 Journal of Cancer Therapy, 2014, 5, 1-10 Published Online January 2014 (http://www.scirp.org/journal/jct) http://dx.doi.org/10.4236/jct.2014.51001 OPEN ACCESS JCT GSTP1 (Ile105Val) Gene Polymorphism: Risk and Treatment Response in Chronic Myeloid Leukemia Shereen Elhosei ny1, Mohamed El-Wakil2, Mohamed Fawzy3, Aya Abdel Rahman1 1Department of Clinical and Chemical Pathology, Beni Suef Teaching Hospital, Beni Suef University, Beni Suef, Egypt; 2Depart- ment of Clinical Oncology, Beni Suef Teaching Hospital, Beni Suef University, Beni Suef, Egypt; 3Department of Pediatric Oncolo- gy, National Cancer Institute, Cairo University, Egypt. Email: wakilmohamed171@gmail.com Received November 14th, 2013; revised December 10th, 2013; accepted December 18th, 2013 Copyright © 2014 Shereen Elhoseiny et al. This is an open access article distributed under the Creative Commons Attribution Li- cense, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In accordance of the Creative Commons Attribution License all Copyrights © 2014 are reserved for SCIRP and the owner of the intel- lectual property Shereen Elhoseiny et al. All Copyright © 2014 are guarded by law and by SCIRP as a guardian. ABSTRACT Background: Genetic variation influencing individual susceptibility to chemical carcinogens is one of the main factors leading to cancer development. The glutathione S-transferases (GSTs) are a family of enzymes belonging to phase II enzymes involved in detoxification of xenobiotics. A significant relationship is observed between the risk of developing cancer and genetic polymorphisms within GSTs. Methods: In this study, we investigated the influence of inherited GSTP1 (Ile105Val) gene polymorphism on the susceptibility to CML in Egypt in 40 CML patients (20 children and 20 adults), together with 40 healthy controls using a [PCR-RFLP] assay. Results: We found that the mutant type (IIe/Val, Val/Val) was significantly higher in CML patients (67.5%) compared to controls (35%) (p = 0.004); [odds ratio 3.9; 95% CI: 1.5 - 9.7]. The mutant type was associated with more ad- vanced phases in disease and with both worse hematological and cytogenetic responses when compared to the wild type (p = 0.03, p = 0.05, and p < 0.001, respectively). Conclusion: GSTP1 (Ile105Val) gene polymorphism conferred a significant association with increased risk of CML and is associated with worse prognosis. Further studies on the functional consequences of this genetic polymorphism would pave the way to declare its role in the pathogenesis of CML or as a p oss ible predictor for resp ons e to ther apy. KEYWORDS CML; GSTs; GSTP1 (Ile105Val) 1. Introduction Chronic myeloid leukemia (CML) is a clo nal myelopro- liferative disease resulting from neoplastic transforma- tion of multipotent stem cell. The disease is characterized by high levels of leukocytes, splenomegaly, myeloid hyperplasia in bone marrow and high levels of mature myeloid cells in peripheral blood [1]. Although clinical and biological aspects are well documented, little is known about individual susceptibility to this disease [2]. Exposure to endogenous or exogenous toxic substances can lead to genetic alterations and hence increased sus- ceptibility to cancer [3]. It is claimed that cytotoxic and genotoxic environmental agents (especially ionization, radiation and similar factors) may increase the risk of CML development [ 4]. Xenobiotic metabolizing en- zymes (XMEs) constitute on e of the first lines of defense against environmental chemicals. They play a central role in the metabolism, elimination, and detoxification of xenobiotics or exogenous compounds introduced into the body [ 5]. Cells have developed an effective mechanism to prevent accumulation of damaging xenobiotics by way of their elimination catalyzed by multiple enzy me system. The enzymes of the multiple enzyme system are classi- fied in two categories namely Phase I and Phase II. Phase I enzymes like Cytochrome P450 can activate the car- cinogens directly and produce active metabolites while Phase II enzymes like glutathione-S-transferase (GSTs) can detoxify and process the activated metabolites for final breakdow n [6].  