 osteosarco- ma patients and found TP53 mutations in 19%. In an- other study, published in 1998, the same group found 15.7% of mutated osteosarcomas which do not correlate with tumor progression [22]. In a large study containing 272 primary osteosarcomas, TP53 mutation was obser- ved in 22% of patients without the correlation of tumor progression [23]. The mutation rate varying from 13.3% [24] (associated with disease free survival), 18% [25], and up to 26.5% [26] has been reported. In a study with a large cohort of 196 osteosarcoma patients, TP53 mu t a- tions were observed in 19.4% of osteosarcoma patients, and no correlation with p53 mutations and prognosis were observed, nevertheless patient age was the only factor that varied with TP53 gene status (p < 0.05) [27]. We confirmed this data. In our study, TP53 mutation status was not predictive of chemoresistance; suggesting that chemotherapy response is independent of the TP53 gene. Nevertheless, the mutation rate could be associated with patient age (p<0.05), as already described [26,27]. A very surprisingly result was observed when we compared the overall survival rate between osteosarcoma patients with TP53 wild type and mutated TP53. In our cohort, mutated patients have the same live span with patients harboring wild type TP53. It is interesting to note that the follow-up over 15 years, as we have des- cribed, is very rare to see in the literature. Nevertheless, the sample size of the current study was too small to draw definitive conclusions regarding the relationship between clinicopathologic features and p53 changes. Studying the expression of IDH1 and TP53 in os- teosarcoma, Hu and coworkers (2010), detected higher levels of IDH1 in the wild type than in TP53 mutant cells. IDH1 correlates with histological Rosen grade and me- tastasis negatively. TP53 correlates with histological Rosen grade, metastasis and overall survival in clinical osteosarcoma biopsies. Osteosarcoma patients with high IDH1 expression have a very high p53 expression, so IDH1 may correlate with p53 and is a candidate bio- marker for osteosarcoma [28]. Finally, a recent whole-genome sequencing study on osteosarcomas and chordomas describes a new process of cancer genome evolution termed chromothripsis. Rather than a multistep accumulation of unbalanced rearrange- ments, there is a single catastrophic genomic instability event that primarily affects a single chromosome [29]; maybe this could explain why TP53 mutations alone do not correlate to tumor progression. Moreover, to establish clinical implications of p53, it is also necessary to con- sider miRNA expression. They are generally deleted and/or disregulated in cancer. Certain microRNAs, (mir- 34a, b, and c) can be transcriptionally transactivated by p53 [30,31]. On the other hand, mir-125b, negatively re- gulates TP53 expression [32]. So, it seems that not only TP53 mutations do affect its downstream functions, but also change in the related miRNAs need to be considered. Furthermore, p53 isoform variants originated from alter- native splicing and the promoter usage can also have clinical implications [33]. In conclusion, the analysis of TP53 mutations status is the first step in the understand- ing of this important tumor suppressor gene in osteosar- comatumorigenesis. Further analysis, at the protein level and miRNA expression, needs to be performed aiming to establish a correlation between p53 and osteosarcoma clinical features. 5. Author’s Contribution LR: carried out the molecular genetic studies and par- ticipated in the design of the study and drafted the manuscript. CDB: participated in the design of the study, participated in the sequence alignment, performed the statistical analysis and coordination and draft the manu- script. MB conceived the study, and participated in its design and coordination. All authors read and approved the final manuscript. 