 Journal of Biosciences and Medicines, 2013, 1, 11-15 JBM http://dx.doi.org/10.4236/jbm.2013.13003 Published Online December 2013 (http://www.scirp.org/journal/jbm/) OPEN ACCESS PGJIFs, new mitochondrial PGJ2 target factors, regulate cell proliferation Makoto Umeda1, Tatsuya Uebi1, Naoya Maekawa1, Hiroshi Handa2, Takeshi Imai1* 1Department of Aging Intervention, National Center for Geriatrics and Gerontology (NCGG), Obu, Japan 2Faculty of Bioscience and Biotechnology, Tokyo Institute of Technology, Yokohama, Japan Email: *dai.ncgg@gmail.com Received January 2013 ABSTRACT Our previous study showed that prostaglandin J2 (PGJ2) interacting factor (PGJIF) was purified and identified with magnetic nanobeads. Farther analysis of PGJ2 function shows that PGJ2 inhibits cell proli- feration and rhodamine 123 incorporation. Using PGJ2- immobilized nanobeads, two target proteins were also purified and identified as PGJIF1 and PGJIF2. PGJIF1 genetic analysis showed that PGJIF1 regu- lates cell proliferation as well as rhodamine 123 in- corporation in mitochondria, indicating that PGJIF1 is one of the PGJ2 target proteins. The other target protein, PGJIF2, changes its intracellular localization in PGJ2-dependent manner. Using nanobeads tech- nology, two PGJ2 target factors were purified and identified. Keywords: Prostaglandin J2; Nanobeads; Mitochondria 1. INTRODUCTION The drug target proteins identification is the way of evi- dence based drug discovery. Small chemical-immobi- lized nanobeads were developed as purifying target fac- tor(s) [1-7]. Prostaglandin J2 (PGJ2) family have been reported to show various kinds of biological activities, including adipogenesis. The PGJ2 target in adipocyte is peroxi- some proliferator-activated receptor γ (PPAR γ ), which promotes the expression of the crucial genes for adipo- genesis [8-11]. However the physiological role of this compound in vivo still remains as intriguing issue. Since PGJ2 exerts its biological effects at least in part through a reaction with cellular proteins, the identification of target mole- cules of PGJ2 may facilitate the understanding of the diverse biological activities of PGJ2 in vi v o [12]. Using PGJ2-immobilized nanobeads the new PGJ2 target protein (PGJ2 interacting factor, PGJIF) was puri- fied and identified voltage dependent anionic channel 1 (VDAC1) as PGJIF1 from crude extracts of HEK293 cells using this affinity purification system [3]. Here first we demonstrate the PGJ2 function in cell culture system. PGJ2 regulates cell proliferation and does not regulate cell death in low dose (0.1 - 1.0 µM), which is natural and is not suitable for PPAR γ interaction as well as apoptosis [3]. Moreover we found other PGJ2- function, PGJ2-reduced rhodamine 123 incorporation. Using PGJ2-immobilzed nanobeads 2 target factors were effectively purified and identified, PGJIF1 and PGJIF2. One of the PGJ2 target factor, PGJIF1, regulated cell proliferation and rhodamine 123 incorporation. Second target factor, PGJIF2, changed its intracellular localiza- tion from nuclear to mitochondria by PGJ2 administra- tion. Taken together multifunctional PGJ2 has at least 2 target factors, PGJIF1 & PGJIF2, regulate cell prolifera- tion and rhodamine 123 incorporation. 