W. Z. YAN, L. BAI
Copyright © 2013 SciRes. ENG
401
and robustness of an ANN used in chromosome classifi-
cation [4].
In order to improve the performance of traditional
multilayer ANNs, a number of other more sophisticated
neural networks have been proposed and tested in this
area.
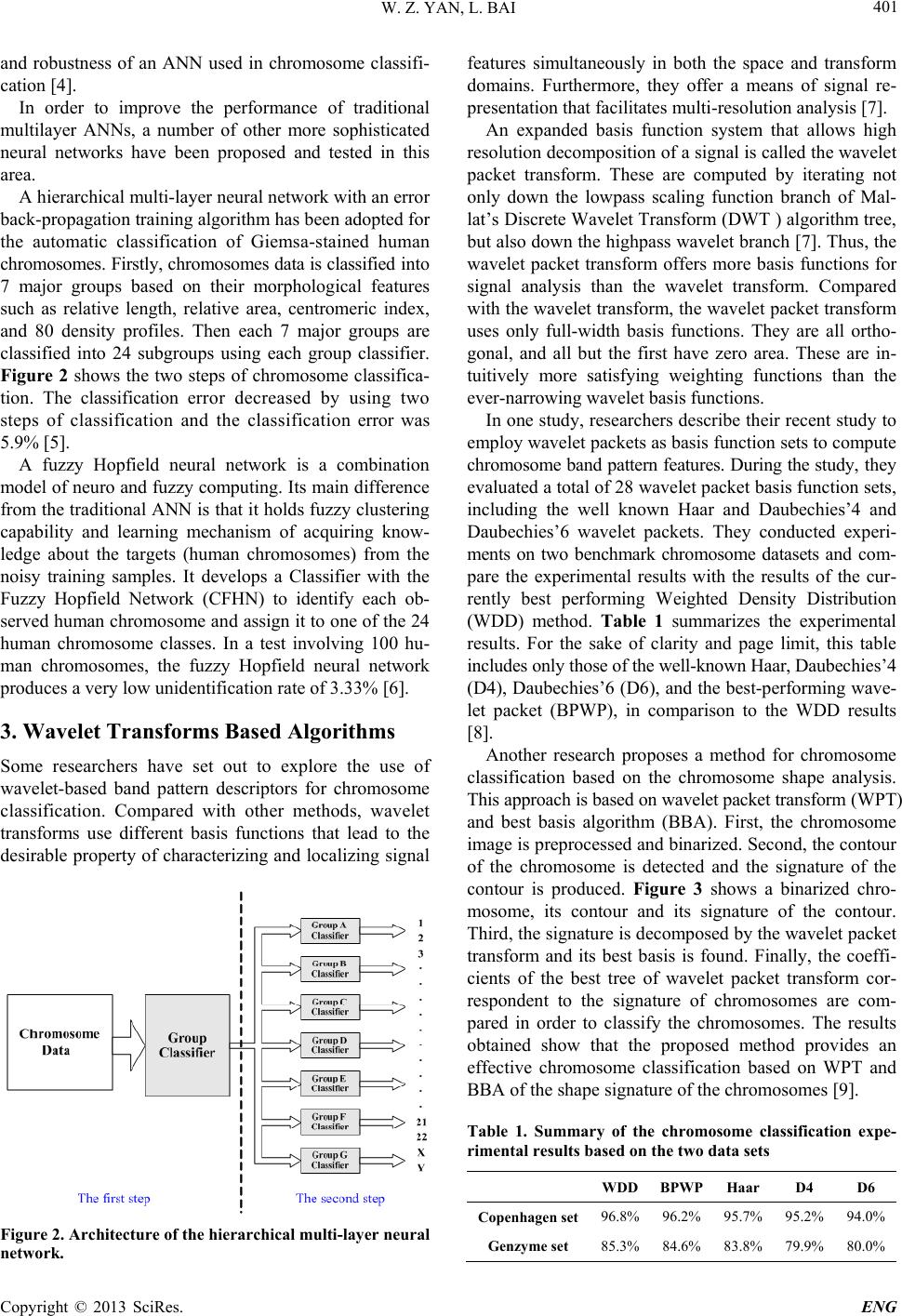
A hierarchical multi-layer neural network with an error
back-propagation training algorithm has been adopted for
the automatic classification of Giemsa-stained human
chromosomes. Firstly, chro mosomes data is classified into
7 major groups based on their morphological features
such as relative length, relative area, centromeric index,
and 80 density profiles. Then each 7 major groups are
classified into 24 subgroups using each group classifier.