GSTP1 (Ile105Val) Gene Polymorphism: Risk and Treatment Response in Chronic Myeloid Leukemia OPEN ACCESS JCT The GSTs are a family of enzymes belonging to phase II enzymes involved in detoxificati on of xe nobiotics (car- cinogens, pesticides, antitumor agents & environmental pollutants). Hence GSTs play a significant role in the cellular defense. GSTs fall into two distinct super fami- lies: membrane bound microsomal GSTs and the soluble or cytosolic GSTs. The cytosolic glutathione S-tr an s- ferase were classified into eight classes on the basis of sequence diversity and designated as Alpha (α), Mu (μ), Pi (л), Kappa (K), Theta (θ), Omega (O), Sigma (ε) and Zeta (Z) [7]. These cytosolic enzymes play a major role in the detoxification of a broad range of compounds, in- cluding xenobiotics, pesticides, environmental carcino- gens, PAH, and some chemotherapeutic drugs (including alkylating agents, Doxorubicin, and Vincristein) [8]. Glu- tathione S-transferase P1 (GSTP1) belongs to the pi class gene family, located on chromosome 11q13 [9]. It spans 2.48 kb of DNA and comprises 7 exons that encode for cytosolic GST enzyme. GSTP1 is considered as a major antioxidant present in both the epidermis and the dermis, overexpressed in a variety of preneoplastic and neoplas- tic tissues [10]. In some cancer models, GSTP1 expres- sion was considered as pre neoplastic tumor marker. In- creased levels of GSTP1 in tumors might account for part of the inherent drug resistance, which was observed in many tumors suggesting its role in cancer etiology and therapy [11]. GST polymorphisms may alter the ability of enzymes to metabolize the chemical carcinogens and mutagens. It had been suggested that these differences in the ability to metabolize carcinogens and mutagens might influence the susceptibility to cancer [4]. The first polymorphism identified was an A-G polymorphism at nucleotide 313 in exon 5 of GST P1 gene which leads to an amino acid substitution of isoleucine (IIe) by valine (Val) at 105 amino acid position (I le105Val). This substitution results in three GSTP1 genotypes: they ar e isoleucine/isoleucine (Ile/Ile) homozygous wild type, isoleucine/valine (Ile/Va l) heterozygote and valine/valine (Val/Val) homozygous variant. GSTP1 codon 105 polymorphism might play an important role in leukemogenesis, as it potentially alters protein function, diminishing its d etoxification ability f or certain mutagens and carcinogens, which could result in increased DNA damage and mutation, and a greater risk of develo ping cance r [12]. To our knowledge, the GSTP1 (Ile105Val) gene has never been studied in Egyptian CML patients. Thus, this study was done to investigate the influence of inherited genetic polymorphism of GSTP1 (Ile105Val) on the sus- ceptibility to CML in Egyptian pediatric and adult pa- tients evaluating its impact on the response to therapy. 2. Patients and Methods Included pediatric and adult patients underwent cross sectional evaluation as regards study parameters while forty unrelated healthy individuals were served as control group. Patients: In the present study 40 Egyptian CML patients were included; 20 children and 20 adults at different phases of disease; newly diagnosed or received treatment. Patients were recruited from the Pediatric Oncology Department, National Cancer Institute (NCI), Cairo University and from Beni Suef University Hospital. Data confidentiality was preserved according to the Revised Helsinki De- claration of Bioethics (2008) [13]. Patients were subjected to full history taking and tho- rough clinical examination. In addition, laboratory in- vestigations as complete blood count (CBC), liver and kidney functions, serum uric acid, LDH and coagulation profile were reviewed. Diagnosis of CML was based on morphologic findings from Giemsa stained smears of peripheral blood sample, cytochemical stains criteria such as LAP score, Philadelphia chromosome detection by conventional cytogenetic study and BCR ABL fusion gene detected by FISH. Treatment and phase of disease: Medical records of all patients were reviewed as re- gards clinical data, diagnostic laboratory information, treatment received, and disease status. Patients in chronic phase (CML-CP) received debulking hydroxyurea and imatinib (STI571/Glivec) tyrosine kinase inhibitor (TKI) at 340 mg/m2/day in children and 400 mg/day for adults. One pediatric patient in chronic phase underwent allo- geneic bone marrow transplantation (BMT). Patients progressed to myeloid blastic crisis within the course of their therapy were candidates for AML-like treatment. Children received ADE (Ara-c, Daunorubicin, Etoposide) as induction therapy followed by postremission consoli- dation in form of MidAC (Mitoxantrone, Ara-c). Adult patient was candidate for high dose ara-c based regimens. Adults in acceleration phase received escalated imatinib dose of 600 - 800 mg daily [14,15]. Response: Patients were evaluated according to hematologic and cytogenetic response where • Complete hematological response (CHR) was de- fined as a WBC