2. MA TERIALS AND METHODS 2.1. Materials Most potent prostaglandin J2 derivative, 15-deoxy- ∆12,14-PGJ2, was used as prostaglandin in this study [13-15]. 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC), triethylamine, N,N-dimethyl-4- aminopyridine (DMAP), N,N-dimethylformamide (DMF), acetic anhydride, dithiothreitol and iodoacetamide were purchased from Nacalai Tesque (Kyoto, Japan). Ethyle- neglycol diglycidyl ether (EGDE) was purchased from Wako Chemicals (Osaka, Japan). Trypsin was obtained from Promega (Madison, WI, USA). HEK293 and MC3T3-E1 cells were obtained from American Type Culture Collection (Manassas, VA, USA). Antibodies of anti-PHB2 (Upstate) and anti-VDAC1 (31HL, Calbio- chem) were used. 2.2. Effective Purification and Identification of PGJIFs FG beads were prepared as previously described [16]. *Corresponding a uthor.  M. Umeda et al. / Journal of Biosciences and Medicines 1 (2013) 11-15 Copyright © 2013 SciRes. OPEN ACCESS Epoxy groups on FG beads were aminolysed by NH4OH and coupled to EGDE to produce FGNEGDE beads [17, 18]. Epoxy groups on FGNEGDE beads were amino- lysed by NH4OH (FGNEGDEN), carboxylated with suc- cinic anhydride (FGNEGDENS) and activated with NHS to produce NHS-activated FGNEGDENS beads [19]. NHS-activated FGNEGDENS beads (5.0 mg) were in- cubated with 15.0 mM PGJ2 in 500 µL of DMSO con- taining 10 (V/V)% triethylamine at 25˚C for 24 hours. Unreacted amino groups on the surface of the beads were masked with acetic anhydride in DMF containing trie- thylamine at 25˚C for 24 hours. PGJ2-immobilized b eads were suspended in distilled water and stored at 4 ˚C until use [2,19]. PGJ2-immobilized beads or control beads (200 μg) were equilibrated with binding buffer and in- cubated with 200 L of the whole cell extracts at 4˚C for 4 hours using RT-50 rotator (15 rpm, TAITEC, Saitama, Japan) [2]. Affinity purified PGJIFs were separated by SDS-PAGE and gels subjected to silver staining. The specific protein bands were excised, reduced with 10 mM DTT followed by alkylation with 55 mM iodoace- tamide. Band slices were digested w ith trypsin ( 12 g/mL) overnight and desalted with ZipTip C18 (Millipore, MA, USA). The extracted peptides were then separated via nano-flow liquid chromatography (LC, Paradigm MS4, Tokyo, Japan) using a reverse phase C18 column (Magic C18, AMR, Tokyo, Japan). The LC eluent was coupled to a micro-ionspray source attached to a LCQ Advantage MAX mass spectrometer (Thermo Electron Corporation, MA, USA). All MS/MS spectra were searched using the TurboSEQUEST algorithm within the BioWorks 3.2 software (Thermo Electron Corporation, MA, USA; [18]) 2.3. Protein-Pr otein Interaction Analysis Protein-protein interaction in cell line analysis with im- muno-precipitated (Ip)-Western blotting was performed as described previously [20]. 