Figure 2 shows the two steps of chromosome classifica-
tion. The classification error decreased by using two
steps of classification and the classification error was
5.9% [5].
A fuzzy Hopfield neural network is a combination
model of neuro and fuzzy computing. Its main difference
from the traditiona l ANN is that it holds fuzzy clustering
capability and learning mechanism of acquiring know-
ledge about the targets (human chromosomes) from the
noisy training samples. It develops a Classifier with the
Fuzzy Hopfield Network (CFHN) to identify each ob-
served human chromosome and assign it to one of the 24
human chromosome classes. In a test involving 100 hu-
man chromosomes, the fuzzy Hopfield neural network
produces a very low u nidenti f ication rate of 3. 33 % [6].
3. Wavelet Transforms Based Algorithms
Some researchers have set out to explore the use of
wavelet-based band pattern descriptors for chromosome
classification. Compared with other methods, wavelet
transforms use different basis functions that lead to the
desirable property of characterizing and localizing signal
Figure 2. Architecture of the hierarchical multi-layer neural
network.
features simultaneously in both the space and transform
domains. Furthermore, they offer a means of signal re-
presentation that facilitates multi-resolution analysis [7].
An expanded basis function system that allows high
resolution decomposition of a signal is called the wavelet
packet transform. These are computed by iterating not
only down the lowpass scaling function branch of Mal-
lat’s Discrete Wavelet Transform (DWT ) algorithm tree,
but also down the highpass wavelet branch [7]. Thus, the
wavelet packet transform offers more basis functions for
signal analysis than the wavelet transform. Compared
with the wavelet transform, the wavelet packet transform
uses only full-width basis functions. They are all ortho-
gonal, and all but the first have zero area. These are in-
tuitively more satisfying weighting functions than the
ever-narrowing wavele t basis f unc t ions.
In one study, researchers describe their recent study to
employ wavelet packets as basis function sets to compute
chromosome band pattern features. During the study, they
evaluated a total of 28 wavelet packet basis function sets,
including the well known Haar and Daubechies’4 and
Daubechies’6 wavelet packets. They conducted experi-
ments on two benchmark chromosome datasets and com-
pare the experimental results with the results of the cur-
rently best performing Weighted Density Distribution
(WDD) method. Table 1 summarizes the experimental
results. For the sake of clarity and page limit, this table
includes only those of the well-known Haar, Daubechies’4
(D4), Daubechies’6 (D6), and the best-performing wave-
let packet (BPWP), in comparison to the WDD results
[8].
Another research proposes a method for chromosome
classification based on the chromosome shape analysis.
This approach is based on wavelet packet transform (WPT)
and best basis algorithm (BBA). First, the chromosome
image is preprocessed and binarized. Second, the contour
of the chromosome is detected and the signature of the
contour is produced. Figure 3 shows a binarized chro-
mosome, its contour and its signature of the contour.
Third, the signature is decomposed by the wavelet packet
transform and its best basis is found. Finally, the coeffi-
cients of the best tree of wavelet packet transform cor-
respondent to the signature of chromosomes are com-
pared in order to classify the chromosomes. The results
obtained show that the proposed method provides an
effective chromosome classification based on WPT and
BBA of the shape signature of the chromosomes [9].
Table 1. Summary of the chromosome classification expe-
rimental results based on the two data sets
WDD BPWP Haar D4 D6
Copenhagen set 96.8% 96.2% 95.7% 95.2% 94.0%
Genzyme set 85.3% 84.6% 83.8% 79.9% 80.0%