count <10 × 109/L, a platelet count <450 × 109/L, basophils <5%, no immature cells (blasts, promyelocytes, myelocytes) in the peripheral blood, and disappearance of all signs and symptoms related to leukemia. • Cytogenetic response was expressed in terms of the ratio of number of Ph+ metaphases in bone marrow divided by initial number of Ph+ metaphases and ca- tegorized as follows: 1) Complete response: 0% Ph+ cells; 2) Partial re- sponse: 1% - 35% Ph+ cells, 3) Minor response: 36% - 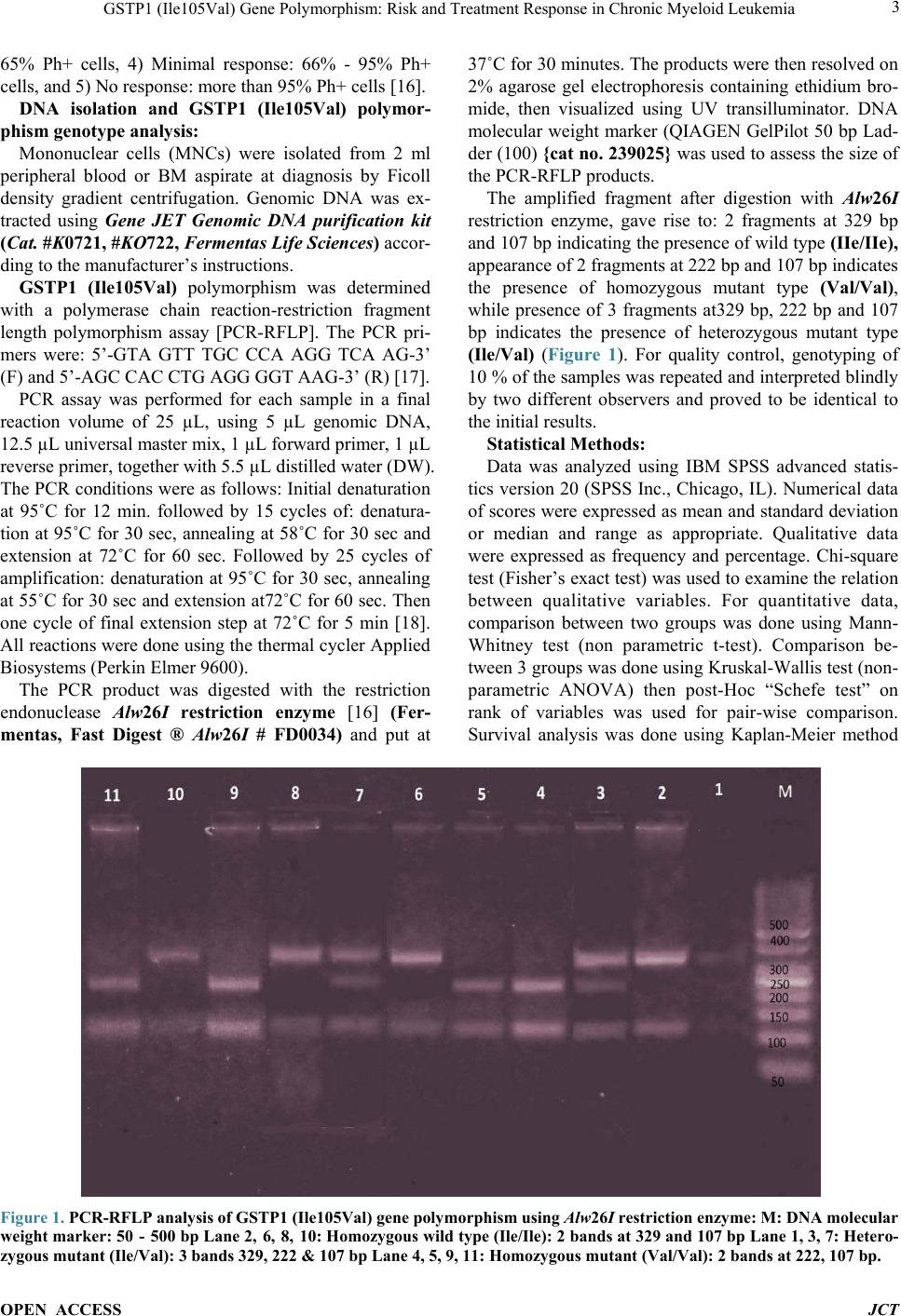 GSTP1 (Ile105Val) Gene Polymorphism: Risk and Treatment Response in Chronic Myeloid Leukemia OPEN ACCESS JCT 65% Ph+ cells, 4) Minimal response: 66% - 95% Ph+ cells, and 5) No response: more than 95% Ph+ cells [16]. DNA isolation and GSTP1 (Ile105Val) polymor- phism genotype analysis: Mononuclear cells (MNCs) were isolated from 2 ml peripheral blood or BM aspirate at diagnosis by Ficoll density gradient centrifugation. Genomic DNA was ex- tracted using Gene JET Genomic DNA purification kit (Cat. #K0721, #KO722, Fermentas Life Sciences) accor- ding to the manufacturer’s instructions. GSTP1 (Ile105Val) polymorphism was determined with a polymerase chain reaction-restriction fragment length polymorphism assay [PCR-RFLP]. The PCR pri- mers were: 5’-GTA GTT TGC CCA AGG TCA AG-3’ (F) and 5’-AGC CAC CTG AG G GGT AAG-3’ (R) [17]. PCR assay was performed for each sample in a final reaction volume of 25 µL, using 5 µL genomic DNA, 12.5 µL universal master mix, 1 µL forward primer, 1 µL reverse primer, together with 5.5 µL distilled water (DW). The PCR conditions w ere as follows: Initial denaturation at 95˚C for 12 min. followed by 15 cycles of: denatura- tion at 95˚C for 30 sec, annealing at 58˚C for 30 sec and extension at 72˚C for 60 sec. Followed by 25 cycles of amplification: denaturation at 95˚C for 30 sec, annealing at 55˚C for 30 sec and extension at72˚C for 60 se c. Then one cycle of final extension step at 72˚C for 5 min [18]. All reactions were done using the thermal cycler App lied Biosystems (Perkin Elmer 9600). The PCR product was digested with the restriction endonuclease Alw26I restriction enzyme [16] (Fer- mentas, Fast Digest ® Alw26I # FD0034) and put at 37˚C for 30 minutes. The products were then resolved on 2% agarose gel electrophoresis containing ethidium bro- mide, then visualized using UV transilluminator. DNA molecular weight marker (QIAGEN GelPilot 50 bp Lad- der (100) {ca t no. 239 025} was used to assess the size of the PCR-RFLP products. The amplified fragment after digestion with Alw26I restriction enzyme, gave rise to: 2 fragments at 329 bp and 107 bp indicating the presence of wild type (IIe/IIe), app earan ce of 2 fragments at 222 bp and 107 bp indicates the presence of homozygous mutant type (Val/Val), while presence of 3 fragments at329 bp, 222 bp and 107 bp indicates the presence of heterozygous mutant type (Ile/Val) (Figure 1). For quality control, genotyping of 10 % of the samples was repeated and interpreted blindly by two different observers and proved to be identical to the initial results. Statistical Methods: Data was analyzed using IBM SPSS advanced statis- tics version 20 (SPSS Inc., Chicago, IL). Numerical data of scores were expressed as mean and standard deviation or median and range as appropriate. Qualitative data were expressed as frequency and percentage. Chi-square test (Fisher’s exact test) was used to examine the relation between qualitative variables. For quantitative data, comparison between two groups was done using Mann- Whitney test (non parametric t-test). Comparison be- tween 3 groups was done using Kruskal-Wallis test (non- parametric ANOVA) then post-Hoc “Schefe test” on rank of variables was used for pair-wise comparison. Survival analysis was done using Kaplan-Meier method Figure 1. PCR-R FLP analysis of GSTP1 (Ile105Val) gene polymorphism using Alw26I restriction enzyme: M: DNA molecular weight marker: 50 - 500 bp Lane 2, 6, 8, 10: Homozygous wild type (Ile/Ile): 2 bands at 329 and 107 bp Lane 1, 3, 7: Hetero- zygous mutant (Ile/Val): 3 bands 329, 222 & 107 bp Lane 4, 5, 9, 11: Homozygous mutant (Val/Val): 2 bands at 222, 107 bp. 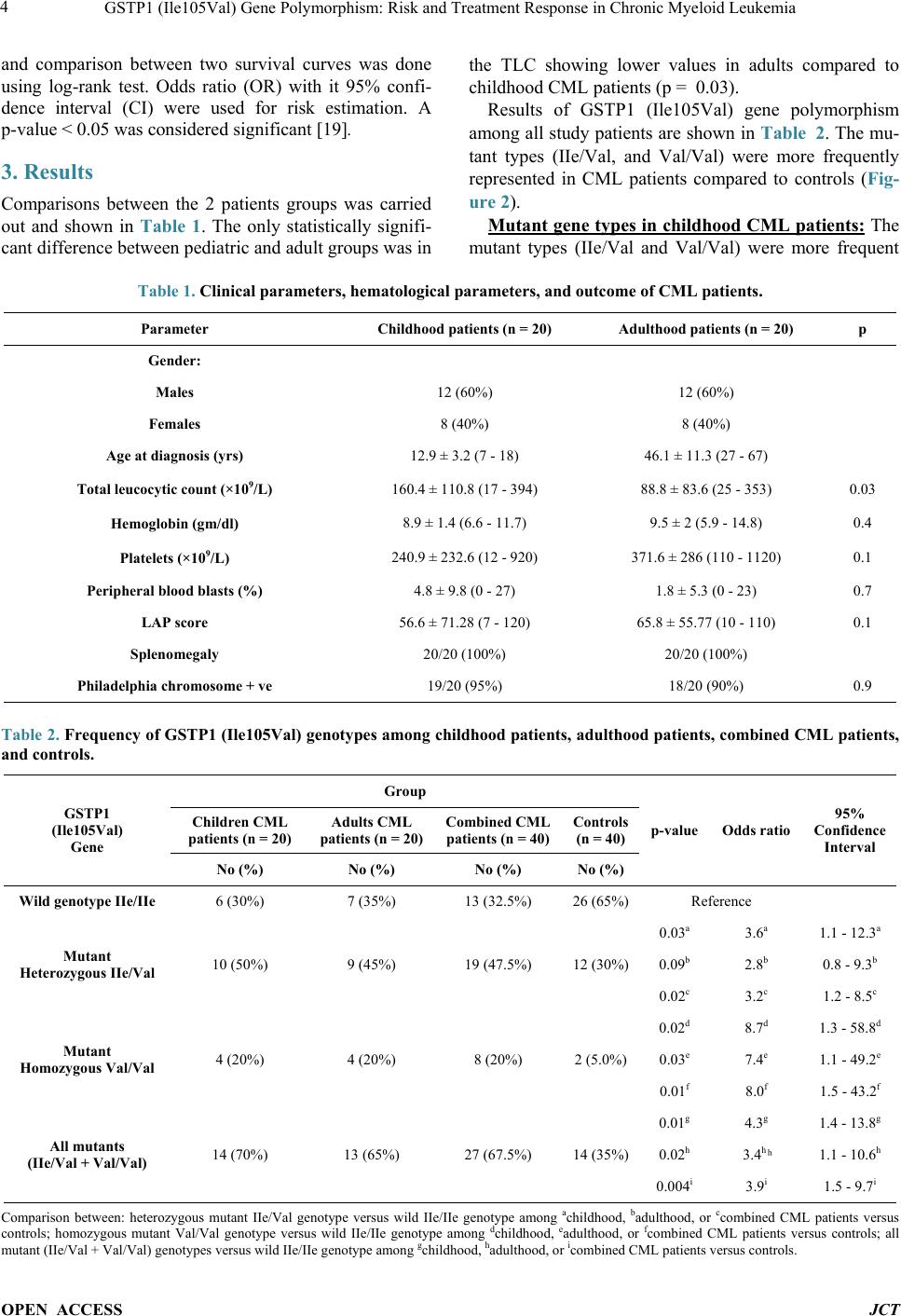 GSTP1 (Ile105Val) Gene Polymorphism: Risk and Treatment Response in Chronic Myeloid Leukemia OPEN ACCESS JCT and comparison between two survival curves was done using log-rank test. Odds ratio (OR) with it 95% confi- dence interval (CI) were used for risk estimation. A p-value < 0.05 was considered significant [19]. 