2.4. Measurement of Cell Numbers and Viability The cells were seeded at 1 × 105 cells per well in 24-well plate. These cells were grown in DMEM supplemented with 5% FBS with or without PGJ2, and cells were har- vested and counted the number after indicated time. Each experiment was performed on triplicate cultures. The values are reported as means + SEM. Statistical signifi- cance (p < 0.05) was determined by unpaired Student’s t-test (STATVIEW). 2.5. Immunocytochemistr y The cells were plated on 35-mm poly-L-lysine-coated glass-bottomed dishes. After mitochondria were stained with 200 nM rhodamine 123 (Wako Chemicals) for 30 min at 37˚C, the cells were fixed for 20 minutes at room temperature with 3.7% paraformaldehyde and 0.4% Tri- ton X-100 in PBS, and then sequentially incubated with anti-PHB2 antibody. Fluorescent images were captured with a fluorescence microscopy (Keyence, Japan) and analyzed. 3. RESULTS AND DISCUSSION 3.1. PGJ2 Regulates Cell Proliferation and Rhodamine 123 Incorporation (Figure 1) Our previous study [3] and Figure 1(A) showed that PGJ2 inhibited HEK293 cell growth. The PGJ2 concen- tration was similar to in vivo (10 n - 1 µM, [11]). The reduction of cell number was not due to the cell death (Fi g ure 1(B)), indicating th at PGJ2 inhibited cell growth significantly. Moreover PGJ2 administration reduced rhodamine 123 incorporation in few hours (Figure 1(C)), suggesting that cellular PGJ2 target proteins (so called receptor) are in the mitochondria. 3.2. Identification of PGJ2 Mitochondrial Receptor PGJIF (Figure 2) PGJ2 is immediately converted from PGD2 which is very unstable prostanoid. In serum mainly PGJ2 is de- tected, suggesting that most of PGJ2 reaches to the target tissue (or cells). Cellular PGJ2 target factor is known to be transcriptional factor PPAR γ [3,8,9], but there is dis- crepancy. The active PGJ2 concentration for PPAR γ is from 10 µM to 1 mM, but in vivo PGJ2 concentration is from 10 nM to 1 µM, which is one-thousa ndth [3,12]. Using PGJ2-immobilzed nanobeads (Figure 2(A)), two factors (PGJIF1 and PGJIF2, respectively) are puri- fied and identified from HEK293 cells. PGJIF1 & PGJIF2 encode VDAC1 & PHB2, respectively (Figures 2(C) and (D)). In our previous study only PGJIF1/ VDAC1 was purified and identified. Our purification system improving made new factor purification be poss- ible. Low affinity PGJ2 target factor PGJIF2/PHB2 was also identified. Using antibody against VDAC1 and PHB2, they bound to PGJ2-immobilized beads (Figure 2(E) and [3]). 