6. Acknowledgments and Funding We are grateful to Dr Ricardo Karan Kalil for technical support and Dr Andréa Carla de Souza Góes for critical reading. The Sarah Network of Hospitals for Rehabilita- tion financed this work. REFERENCES [1] Dorfman, H.D. and Czerniak, B. (1995) Bone cancers. Open Access IJCM  TP53 Mutations and Chemotherapy Response to Neoadjuvant Metotrexate, Cisplatin and Adryamicin Chemotherapy in Resected Osteosarcoma 49 Cancer, 75, 203-210. http://dx.doi.org/10.1002/1097-0142(19950101)75:1+<20 3::AID-CNCR2820751308>3.0.CO;2-V [2] Dahlin, D.C. and Unni, K.K. (1977) Osteosarcoma of bone and its important recognizable varieties. The Ameri- can Journal of Surgical Pathology, 1, 61-72. http://dx.doi.org/10.1097/00000478-197701010-00007 [3] Marina, N., Gebhardt, M., Teot, L. and Gorlick, R. (2004) Biology and therapeutic advances for pediatric osteosar- coma. The Oncologist, 9, 422-441. [4] Group, E.G.W. and Saeter, G. (2007) Osteosarcoma: ESMO clinical recommendations for diagnosis, treatment and follow-up. Annals of Oncology: Official Journal of the European Society for Medical Oncology/ESMO, 18, ii77-78. [5] Simons, A., Schepens, M., Forus, A., Godager, L., van Asseldonk, M., Myklebost, O., et al. (1999) A novel chromosomal region of allelic loss, 4q32-q34, in human osteosarcomas revealed by representational difference analysis. Genes, Chromosomes & Cancer, 26, 115-124. http://dx.doi.org/10.1002/(SICI)1098-2264(199910)26:2< 115::AID-GCC3>3.0.CO;2-E [6] Baker, S.J., Markowitz, S., Fearon, E.R., Willson, J.K. and Vogelstein, B. (1990) Suppression of human colo- rectal carcinoma cell growth by wild-type p53. Science, 249, 912-915. http://dx.doi.org/10.1126/science.2144057 [7] Ozaki, T. and Nakagawara, A. (2011) p53: The attractive tumor suppressor in the cancer research field. Journal of Biomedicine & Biotechnology, 2011, 603925. [8] Vousden, K.H. and Lu, X. (2002) Live or let die: The cell’s response to p53. Nature reviews. Cancer, 2, 594- 604. http://dx.doi.org/10.1038/nrc864 [9] Petitjean, A., Mathe, E., Kato, S., Ishioka, C., Tavtigian, S.V., Hainaut, P., et al. (2007) Impact of mutant p53 functional properties on TP53 mutation patterns and tu- mor phenotype: Lessons from recent developments in the IARC TP53 database. Human Mutation, 28, 622-629. http://dx.doi.org/10.1002/humu.20495 [10] Olivier, M., Goldgar, D.E., Sodha, N., Ohgaki, H., Klei- hues, P., Hainaut, P. and Eeles, R.A. (2003) Li-Fraumeni and related syndromes: Correlation between tumor type, family structure, and TP53 genotype. Cancer Research, 63, 6643-6650. [11] Staples, O.D., Steele, R.J. and Lain, S. (2008) p53 as a therapeutic target. The Surgeon: Journal of the Royal Col- leges of Surgeons of Edinburgh and Ireland, 6, 240-243. http://dx.doi.org/10.1016/S1479-666X(08)80034-0 [12] Vousden, K.H. and Lane, D.P. (2007) p53 in health and disease. Nature reviews. Molecular Cell Biology, 8, 275- 283. http://dx.doi.org/10.1038/nrm2147 [13] Staib, F., Robles, A.I., Varticovski, L., Wang, X.W., Zeeberg, B.R., Sirotin, M., et al. (2005) The p53 tumor suppressor network is a key responder to microenviron- mental components of chronic inflammatory stress. Can- cer Research, 65, 10255-10264. http://dx.doi.org/10.1158/0008-5472.CAN-05-1714 [14] Olivier, M., Langerod, A., Carrieri, P., Bergh, J., Klaar, S., Eyfjord, J., et al. (2006) The clinical value of somatic TP53 gene mutations in 1794 patients with breast cancer. Clinical Cancer Research, 12, 1157-1167. http://dx.doi.org/10.1158/1078-0432.CCR-05-1029 [15] Perrone, F., Bossi, P., Cortelazzi, B., Locati, L., Quat- trone, P., Pierotti, M.A., et al. (2010) TP53 mutations and pathologic complete response to neoadjuvant cisplatin and fluorouracil chemotherapy in resected oral cavity squamous cell carcinoma. Journal of Clinical Oncology: Official Journal of the American Society of Clinical On- cology, 28, 761-766. http://dx.doi.org/10.1200/JCO.2009.22.4170 [16] Picci, P., Bacci, G., Campanacci, M., Gasparini, M., Pilo- tti, S., Cerasoli, S., et al. (1985) Histologic evaluation of necrosis in osteosarcoma induced by chemotherapy. Re- gional mapping of viable and nonviable tumor. Cancer, 56, 1515-1521. http://dx.doi.org/10.1002/1097-0142(19851001)56:7<151 5::AID-CNCR2820560707>3.0.CO;2-6 [17] Kurvinen, K., Hietanen, S., Syrjanen, K. and Syrjanen, S. (1995) Rapid and effective detection of mutations in the p53 gene using nonradioactive single-strand conformation polymorphism (SSCP) technique applied on PhastSystem. Journal of Virological Methods, 51, 43-53. http://dx.doi.org/10.1016/0166-0934(94)00099-3 [18] Masuda, H., Miller, C., Koeffler, H.P., Battifora, H. and Cline, M.J. (1987) Rearrangement of the p53 gene in hu- man osteogenic sarcomas. Proceedings of the National Academy of Sciences of the United States of America, 84, 7716-7719. http://dx.doi.org/10.1073/pnas.84.21.7716 [19] Orita, M., Iwahana, H., Kanazawa, H., Hayashi, K. and Sekiya, T. (1989) Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proceedings