3. Results Comparisons between the 2 patients groups was carried out and shown in Table 1. The only statistically signifi- cant difference between pediatric and adult groups was in the TLC showing lower values in adults compared to childhood CML patients (p = 0.03). Results of GSTP1 (Ile105Val) gene polymorphism among all study patients are shown in Table 2. The mu- tant types (IIe/Val, and Val/Val) were more frequently represented in CML patients compared to controls (Fig- ure 2). UMutant gene types in childhood CML patients:U The mutant types (IIe/Val and Val/Val) were more frequent Table 1. Clinical parameters, hematological parameters, and outcome of CML patients. p Adulthood patients (n = 20) Childhood patients (n = 20) Parameter Gender: 12 (60%) 12 (60%) Males 8 (40%) 8 (40%) Fema l e s 46.1 ± 11.3 (27 - 67) 12.9 ± 3.2 (7 - 18) Age at diagnosis (yrs) 0.03 88.8 ± 83.6 (25 - 353) 160.4 ± 110.8 (17 - 394) Total leucocytic count (×109/L) 0.4 9.5 ± 2 (5.9 - 14.8) 8.9 ± 1.4 (6.6 - 11.7) Hemoglobin (gm/dl) 0.1 371.6 ± 286 (110 - 1120) 240.9 ± 232.6 (12 - 920) Platelets (×109/L) 0.7 1.8 ± 5.3 (0 - 23) 4.8 ± 9.8 (0 - 27) Peripheral blood blasts (%) 0.1 65.8 ± 55.77 (10 - 110) 56.6 ± 71.28 (7 - 120) LAP score 20/20 (100%) 20/20 (100%) Splenomegaly 0.9 18/20 (90%) 19/20 (95%) Philadelphia chromosome + ve Table 2. Frequency of GSTP1 (Ile105Val) genotypes among childhood patients, adulthood patients, combined CML patients, and controls. GSTP1 (Ile105Val) Gene Group p-value Odds ratio 95% Confidence Interval Children CML patients (n = 20) Adults CML patients (n = 20) Com bined CML patients (n = 40) Controls (n = 40) No (%) No (%) No (%) No (%) Wild genotype IIe/IIe 6 (30%) 7 (35%) 13 (32.5%) 26 (65%) Reference Mutant Heterozygous IIe/Val 10 (50%) 9 (45%) 19 (47.5%) 12 (30%) 0.03a 3.6a 1.1 - 12.3a 0.09b 2.8b 0.8 - 9.3b 0.02c 3.2c 1.2 - 8.5c Mutant Homozygous Val/Val 4 (20%) 4 (20%) 8 (20%) 2 (5.0%) 0.02d 8.7d 1.3 - 58.8d 0.03e 7.4e 1.1 - 49.2e 0.01f 8.0f 1.5 - 43.2f All mutants (IIe/Val + Val/Val) 14 (70%) 13 (65%) 27 (67.5%) 14 (35%) 0.01g 4.3g 1.4 - 13.8g 0.02h 3.4h ͪ 1.1 - 10.6h 0.004i 3.9i 1.5 - 9.7i Comparison between: heterozygous mutant IIe/Val genotype versus wild IIe/IIe genotype among achildhood, badulthood, or ccombined CML patients versus controls; homozygous mutant Val/Val genotype versus wild IIe/IIe genotype among dchildhood, eadulthood, or fcombined CML patients versus controls; all mutant (IIe/Val + Val/Val) genotypes versus wild IIe/IIe genotype among gchildhood, ha dultho od, or icombined CML patients versus controls.  GSTP1 (Ile105Val) Gene Polymorphism: Risk and Treatment Response in Chronic Myeloid Leukemia OPEN ACCESS JCT Figure 2. GSTP1 (Ile105Val) gene polymorphism in childhood CML patients, adulthood CML patients and control groups. among childhood CML patients (70%) compared to con- trols (35%), (p = 0.01). Calculated odds ratio revealed fourfold increased risk of CML with mutant types. The homozygous mutant type (Val/Val) was 20% in CML pediatric patients compared to only 5% among controls (p = 0.02), with calculated odds ratio revealed eightfold increased risk of CML. On the other hand, the heterozy- gous mutant type (IIe/Val) found in 50% of pediatric patients compared to 30% in controls (p = 0.03) with calculated odds ratio revealed almost fourfold increased risk of CML (Table 2 an d Figure 2). Mutant gene types in adulthood CML patients: Table 2 The mutant types (IIe/Val and Val/Val) were more frequent among adult CML patients (65%) compared to controls (35%), (p = 0.02). Calculated odds ratio revealed more than threefold increased risk of CML. The homozygous mutant type (Val/Val) like in childhood group, was 20% in CML patients compared to only 5% in controls (p = 0.03), and calculated odds ratio revealed more than se- venfold increased risk of CML. As for the heterozygous mutant type (IIe/Val), although it was more frequently represented among patients (45%) compared to 30% in controls, yet the difference was not of statistical signi- ficance (p = 0.09). However, calculated odds ratio re- vealed almost threefold increased risk of CML for hete- rozygous mutants ( and Figure 2). Characteristics and outcome of childhood (Table 3 ) and adulthood (Table 4) CML patients as regards GSTP1 (Ile105Val) gene polymorphism: In patients with CML-CP, mutant gene types wer e found in 11/16 of children (68.8%) versus 58.8% in adults. Among other phases of disease, 3/4 (75%) of childhood patients had blastic crisis during their treatment course found to be harboring the mutant type, compared to only 1/4 (25%) harboring the wild type. Another 2 adulthood patients with accelerated phase of disease were harboring the