3.3. PGJIF1 Regulates Cell Proliferation and Rhodamine 123 Incorporation (Figure 3) With genetic approach in cells PGJIF1/VDAC1 was evaluated for cell proliferation and rhodamine 123 in- corporation. First, PGJIF1/VDAC1 overexpression vec- tor was synthesized, and confirmed its expression by Western blotting (Figure 3(A)). Using this expression vector, PGJIF1/VDAC1 function in HEK293 cells was evaluated. As expected, PGJIF1/VDAC1 regulates both cell proliferation and Rhodamine 123 incorporation 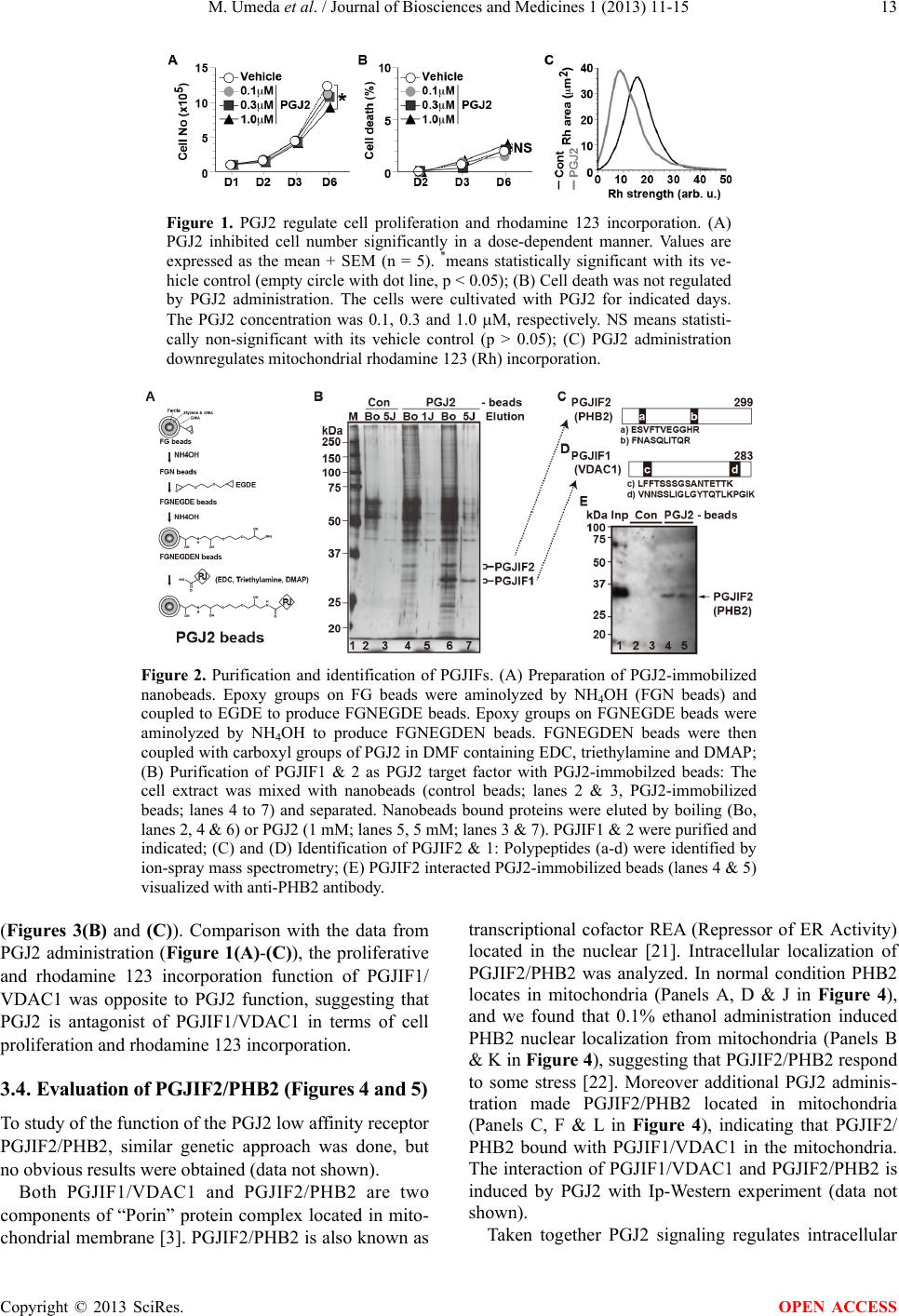 M. Umeda et al. / Journal of Biosciences and Medicines 1 (2013) 11-15 Copyright © 2013 SciRes. OPEN ACCESS Figure 1. PGJ2 regulate cell proliferation and rhodamine 123 incorporation. (A) PGJ2 inhibited cell number significantly in a dose-dependent manner. Values are expressed as the mean + SEM (n = 5). *means statistically significant with its ve- hicle control (empty circle with dot line, p < 0.05); (B) Cell death was not regulated by PGJ2 administration. The cells were cultivated with PGJ2 for indicated days. The PGJ2 concentration was 0.1, 0.3 and 1.0 µM, respectively. NS means statisti- cally non-significant with its vehicle control (p > 0.05); (C) PGJ2 administration downregulates mitochondrial rhodamine 123 (Rh) incorporation. Figure 2. Purification and identification of PGJIFs. (A) Preparation of PGJ2-immobilized nanobeads. Epoxy groups on FG beads were aminolyzed by NH4OH (FGN beads) and coupled to EGDE to produce FGNEGDE beads. Epoxy groups on FGNEGDE beads were aminolyzed by NH4OH to produce FGNEGDEN beads. FGNEGDEN beads were then coupled with carboxyl groups of PGJ2 in DMF containing EDC, triethylamine and DMAP; (B) Purification of PGJIF1 & 2 as PGJ2 target factor with PGJ2-immobilzed beads: The cell extract was mixed with nanobeads (control beads; lanes 2 & 3, PGJ2-immobilized beads; lanes 4 to 7) and separated. Nanobeads bound proteins were eluted by boiling (Bo, lanes 2, 4 & 6) or PGJ2 (1 mM; lanes 5, 5 mM; lanes 3 & 7). PGJIF1 & 2 were purified and indicated; (C) and (D) Identification of PGJIF2 & 1: Polypeptides (a-d) were identified by ion-spray mass spectrometry; (E) PGJIF2 interacted PGJ2-immobilized beads (lanes 4 & 5) visualized with anti-PHB2 antibody. (Figures 3(B) and (C)). Comparison with the data from PGJ2 administration (Figure 1(A)-(C)), the proliferative and rhodamine 123 incorporation function of PGJIF1/ VDAC1 was opposite to PGJ2 function, suggesting that PGJ2 is antagonist of PGJIF1/VDAC1 in terms of cell proliferation and rhodamine 123 incorporation. 3.4. Evaluation of PGJIF2/PHB2 (Figures 4 and 5) To study of the function of the PGJ2 low affinity receptor PGJIF2/PHB2, similar genetic approach was done, but no obvio us re sults were obtained (data not shown). Both PGJIF1/VDAC1 and PGJIF2/PHB2 are two components of “Porin” protein complex located in mito- chondrial membrane [3]. PGJIF2/PHB2 is also known as transcriptional cofactor REA (Repressor of ER Activity) located in the nuclear [21]. Intracellular localization of PGJIF2/PHB2 was analyzed. In normal condition PHB2 locates in mitochondria (Panels A, D & J in