Journal of Cancer Therapy
Vol.07 No.10(2016), Article ID:70929,12 pages
10.4236/jct.2016.710072
Telomere Elongation in the Breast cancer cell Line 21NT after Treatment with an epigenetic modifying Drug
Azadeh Motevalli1,2#, Hemad Yasaei1,3#, Sara Anjomani Virmouni1,4, Morteza Mirabdulhagh5, Predrag Slijepcevic1, Terry Roberts1,6*
1Department of Life Sciences, College of Health and Life Sciences, Brunel University London, Uxbridge, UK
2Present Address: Department of Cancer Immunotherapeutics and Tumour Immunology, City of Hope Comprehensive Cancer Centre, Beckman Centre, Duarte, CA, USA
3Present Address: School of Cellular and Molecular Medicine, University of Bristol, Bristol, UK
4Present Address: The Institute of Cancer Research, Chester Beatty Laboratories, London, UK
5Faculty of Life Sciences and Medicine, School of Medical Education, Kings College London, London, UK
6Health and Environment Theme, Institute of Environment, Health and Societies, Brunel University London, Uxbridge, UK

Copyright © 2016 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: July 21, 2016; Accepted: September 25, 2016; Published: September 28, 2016
ABSTRACT
Background: Telomere length dysregulation plays a major role in cancer development and aging. Telomeres are maintained by a group of specialized genes known as shelterin and shelterin-associated proteins. In breast cancer lines it has been shown that shelterin proteins are dysregulated thereby affecting the telomere stability and contributing to the neoplastic conversion of the mammary epithelial cells. Interestingly, the regulation of some of the shelterin genes is thought to be controlled epigenetically. Methods and Results: In this study, we set out to measure the effect of increased shelterin gene expression on telomere length in breast cancer cell line 21NT treated with 5-aza-2-deoxycytidine (5-aza-CdR) using known telomere length assays. We measured telomere lengths using: Telomere Restriction Fragment length (TRF), absolute quantitative-PCR and cytogenetic Interphase Quantitative Fluorescent in situ Hybridization (iQ-FISH). We found that non-cytotoxic levels of 5-aza-CdR affect telomere lengths by causing a significant and stable increase in telomere lengths of the breast cancer cell line. The increase in telomere lengths was consistently observed when various telomere length methods were used. Conclusions: Further investigation is required to understand the underlying mechanism involved, and the significance of telomere length elongation in relation to clinical outcome when epigenetic modifying drugs are utilized.
Keywords:
Telomere, 5-Aza-2-deoxycytidine (5-aza-CdR), Trichostatin A (TSA), Shelterin, iQ-FISH, Breast Cancer

1. Introduction
The importance of telomere structural integrity in cancer is becoming more evident [1] . Telomeres are implicated in cellular aging in vitro [2] and are thought to be a biomarker for aging in vivo [3] . Telomere shortening occurs as a result of each cellular division that can trigger cellular senescence leading to irreversible cell cycle arrest [4] . Telomere dysfunction can be governed by telomere length shortening or perturbation of the protective shelterin proteins, triggering DNA-damage response, resulting in formation of chromosome end-to-end fusions and genomic instability [5] . Induction of genomic instability as a result of telomere dysfunction is thought to be critical in initiation of clonal evolution in cancer [4] [6] . Cancer cells maintain critically short telomere lengths by activating the two mechanisms of telomere length maintenance: 1) a telomere-specific DNA polymerase called telomerase (the primary mechanism), and 2) the secondary Alternative Lengthening of Telomeres mechanism known as ALT (a rare mechanism found in 10% - 15% of human tumors as well as immortalized cell lines [7] ). The reverse transcriptase enzyme telomerase is responsible for the addition of hexanucleotide repeats TTAGGG, onto the 3’-end of a telomere, and consequently counters the process of replication-associated telomere shortening. In view of the fact that the capacity for limitless replication is a sign of cancer, telomerase or ALT must be activated to overcome the process of telomere erosion and to enable replicative immortality [6] . Contrary to normal diploid somatic cells, 85% - 90% of tumor cells express high levels of telomerase which is responsible for maintaining the 2 - 4 kb telomere length of most cancer epithelial cells (carcinomas) [8] .
