J. J. XU ET AL.
Copyright © 2013 SciRes. ENG
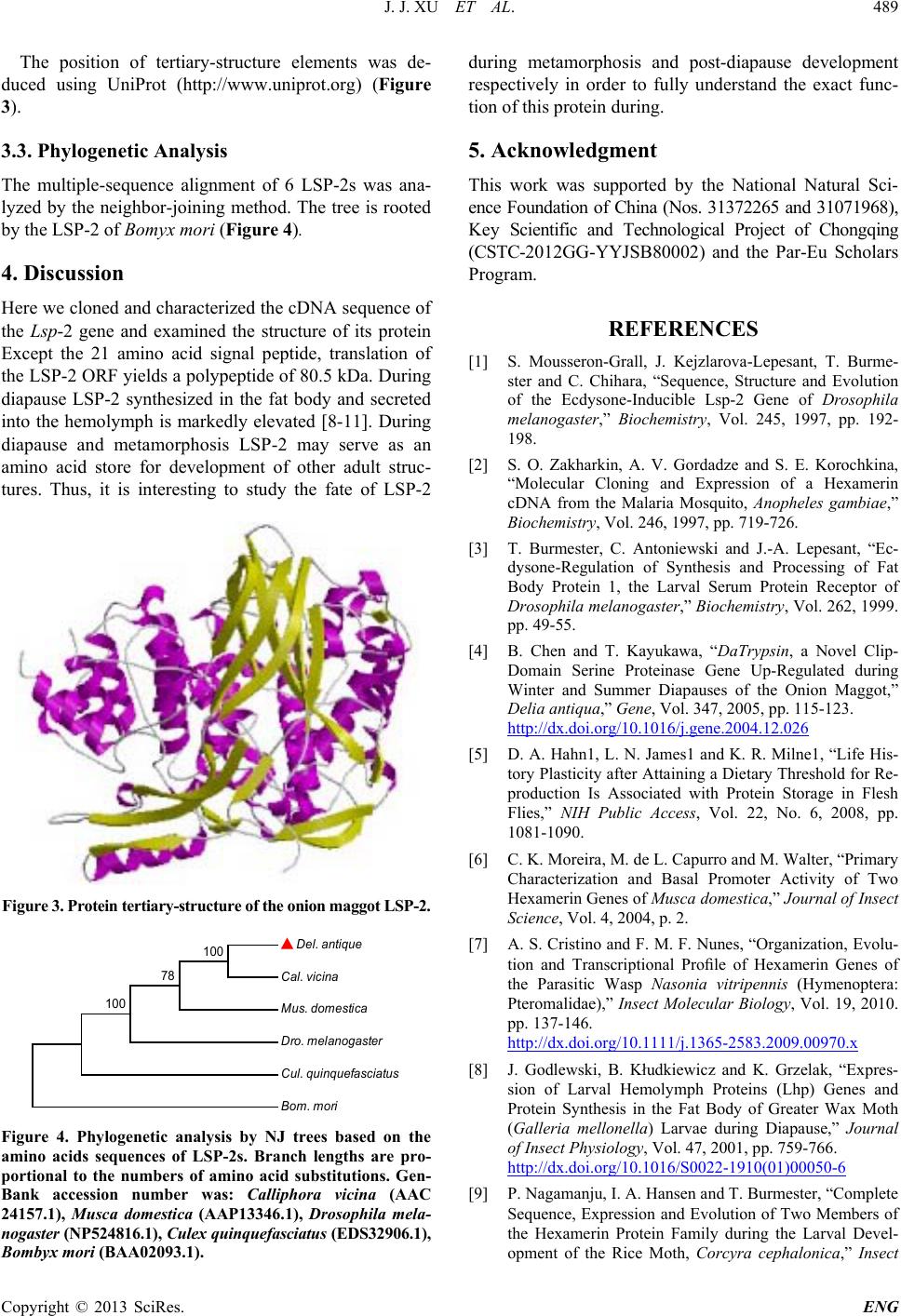
The position of tertiary-structure elements was de-
duced using UniProt (http://www.uniprot.org) (Figure
3).
3.3. Phylogenetic Analysis
The multiple-sequence alignment of 6 LSP-2s was ana-
lyzed by the neighbor-joining method. The tree is rooted
by the LSP-2 of Bomyx mori (Figure 4).