Figure 4), and we found that 0.1% ethanol administration induced PHB2 nuclear localization from mitochondria (Panels B & K in Figure 4), suggesting that PGJIF2/P HB2 respond to some stress [22]. Moreover additional PGJ2 adminis- tration made PGJIF2/PHB2 located in mitochondria (Panels C, F & L in Figure 4), indicating that PGJIF2/ PHB2 bound with PGJIF1/VDAC1 in the mitochondria. The interaction of PGJIF1/VDAC1 and P GJIF2/PHB2 is induced by PGJ2 with Ip-Western experiment (data not shown). Taken together PGJ2 signaling regulates intracellular  M. Umeda et al. / Journal of Biosciences and Medicines 1 (2013) 11-15 Copyright © 2013 SciRes. OPEN ACCESS Figure 3. Evaluation of PGJIF1 (VDAC1) on cell number and rhodamine 123 incorpora- tion. (A) Evaluation of PGJIF1 (VDAC1) over expression vector. PGJIF1 (VDAC1) over expression vector was transfected and PGJIF1 (VDAC1) proteins were detected by West- ern blotting with anti-VDAC1 antibody; (B) Significant higher cell growth in the PGJIF1 (VDAC1) over expressed cells (filled triangle). *means statistically significant with its control (empty circle, p < 0.05); (C) PGJIF1 (VDAC1) over-expressed cells are higher rho- damine 123 (Rh) incorporation. Figure 4. Evaluation of PGJIF2 (PHB2). PGJ2 inhibited EtOH-in- duced PHB2 nuclear localization. The cells treated with 0.1% EtOH (panels B, C, E, F, H, I, K & L) and 1 µM PGJ2 (panels C, F, I & L) were visualized with anti-PHB2 antibody (red color, panels A-C & J-L), rho- damine 123 (green color, panels D- F), DAPI (blue color, panels G-L). The original size of each panel is 30 µm × 30 µm. Figure 5. Possible model of PGJ2 action. New mitochondrial PGJ2 target proteins (PGJIF1/VDAC1, PGJIF2/PHB2) make complex in a PGJ2-dependent manner. EtOH induces dissociation of the com- plex and PHB2 changes its intracellular localization from mitochondria (green color) to nuclear (blue color). localization of PGJIF2/PHB2 (Figure 5). The PGJ2 re- gulates cell proliferation through binding to the PGJIF1/ VDAC1 and PGJ I F2/ PHB2. 4. CONCLUSION The nanobeads technology enable to efficient purify Pro- staglandin J2 targe t fa c t ors for new drug discovery. 5. ACKNOWLEDGEMENTS We are grateful to our department members in NCGG for helpful dis- cussions. This work was supported by a Grant-in-Aid for the Ministry of Education, Culture, Sports, Science and Technology. REFERENCES [1] Ito, T., Ando, H., Suzuki, T., Ogura, T., Hotta, K., Im- amura, Y., Yamaguchi, Y. and Handa, H. (2010) Identi- fication of a primary target of thalidomide teratogenicity. Science, 327, 1345-1350. http://dx.doi.org/10.1126/science.1177319 [2] Hiramoto, M., Maekawa, N., Kuge, T., Ayabe, F., Wata- nabe, A., Masaike, Y., Hatakeyama, M., Handa, H. and Imai, T. (2010) High-performance affinity chromatogra- phy method for identification of L-arginine interacting factors using magnetic nanobeads. Biomedical Chroma- tography, 24, 