mutant type (100%) as well, whereas the only adult pa- tient with blastic crisis was harboring the mutant type (100%). GSTP1 (Ile105Val) gene polymorphism pattern in different CML Phases and its impact on response among combined study patients (Table 5): Among all study patients of different age groups there was a sta- tistically significant difference between the wild gene type IIe/IIe and mutant types (IIe/Val or Val/Val) as re- gards CML phases. Both accelerated phase and blastic crisis were more common in patients harboring the mu- tant type (57.1% homozygous mutant, 28.6% heterozy- gous mutant, and only 14.3% with wild type; p = 0.03). The mutant type was associated with poorer hematolo- gical response as all p atients who didn’t achieve CHR (n = 6) were harboring the mutant type; 3 homozygous, and 3 heterozygous mutants (p = 0.05). Again, the mutant type was also associated with poorer cytogenetic response. While 9/9 patients (100%) with minimal cytogenetic response had the mutant type; the homozygous mutant gene found in 6/9 (66.7%) was more com mo n in comparison to heterozygous mutant gene pattern found in 3/9 (33.3%) of those patients. On the other hand, complete cytogenetic response was more in patients harboring the wild type (75%) compared to the heterozygous (25%), and homozygous (0%) mutant types, (p < 0.001). Allele frequency among combined study patients versus controls (Table 6 and Figure 3): The Val allele was significantly higher in CML patients (43.7%) when results of all patients were compared collectively to con- trols (20% ) (p = 0.001). \ 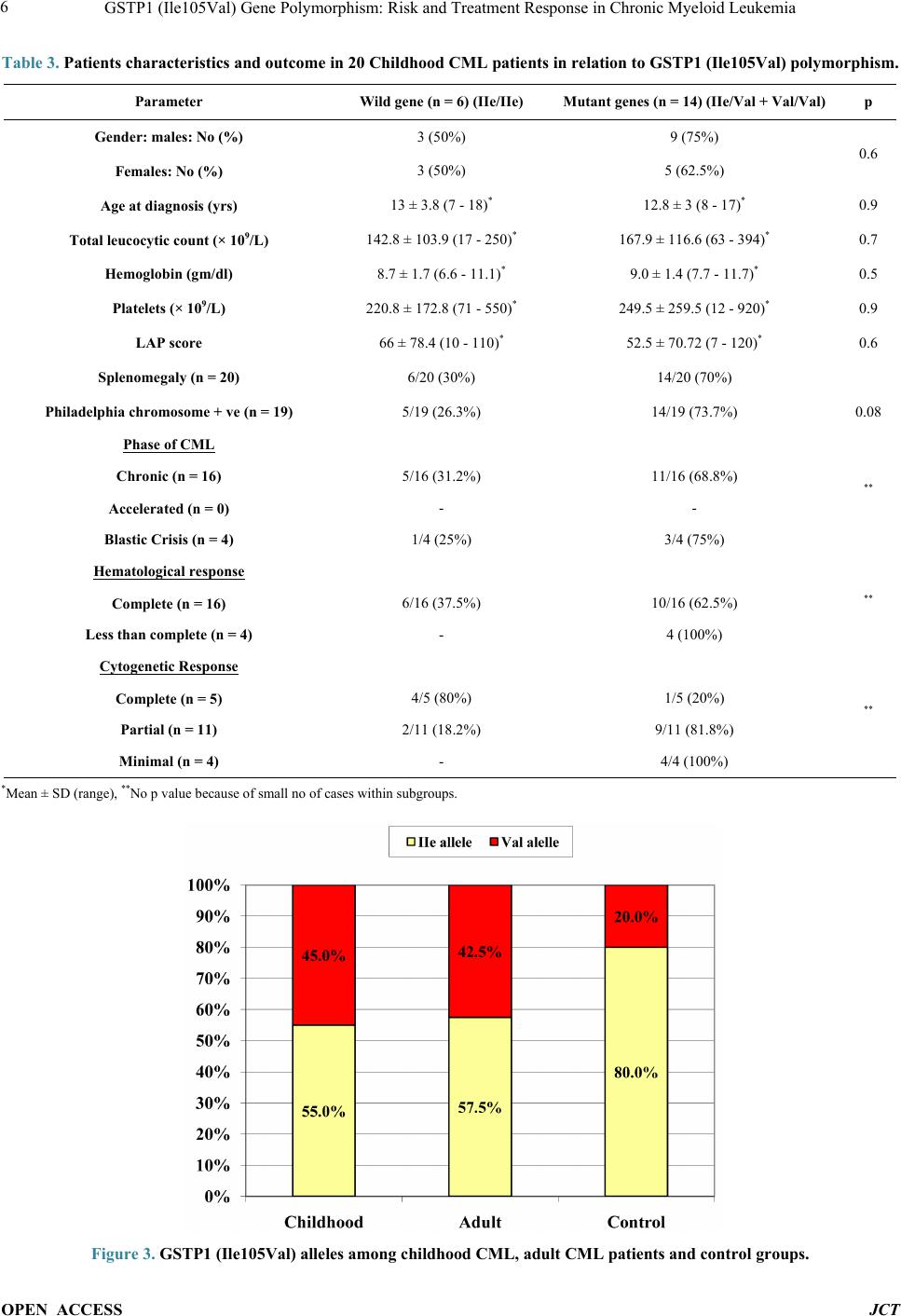 GSTP1 (Ile105Val) Gene Polymorphism: Risk and Treatment Response in Chronic Myeloid Leukemia OPEN ACCESS JCT Table 3. Patients characteristics and outcome in 20 Childhood CML patients in relation to GSTP1 (Ile105Val) polymorphism. p Mutant genes (n = 14) (IIe/Val + Val/Val) Wild gene (n = 6) (IIe/IIe) Parameter 0.6 9 (75%) 3 (50%) Gender: males: No (%) 5 (62.5%) 3 (50%) Females: No (%) 0.9 12.8 ± 3 (8 - 17)* 13 ± 3.8 (7 - 18)* Age at diagnosis (yrs) 0.7 167.9 ± 116.6 (63 - 394)* 142.8 ± 103.9 (17 - 250)* Total leucocytic count (× 109/L) 0.5 9.0 ± 1.4 (7.7 - 11.7)* 8.7 ± 1.7 (6.6 - 11.1)* Hemoglobin (gm/dl) 0.9 249.5 ± 259.5 (12 - 920)* 220.8 ± 172.8 (71 - 550)* Platelets (× 109/L) 0.6 52.5 ± 70.72 (7 - 120)* 66 ± 78.4 (10 - 110)* LAP score 14/20 (70%) 6/20 (30%) Splenomegaly (n = 20) 0.08 14/19 (73.7%) 5/19 (26.3%) Philadelphia chromosome + ve (n = 19) ** 11/16 (68.8%) Phase of CML 5/16 (31.2%) Chronic (n = 16) - - Accelerated (n = 0) 3/4 (75%) 1/4 (25%) Blastic Crisis (n = 4) ** 10/16 (62.5%) Hematological response 6/16 (37.5%) Complete (n = 16) 4 (100%) - Less than complete (n = 4) ** UCytogenetic Response 1/5 (20%) 4/5 (80%) Complete (n = 5) 9/11 (81.8%) 2/11 (18.2%) Partial (n = 11) 4/4 (100%) - Minimal (n = 4) *Mean ± SD (range), **No p value because of small no of cases within subgroups. Figure 3. GSTP1 (Ile105Val) alleles among childhood CML, adult CML patients and control groups. 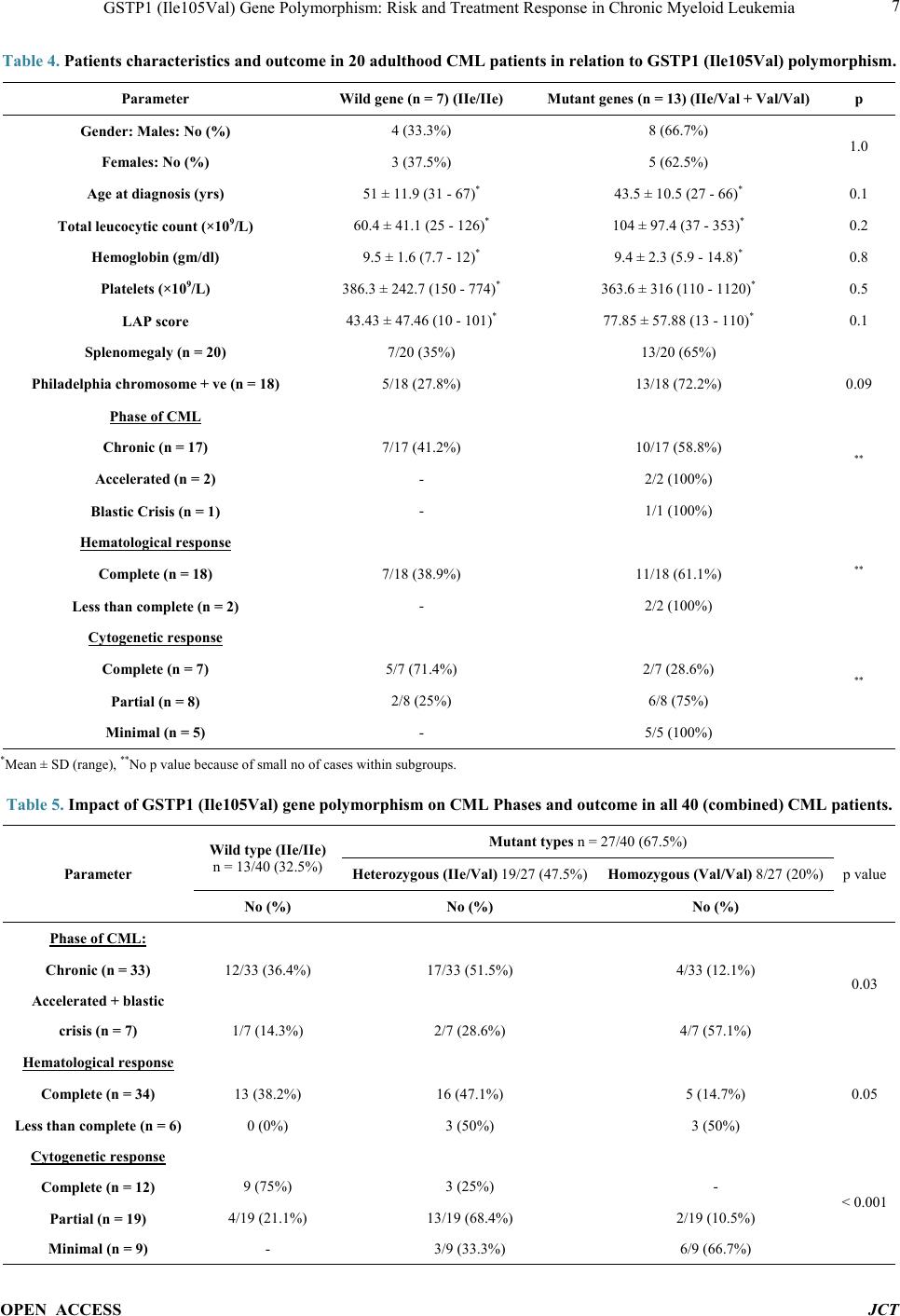 GSTP1 (Ile105Val) Gene Polymorphism: Risk and Treatment Response in Chronic Myeloid Leukemia OPEN ACCESS JCT Table 4. Patients characteristics and outcome in 20 adulthood CML patients in relation to GSTP1 (Ile105Val) polymorphism. p Mutant genes (n = 13) (IIe/Val + Val/Val) Wild gene (n = 7) (IIe/IIe) Parameter 1.0 8 (66.7%) 4 (33.3%) Gender: Males: No (%) 5 (62.5%) 3 (37.5%) Females: No (%) 0.1 43.5 ± 10.5 (27 - 66)* 51 ± 11.9 (31 - 67)* Age at diagnosis (yrs) 0.2 104 ± 97.4 (37 - 353)* 60.4 ± 41.1 (25 - 126)* Total leucocytic count (×109/L) 0.8 9.4 ± 2.3 (5.9 - 14.8)* 9.5 ± 1.6 (7.7 - 12)* Hemoglobin (gm/dl) 0.5 363.6 ± 316 (110 - 1120)* 386.3 ± 242.7 (150 - 774)* Platelets (×109/L) 0.1 77.85 ± 57.88 (13 - 110)* 43.43 ± 47.46 (10 - 101)* LAP score 13/20 (65%) 7/20 (35%) Splenomegaly (n = 20) 0.09 13/18 (72.2%) 5/18 (27.8%) Philadelphia chromosome + ve (n = 18) ** 10/17 (58.8%) Phase of CML 7/17 (41.2%) Chronic (n = 17) 2/2 (100%) - Accelerated (n = 2) 1/1 (100%) - Blastic Crisis (n = 1) ** 11/18 (61.1%) Hematological response 7/18 (38.9%) Complete (n = 18) 2/2 (100%) - Less than complete (n = 2) ** UCytogenetic response 2/7 (28.6%) 5/7 (71.4%) Complete (n = 7) 6/8 (75%) 2/8 (25%) Partial (n = 8) 5/5 (100%) - Minimal (n = 5) *Mean ± SD (range), **No p value because of small no of cases within subgroups. Table 5. Impact of GSTP1 (Ile105Val) gene polymorphism on CML Phases and outcome in all 40 (combined) CML patients. p value Mutant types n = 27/40 (67.5%) Wild type (IIe/IIe) n = 13/40 (32.5%) Parameter Homozygous (Val/Val) 8/27 (20%) Heterozygous (IIe/Val) 19/27 (47.5%) No (%) No (%) No (%) 0.03 UPhase of CML: 4/33 (12.1%) 17/33 (51.5%) 12/33 (36.4%) Chronic (n = 33) Accelerated + blastic 4/7 (57.1%) 2/7 (28.6%) 1/7 (14.3%) crisis (n = 7) 0.05 UHematological response 5 (14.7%) 16 (47.1%) 13 (38.2%) Complete (n = 34) 3 (50%) 3 (50%) 0 (0%) Less than complete (n = 6) < 0.001 UCytogenetic response - 3 (25%) 9 (75%) Complete (n = 12) 2/19 (10.5%) 13/19 (68.4%) 4/19 (21.1%) Partial (n = 19) 6/9 (66.7%) 3/9 (33.3%) - Minimal (n = 9)  GSTP1 (Ile105Val) Gene Polymorphism: Risk and Treatment Response in Chronic Myeloid Leukemia OPEN ACCESS JCT Table 6. Allele frequency in different CML patients and controls. (95% confidence interval) Odds ratio p value GSTP1 allele Group Val Allele Ile Allele 20% 80% Controls (n = 40) 1.428 - 7.502a 3.273a 45% 0.004a 55% Childhood CML patients (n = 20) 1.286 - 6.797b 2.957b 42.5% 0.009b 57.5% Adulthood CML patients (n = 20) 1.539c - 6.288c 3.111c 43.7% 0.001c 56.3% Combined CML patients (n = 40) 0.373 - 2.186d 0.903d 0.8d Childhood versus adulthood CML patients Allele frequency among aChildhood, bAdulthood or, cAll (combined) CML patients versus controls. dAllele frequency among childhood CML versus adult patients. 4. Discussion CML is a myeloproliferative disorder but definite me- chanism leading to this carcinogenesis is not completely understood ye t [6]. Genetic susceptibility studies of CML may serve to identify populations at risk and clarify im- portant disease mechanisms. Genetic variants within genes that encode enzymes involved with metabolism such as GST have been shown to increase the likelihood of developing various forms of cancers [20]. An associa- tion between the polymorphic forms of the XMEs and the altered risk to various cancers including CML was reported [3]. Several studies investigated the relationship between GST polymorphisms and acute leukemia [12,21]. However, there is very little information on the role of GST polymorphisms in CML development. To the best of our knowledge, Taspinar et al. 2008, studied the GSTM1 and GSTT1 polymorphisms [4], while in two other studies GSTP1 (Ile105Val) gene polymorphism was studied in CML patients [17, 22]. In the current study, frequency of the mutant gene types (IIe/Val and Val/Val) was significantly higher in CML patients compared to controls (respectively 67.5% v 35%; p = 0.004), with fourfold increased risk of CML. This is in accordance with some other investigators re- ported that there was an association between the GSTP1 (Ile105Val) polymorphism and the occurrence of CML [17]. On contrary, Karkucah et al. 2012 didn’t find any statistically significant difference between CML patients and the control group as regards the GSTP1 (Ile105Val) gene polymorphism [22]. It was also reported that heterozygote mutant type Ile/ Val was elevated in a group of patients below 20 years when compared to patients in higher age groups. This agrees with our findings as the heterozygous mutant type was 50% in childhood compared to 45% in adulthood patients. Authors suggested that the presence of valine allele confers increased risk to develop CML at early age. This was attributed to the reduced rate of detoxification of metabolites derived due to UVR-derived oxidative stress and other environmental carcinogens. We also found that the homozygous mutant type (Val/Val) was significantly higher in our CML patients (20%) com- pared to controls (5%). This was in agreement to same investigators data but at different frequencies as homo- zygous mutant type was significantly elevated among their study patients compared to controls (6.5% v 1.2%, respectively) [17]. In previous studies, individuals with at least one Val allele at codon 105 of GSTP1 enzyme were thought to have an underlying predisposition to cancer when ex- posed to environmentally derived or endogenously for- med GSTP1 substrates [23]. Indeed, the GSTP1 codon 105Val allele was associated with a significantly in- creased risk of lung, bladder, testicular cancer, cancer breast, and multiple myeloma [23-26]. Valin e genotype has decreased enzyme activity which might be due to altered catalytic activity and thermal stability of the en- zyme. This could lead to less detoxifying efficiency for the ultimate carcinogens like polycyclic aromatic hydro- carbons (PAH) which can induce DNA adducts and ul- timately lead to carcinogenesis [27]. The Val allele was significantly higher in CML patients compared to con- trols in our study which similarly found by other’s data [16]. In discrepancy, the Val allele was higher in controls