4. Discussion
Here we cloned and characterized the cDNA sequence of
the Lsp-2 gene and examined the structure of its protein
Except the 21 amino acid signal peptide, translation of
the LSP-2 ORF yields a polypeptide of 80.5 kDa. During
diapause LSP-2 synthesized in the fat body and secreted
into the hemolymph is markedly elevated [8-11]. During
diapause and metamorphosis LSP-2 may serve as an
amino acid store for development of other adult struc-
tures. Thus, it is interesting to study the fate of LSP-2
Figure 3. Protein tertiary-structure of the on ion maggot LS P-2.
Figure 4. Phylogenetic analysis by NJ trees based on the
amino acids sequences of LSP-2s. Branch lengths are pro-
portional to the numbers of amino acid substitutions. Gen-
Bank accession number was: Calliphora vicina (AAC
24157.1), Musca domestica (AAP13346.1), Drosophila mela-
nogaster (NP524816.1), Culex qui nquefa sciatu s (EDS32906.1),
Bombyx mori (BAA02093.1).
during metamorphosis and post-diapause development
respectively in order to fully understand the exact func-
tion of thi s protei n during.
5. Acknowledgment
This work was supported by the National Natural Sci-
ence Foundation of China (Nos. 31372265 and 31071968),
Key Scientific and Technological Project of Chongqing
(CSTC-2012GG-YYJSB80002) and the Par -Eu Scholars
Program.
REFERENCES
[1] S. Mousseron-Grall, J. Kejzlarova-Lepesant, T. Burme-
ster and C. Chihara, “Sequence, Structure and Evolution
of the Ecdysone-Inducible Lsp-2 Gene of Drosophila
melanogaster,” Biochemistry, Vol. 245, 1997, pp. 192-
198.
[2] S. O. Zakharkin, A. V. Gordadze and S. E. Korochkina,
“Molecular Cloning and Expression of a Hexamerin
cDNA from the Malaria Mosquito, Anopheles gambiae,”
Biochemistry, Vol. 246, 1997, pp. 719-726.
[3] T. Burmester, C. Antoniewski and J.-A. Lepesant, “Ec-
dysone-Regulation of Synthesis and Processing of Fat
Body Protein 1, the Larval Serum Protein Receptor of
Drosophila melanogaster,” Biochemistry, Vol. 262, 1999.
pp. 49-55.
[4] B. Chen and T. Kayukawa, “DaTrypsin, a Novel Clip-
Domain Serine Proteinase Gene Up-Regulated during
Winter and Summer Diapauses of the Onion Maggot,”
Delia antiqua,” Gene, Vol. 347, 2005, pp. 115-123.
http://dx.doi.org/10.1016/j.gene.2004.12.026
[5] D. A. Hahn1, L. N. James1 and K. R. Milne1, “Life His-
tory Plasticity after Attaining a Dietary Threshold for Re-
production Is Associated with Protein Storage in Flesh
Flies,” NIH Public Access, Vol. 22, No. 6, 2008, pp.
1081-1090.
[6] C. K. Moreira, M. de L. Capurro and M. Walter, “Prima ry
Characterization and Basal Promoter Activity of Two
Hexamerin Genes of Musca domestica,” Journal of Insec t
Science, Vol. 4, 2004, p. 2.
[7] A. S. Cristino and F. M. F. Nunes, “Organization, Evolu-
tion and Transcriptional Profile of Hexamerin Genes of
the Parasitic Wasp Nasonia vitripennis (Hymenoptera:
Pteromalidae),” Insect Molecular Biology, Vol. 19, 2010.
pp. 137-146.
http://dx.doi.org/10.1111/j.1365-2583.2009.00970.x
[8] J. Godlewski, B. Kłudkiewicz and K. Grzelak, “Expres-
sion of Larval Hemolymph Proteins (Lhp) Genes and
Protein Synthesis in the Fat Body of Greater Wax Moth
(Galleria mellonella) Larvae during Diapause,” Journal
of Insect Physiology, Vol. 47, 2001, pp. 759-766.
http://dx.doi.org/10.1016/S0022-1910(01)00050-6
[9] P. Nagamanju, I. A. Hansen and T. Burmester, “Complete
Sequence, Expression and Evolution of Two Members of
the Hexamerin Protein Family during the Larval Devel-
opment of the Rice Moth, Corcyra cephalonica,” Insect
Del. a n tiq ue
Cal. vi c i n a
Mus. d o mestic a
Dr o . melan og a s ter
Cul. q u inq u efasc ia tu s
Bom. mori
100
78
100