606-612. [3] Maekawa, N., Hiramoto, M., Sakamoto, S., Azuma, M., Ito, T., Ikeda, M., Naitou, M., Acharya, H.P., Kobay ashi, Y., Suematsu, M., Handa, H. and Ima i, T. (2011) High- performance affinity purification for identification of 15- deoxy-12,14-PGJ2 interacting factors using magnetic na- nobeads. Biomedical Chromatography, 25, 466-471. http://dx.doi.org/10.1002/bmc.1469 [4] Karasawa, S., Azuma, M., Kasama, T., Sakamoto, S., Kabe, Y., Imai, T., Yamaguchi, Y., Miyazawa, K. and Handa, H. (2013) Vitamin K2 covalently binds to Bak and induces Bak-mediated apoptosis. Molecular Phar- maceutics, 83, 613-620. http://dx.doi.org/10.1124/mol.112.082602 [5] Hotta, K., Nashimoto, A., Yasumura, E., Suzuki, M., Azuma, M., Shima, D., Nabeshima, R., Hiramoto, M., Okada, A., Sakata-Sogawa, K., Tokunaga, M., Ito, T., Sakamoto, S., Kabe, Y., Aizawa, S., Imai, T., Yamagu- chi, Y., Wat anabe, H. and Handa, H. (2103) Vesnarinone suppresses TNFα mRNA expression by inhibiting valosin-  M. Umeda et al. / Journal of Biosciences and Medicines 1 (2013) 11-15 Copyright © 2013 SciRes. OPEN ACCESS containing proteins. Molecular Pharmaceutics, 83, 930- 938. [6] Umeda, M., Uebi, T., Maekawa, N., Isaki, M., Miyama, Y., Masaike, Y., Handa, H. and Imai, T. (2013) Effective cofactor complex purification using nanobeads. Journal of Bioscience and Medicine, in Press. [7] Uebi, T., Umeda, M., Maekawa, N., Karasawa, S., Handa, H. and Imai, T. (2013) Prohibitins, novel vitamin K2 tar- get factors in osteoblast. Journal of Bioscience and Medi- cine, in Press. [8] Kliewer, S.A., Lenhard, J.M., Willson, T.M., Patal, I., Morris, D.C. and Lehmann, J.M. (1995) A prostaglandin J2 metabolite binds peroxisome proliferator-activated re- ceptor gamma and promotes adipocyte differentiation. Cell, 83, 813-819. http://dx.doi.org/10.1016/0092-8674(95)90194-9 [9] Bell-Parikh, L.C., Ide, T., Lawson, J.A., McNamara, P., Reilly, M. and FitzGerald, G.A. (2003) Biosynthesis of 15-deoxy-12,14-PGJ2 and the ligation of PPAR. Journal of Clinical Investigation, 112, 945-955. [10] Heikkinen, S., Auwerx, J. and Argmann, C.A. (2007) PPARγ in human and mouse physiology. Biochimica et Biophysica Acta, 1771, 999-1013. http://dx.doi.org/10.1016/j.bbalip.2007.03.006 [11] Imai, T. (2013) Adipogensis and osteoblastgenesis in aging. Adipogensis: Signaling pathways, Molecular reg- ulation and impact on human disease. In: Lin, Y.F. and Cai, X.X., Eds., Cell Biology Research Progress, Nova Science Publishers Inc., New York, 95-108. [12] Aldini, G., Carini, M., Vistoli, G., Shibata, T., Kusano, Y., Gamberoni, L., Dalle-Dounne, I., Milzani, A. and Uchida, K. (2007) Identification of actin as a 15-deoxy-12,14-PGJ2 target in Neuroblastoma cells: Mass spectrometric, com- putational, and functional approaches to investigate the effect on cytoskeletal derangement. Biochemistry, 46, 2707-2718. http://dx.doi.org/10.1021/bi0618565 [13] Acharya, H.P. and Kobayashi, Y. (2005). Synthesis of