compared to CML patients in another study, yet it was of no statistically significant difference [22]. According to our results, CML patients in advanced phases (acceleration/blastic crisis) had higher frequency of mutant gene types. Homozygous mutant was higher (57.1%) than the heterozygous one (28.6%) and they were both higher than wild type (14.3%). These findings could be explained according to other studies suggesting that valine allele predispose the individuals to develop advance d di sease [17]. When response to treatment was considered, the mu- tant type was associated with poorer hematological re- sponse. All 6 patients who didn’t achieve complete re- mission were harboring the mutant type (50% homozyg- ous and 50% heterozygous). On the contrary, Sailaja et al.  GSTP1 (Ile105Val) Gene Polymorphism: Risk and Treatment Response in Chronic Myeloid Leukemia OPEN ACCESS JCT 2010 did not find any association between hematological response and GSTP1 polymorphism. However, as re- gards cytogenetic response, the frequency of combined genotypes (Ile/Val and Val/Val) was elevated in patients with minor cytogenetic response compared to major res- ponders which is in agreement to our data [17]. These results had suggested that GSTP1 Ile105Val polymor- phism with reduced GSTP1 enzyme activity might result in accumulation of intermediate metabolites in the body leading to additional mutations which might influence disease progression and response rates. Yet, the limitation of our study was small sample size and lack of sufficient information about the environmen- tal factors which limited the analysis of the interaction between the genetic and environmental factors. Therefore, these results must be verified by further studies with larger patient populations as well as larger control popu- lation. 5. Conclusion In conclusion, this is the first report highlighting the ge- netic susceptibility due to GSTP1 (Ile105Val) polymer- phism and the risk of CML in the Egyptian patients. The current study revealed that GSTP1 (Ile105Val) polymer- phism might contribute to the risk of CML development. The mutant genotype is linked to poor treatment response and worse prognosis. So, better understanding of the functional consequences of GSTP1 (Ile105Val) gene po- lymorphism would provide a basis for future studies of the role of this polymorphism in the pathogenesis of CML. It can also be used in predicting clinical outcome and prognosis in CML patients. REFERENCES [1] M. E. O’Dwyer, M. J. Mauro and B. J. Druker, “Recent Advancements in th e Treatment of Chronic Myelogenous Leukemia,” Annual Review of Medicine, Vol. 53, 2002, pp. 369-381. http://dx.doi.org/10.1146/annurev.med.53.082901.103853 [2] G. S. Lordelo, A. L. Miranda-Vilela, A. K. Akimoto, et al., “Association between Methylene Tetrahydrofolate Reductase and Glutathione S-Transferase M1 Gene Po- lymorphisms and Chronic Myeloid Le ukemia in a Brazil- ian Population,” Genetics and Molecular Research, Vol. 11, No. 2, 2012, pp. 1013-1026. http://dx.doi.org/10.4238/2012.April.19.6 [3] A. Hishida, S. Terakura, N. Emi, et al., “GSTT1 and GSTM1 Deletions, NQO1 C609T Polymorphism and Risk of Chronic Myelogenous Leukemia in Japanese,” Asian Pacific Journal of Cancer Prevention, Vol. 6, No. 3, 2005, pp. 251-255. [4] M. Taspinar, S. E. Aydos, O. Comez, et al., “CYP1A1, GST Gene Polymorphisms and Risk of Chronic Myeloid Leukemia,” Swiss Medical Weekly, Vol. 138, No. 1-2, 2008, pp. 12-17. [5] C. J. Omiecinski, J. P. Vanden Heuvel, G. H. Perdew, et al., “Xenobiotic Metabolism, Disposition, and Regulation by Receptors: From Biochemical Phenomenon to Predic- tors of Major Toxicities,” Toxicological Sciences, Vol. 120, No. S1, 2011, pp. S49-S75. http://dx.doi.org/10.1093/toxsci/kfq338 [6] G. Bhat, A. Bhat, A. Wani, et al., “Polymorphic Variation in Glutathione-S-Transferase Genes and Risk of Chronic Myeloid Leukaemia in the Kashmiri Population,” Asian Pacific Journal of Cancer Prevention, Vol. 13, No. 1, 2012, pp. 69-73. http://dx.doi.org/10.7314/APJCP.2012.13.1.069 [7] B. Mannervik, “The Isozymes of Glutathione S-Trans- ferase,” Adv ances in Enzymology, Vol. 57, 1985, pp. 357- 417. [8] R. C. Strange, M. A. Spiteri, S. Ramachandran, et al., “Glutathione-S-Transferase Family of Enzymes,” Muta- tion Research/Fundamental and Molecular Mechanisms of Mutagenesis, Vol. 482, No. 1-2, 2001, pp. 21-26. http://dx.doi.org/10.1016/S0027-5107(01)00206-8 [9] H. Autrup, “Genetic Polymorphisms in Human Xenobio- tica Metabolizimg Enzymes as Susceptibility Factors in Toxic Response,” Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis, Vol. 