of the National Academy of Sciences of the United States of America, 86, 2766-2770. http://dx.doi.org/10.1073/pnas.86.8.2766 [20] Goto, A., Kanda, H., Ishikawa, Y., Matsumoto, S., Kawa- guchi, N., Machinami, R., et al. (1998) Association of loss of heterozygosity at the p53 locus with chemoresis- tance in osteosarcomas. Japanese Journal of Cancer Re- search: Gann, 89, 539-547. http://dx.doi.org/10.1111/j.1349-7006.1998.tb03295.x [21] Rosen, G., Loren, G.J., Brien, E.W., Ramana, L., Wax- man, A., Lowenbraun, S., et al. (1993) Serial thallium- 201 scintigraphy in osteosarcoma. Correlation with tumor necrosis after preoperative chemotherapy. Clinical Ortho- paedics and Related Research, 293, 302-306. [22] Radig, K., Schneider-Stock, R., Oda, Y., Neumann, W., Mittler, U. and Roessner, A. (1996) Mutation spectrum of p53 gene in highly malignant human osteosarcomas. Gen- eral & Diagnostic Pathology, 142, 25-32. [23] Gokgoz, N., Wunder, J.S., Mousses, S., Eskandarian, S., Bell, R.S. and Andrulis, I.L. (2001) Comparison of p53 mutations in patients with localized osteosarcoma and metastatic osteosarcoma. Cancer, 92, 2181-2189. http://dx.doi.org/10.1002/1097-0142(20011015)92:8<218 1::AID-CNCR1561>3.0.CO;2-3 [24] Tsuchiya, T., Sekine, K., Hinohara, S., Namiki, T., No- Open Access IJCM  TP53 Mutations and Chemotherapy Response to Neoadjuvant Metotrexate, Cisplatin and Adryamicin Chemotherapy in Resected Osteosarcoma Open Access IJCM 50 bori, T. and Kaneko, Y. (2000) Analysis of the p16INK4, p14ARF, p15, TP53, and MDM2 genes and their prog- nostic implications in osteosarcoma and Ewing sarcoma. Cancer Genetics and Cytogenetics, 120, 91-98. http://dx.doi.org/10.1016/S0165-4608(99)00255-1 [25] Patino-Garcia, A. and Sierrasesumaga, L. (1997) Analysis of the p16INK4 and TP53 tumor suppressor genes in bone sarcoma pediatric patients. Cancer Genetics and Cy- togenetics, 98, 50-55. http://dx.doi.org/10.1016/S0165-4608(96)00397-4 [26] Lonardo, F., Ueda, T., Huvos, A.G., Healey, J. and Ladanyi, M. (1997) p53 and MDM2 alterations in os- teosarcomas: Correlation with clinicopathologic features and proliferative rate. Cancer, 79, 1541-1547. http://dx.doi.org/10.1002/(SICI)1097-0142(19970415)79: 8<1541::AID-CNCR15>3.0.CO;2-Y [27] Wunder, J.S., Gokgoz, N., Parkes, R., Bull, S.B., Eskan- darian, S., Davis, A.M., et al. (2005) TP53 mutations and outcome in osteosarcoma: A prospective, multicenter study. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology, 23, 1483- 1490. http://dx.doi.org/10.1200/JCO.2005.04.074 [28] Hu, X., Yu, A.X., Qi, B.W., Fu, T., Wu, G., Zhou, M., et al. (2010) The expression and significance of IDH1 and p53 in osteosarcoma. Journal of Experimental & Clinical Cancer Research: CR, 29, 43. http://dx.doi.org/10.1186/1756-9966-29-43 [29] Stephens, P.J., Greenman, C.D., Fu, B., Yang, F., Bignell, G.R., Mudie, L.J., et al. (2011) Massive genomic rear- rangement acquired in a single catastrophic event during cancer development. Cell, 144, 27-40. http://dx.doi.org/10.1016/j.cell.2010.11.055 [30] Chang, T.C., Wentzel, E.A., Kent, O.A., Ramachandran, K., Mullendore, M., Lee, K.H., Feldmann, G., Yamakuchi, M., Ferlito, M., Lowenstein, C.J., et al. (2007) Transacti- vation of miR-34a by p53 broadly influences gene ex- pression and promotes apoptosis. Molecular Cell, 26, 745-752. http://dx.doi.org/10.1016/j.molcel.2007.05.010 [31] He, L., He, X., Lim, L.P., de Stanchina, E., Xuan, Z., Liang, Y., Xue, W., Zender, L., Magnus, J., Ridzon, D., et al. (2007) A microRNA component of the p53 tumour suppressor network. Nature, 447, 1130-1134. http://dx.doi.org/10.1038/nature05939 [32] Le, M.T., The, C., Shyh-Chang, N., Xie, H., Zhou, B., Korzh, V., Lodish, H.F. and Lim, B. (2009) Micro- RNA-125b is a novel negative regulator of p53. Genes & Development, 23, 862-876. http://dx.doi.org/10.1101/gad.1767609 [33] Bourdon, J.C., Fernandes, K., Murray-Zmijewski, F., Liu, G., Diot, A., Xirodimas, D.P., Saville, M.K. and Lane, D.P. (2005) p53 isoforms can regulate p53 transcriptional activity. Ge nes & Development, 19, 2122-2137. http://dx.doi.org/10.1101/gad.1339905 List of Abbreviations TP53: tumor protein p53; IARC International Agency for Research on Cancer; DNA: Deoxyribonucleic acid; HNSCC: head and neck squamous cell carcinoma; PCR: polymerase chain reaction; SSCP: Single Strand Conformation Polymorphism; Avh: accumulated volts per hour; IDH1: isocitrate dehydrogenase.
|