phosphorylcholines possessing 5,6- or 14,15-epoxyiso- prostane A2 at sn-2 position. Tetrahedron Letters, 46, 8435-8438. http://dx.doi.org/10.1016/j.tetlet.2005.09.193 [14] Acharya, H.P. and Kobayashi, Y. (2005) Total synthesis of 2-(5,6-epoxyisoprotane A2) phosphorylcholine and elucidation of the relative configuration of the isoprostane moiety. Angewandte Chemie, 44, 3481-3484. http://dx.doi.org/10.1002/anie.200500534 [15] Acharya, H.P. and Kobayashi, Y. (2006) Highly e fficient total synthesis of Δ12-PGJ2,15-deoxy-Δ12,14-PGJ2, and their analogues. Tetrahedron, 62, 3329-3343. http://dx.doi.org/10.1016/j.tet.2006.01.051 [16] Nishio, K., Masaike, Y., Ikeda, M., Na rimat su, H., Goken, N., Tsubouchi, S., Hatakeya ma, M., Sakamoto, S., Hanyu, N., Sandhu, A., Kawaguchi, H., Abe, M. and Handa, H. (2008) Development of novel magnetic nano-carriers for high-performance affinity purification. Colloids and Sur- faces B, Biointerfaces, 64, 162-169. http://dx.doi.org/10.1016/j.colsurfb.2008.01.013 [17] Imai, T., Sumi, Y., Hatakeyama, M., Fujimoto, K., Ka- waguchi, H., Yajima, H. and Handa, H. (1996) Selective isolation of DNA or RNA using single-stranded DNA af- finity latex particles. Journal of Colloid and Interface Science, 177, 245-249. http://dx.doi.org/10.1006/jcis.1996.0027 [18] Shimizu, N., Sugimoto, K., Tang, J., Nishi, T., Sato, I., Hiramoto, M., Aizawa, S., Hatakeyama, M., Ohba, R., Hatori, H., Yoshi kawa, T., Suzuki, F., Oomori, A., Tana- ka, H., Kawaguchi, H., Watanabe, H. and Handa, H. (2000) High-performance affinity beads for identifying drug receptors. Nature Biotechnology, 18, 877-881. http://dx.doi.org/10.1038/78496 [19] Ohtsu, Y., Ohba, R., Imamura, Y., Kobayashi, M., Hatori, H., Zenkoh, T., Hatakeyama, M., Manabe, T., Hino, M., Yamaguchi, Y., Kataoka, K., Kawaguchi, H., Watanabe, H. and Handa, H. (2005) Selective ligand purification using high-performance affinity beads. Analytical Bio- chemistry, 338, 245-252. http://dx.doi.org/10.1016/j.ab.2004.10.006 [20] Imai, T., Matsuda, K., Shimojima, T., Muramatsu, M., Handa, H. and Kato, S. (1997) ERC-55, a binding protein for the papilloma virus E6 oncoprotein, specifically inter- acts with vitamin D receptor among nuclear receptors. Biochemical and Biophysical Research Communications, 233, 765-769. http://dx.doi.org/10.1006/bbrc.1997.6531 [21] He, B., Feng , Q., Mukherjee, A., Lonard, D.M., DeMayo, F.J., Katzenellenbogen, B.S., Lydon, J.P. and O’Malley, B.W. (2008) A repressive role for prohibitin in estrogen signaling. Molecular Endocrinology, 22, 344-360. http://dx.doi.org/10.1210/me.2007-0400 [22] Kuramori, C., Az uma, M., Kume, K., Kaneko, Y., Inoue, A., Yamaguchi, Y., Kabe, Y., Hosoya, T., Kizaki, M., Suematsu, M. and Handa, H. (2009) Capsaicin binds to prohibitin 2 and displaces it from the mitochondria to the nucleus. Biochemical and Biophysical Research Commu- nications, 379, 519-525. http://dx.doi.org/10.1016/j.bbrc.2008.12.103
|