464, No. 1, 2000, pp. 65-76. http://dx.doi.org/10.1016/S1383-5718(99)00167-9 [10] S. Landi, “Mammalian Class Theta GST and Differential Susceptibility to Carcinogens: A Review,” Mutation Re- search/Fundamental and Molecular Mechanisms of Mu- tagenesis, Vol. 463, No. 3, 2000, pp. 247-283. http://dx.doi.org/10.1016/S1383-5742(00)00050-8 [11] S. Tsuchida and K. Sa t o , “Glutathione Transferases and Cancer,” Critical Reviews in Biochemistry and Molecular Biology, Vol. 27, No. 4-5, 1992, pp. 337-384. http://dx.doi.org/10.3109/10409239209082566 [12] N. R. Dunna, S. Vuree, S. Kagita, et al., “Association of GSTP1 Gene (I105V) Polymorphism with Acute Leukae- mia,” Journal of Genetics, Vol. 91, 2012, pp. e60-e63. [13] World Medical Association, “Declaration of Helsinki: Ethical Principles for Medical Research Involving Human Subjects,” The 59th WMA General Assembly, Seoul, 2008. [14] F. Millot, A. Baruchel, J. Guilhot, et al., “Imatinib Is Effective in Children with Previously Untreated Chronic Myelogenous Leukemia in Early Chronic Phase: Results of the French National Phase IV Trial,” Journal of Clini- cal Oncology, Vol. 29, No. 20, 2011, pp. 2827-2832. http://dx.doi.org/10.1200/JCO.2010.32.7114 [15] S. Saussele, M. Lauseker, A. Gratwohl, e t al., “Allogene- ic Hematopoietic Stem Cell Transplantation (Allo SCT) for Chronic Myeloid Leukemia in the Imatinib Era: Eva- luation of Its Impact within a Subgroup of the Rando- mized German CML Study IV,” Blood, Vol. 115, No. 10, 2010, pp. 1880-1885. http://dx.doi.org/10.1182/blood-2009-08-237115 [16] J. R. Andolina, S. M. Neudorf and S. J. Corey, “How I Treat Childhood CML,” Blood, Vol. 119, No. 8, 2012, pp. 1821-1830. http://dx.doi.org/10.1182/blood-2011-10-380774  GSTP1 (Ile105Val) Gene Polymorphism: Risk and Treatment Response in Chronic Myeloid Leukemia OPEN ACCESS JCT [17] K. Sailaja, D. Surekha, D. N. Rao, et al., “Association of the GSTP1 Gene (Ile105Val) Polymorphism with Chron- ic Myeloid Leukemia,” Asian Pacific Journal of Cancer Prevention, Vol. 11, No. 2, 2010, pp. 461-464. [18] T. Ishii, T. Matsuse, S. Teramoto, et al., “Glutathione S-Transferase P1 (GSTP1) Polymorphism in Patients with Chronic Obstructive Pulmonary Disease,” Thorax, Vol. 54, No. 8, 1999, pp. 693-696. http://dx.doi.org/10.1136/thx.54.8.693 [19] B. Dawson and R. G. Trapp, “Basic and Clinical Biosta- tistics,” 3rd Edition, McGraw-Hill Inc., 2001. [20] B. Kiran, M. Karkucak, H. Ozan, et al., “GST (GSTM1, GSTT1 and GSTP1) Polymorphisms in the Genetic Sus- ceptibility of Turkish Patients to Cervical Cancer,” Jour- nal of Gynecologic Oncology, Vol. 21, No. 3, 2010, pp. 169-173. http://dx.doi.org/10.3802/jgo.2010.21.3.169 [21] Z. Ye and H. Song, “Glutathione s-Transferase Polymor- phisms (GSTM1, GSTP1 and GSTT1) and the Risk of Acute Leukaemia: A Systematic Review and Me ta -Ana- lysis,” European Journal of Cancer, Vol. 41, No. 7, 2005, pp. 980-989. http://dx.doi.org/10.1016/j.ejca.2005.01.014 [22] M. Karkucak, T. Yakut, T. Gulte n , et al., “Investigation of GSTP1 (Ile105Val) Gene Polymorphism in Chronic Myeloid Leukaemia Patients,” International Journal of Human Genetics, Vol. 12, No. 3, 2012, pp. 145-149. [23] L. W. Harries, M. J. Stubbins, D. Forman, et al., “Identi- fication of Gentic Polymorphisms at the Glutathione S- Tranferase Pilocus and Association with Susceptibility to Bladder, Testicular and Prostate C anc er,” Carcinogenesis, Vol. 18, No. 4, 1997, pp. 641-644. http://dx.doi.org/10.1093/carcin/18.4.641 [24] D. Ryberg, V. Skaug, A. Hewer, et al., “Genotypes of Glutathione Transferase M1 and P1 and Their Signific- ance for Lung DNA Adduct Levels and Cancer Risk,” Carcinogenesis, Vol. 18, No. 7, 1997, pp. 1285-1289. http://dx.doi.org/10.1093/carcin/18.7.1285 [25] K. J. Helzlsouer, O. Selmin, H. Y. Huang, et al., “Assoc i - ation between Glutathione S-Transferase M1, P1, and T1 Genetic Polymorphisms and Development of Breast Can- cer,” Journal of the National Cancer Institute (Bethesda), Vol. 90, No. 7, 1998, pp. 512-518. http://dx.doi.org/10.1093/jnci/90.7.512 [26] V. Maggini, G. Buda, S. Galimberti, et al., “Lack of As- sociation of NQO1 and GSTP1 Polymorphisms with Mul- tiple Myeloma Risk,” Leukemia Research, Vol. 32, No. 6, 2008, pp. 988-990. http://dx.doi.org/10.1016/j.leukres.2007.10.008 [27] J. D. Hayes and D. J. Pulford, “The Glutathione S- Transferase Supergene Family: Regulation of GST and the Contributions of the Isoenzymes to Cancer Chemo- protection and Drug Resistance,” Critical Reviews in Bio- chemistry and Molecular Biology, Vol. 30, No. 6, 1995, pp. 445-600. http://dx.doi.org/10.3109/10409239509083491
|