The role of telomerase activation in human cancer development has been widely studied [9] and it is important to understand how telomerase activation occurs in breast cancer. It has been reported that the up-regulation of telomerase is associated with cell immortalization and malignancy [10] . There is a strong body of evidence suggesting that short telomeres in breast cancer cells precipitate telomere dysfunction and this may be in part related to shelterin proteins and their expression levels in breast cancer cells [11] . Our previous study revealed that in a large panel of breast cancer cell lines the expression of the shelterin proteins was down-regulated due to the promoter methylation [12] . Similarly, when breast cancer cell lines were treated with 5-aza-CdR alone or in combination with trichostatin A (TSA), the levels of POT1, TIN2 and TPP1 shelterin proteins recovered to normal status [12] . Interestingly, we also reported the first observation of small but significant elongation of telomere lengths (1 - 2 kb increases) in the breast cancer cell lines when treated with the epigenetic modifying drugs 5-aza-CdR and TSA that is also observed in clinical samples of sporadic and familial breast cancer patients [13] . Here, we set out to further investigate the effect of 5-aza- CdR and TSA on telomere lengths in breast cancer cell line, 21NT, by measuring the telomere lengths using various telomere lengths techniques. Our data shows consistent increase in telomere lengths of 21NT cells when treated with both epigenetic modifying drugs. Consequently, we think this increase in telomere length in breast cancer cells may have important clinical consequence when epigenetic modifying chemotherapies is used and therefore requires further investigation in identifying the underlying mechanism.
2. Materials and Methods
2.1. Cell Culture conditions, maintenance and Treatment
Breast cancer cell line, 21NT [14] , normal mammary human epithelial cells (HMEC) (Lonza) and Murine lymphoma cells lines LY-R and LY-S [15] were grown in Modified eagle’s alpha (MEM) containing 10% foetal calf serum (Invitrogen), mammary epithelial growth medium (MEGM basal medium and the single shot kit supplements and growth factors, Lonza) and in RPMI/1640 containing 10% foetal calf serum respectively as described previously [12] . The 21NT cells (1 × 105) were treated with 5-aza-CdR (5 µM, Sigma, St. Louis, MO) and TSA (50 ng/ml, Sigma) for 24 hours initially, the medium was changed and samples were taken at 72 hours (which included the first 24 hours), 3 weeks, 6 weeks and 2 months. This treatment was based on the findings reported previously [12] . The control cells were treated with 0.02% DMSO.
2.2. Telomere length Measurement by Interphase Quantitative Fluorescent In situ Hybridization (iQ-FISH)
Samples were produced according to standard cytogenetic methods, with exception of Colcemid treatment. Briefly, cells were treated with 10ml of hypotonic buffer (0.075 M KCl) for 30 minutes at 37˚C, and were subsequently fixed with methanol and glacial acidic acid (3:1) solution. Cell suspensions were dropped onto slides and aged overnight at 55˚C. The telo-FISH method was applied as previously described [16] .
Images of interphase nuclei were acquired on a digital fluorescence microscope (Zeiss Axioskop 2) equipped with a CCD camera (Photometrics) and Smart Capture software (Digital Scientific, Cambridge, UK) using fixed time exposure of 0.5 seconds and magnification of 63×. IP lab software (Digital Scientific) was used to analyze telomere fluorescence intensity per cell. The signals from the average unmodified telomere fluorescence intensity (uTFI) were calculated as the total intensity of the telomeric signal utilizing the area under curve minus the background signal. The term unmodified fluorescence refers to the fact that no internal control was used to adjust for variability in fluorescence microscope. The experiment was repeated at least twice and each time 100 cells were quantified for each sample. 100× magnification was used for capturing DMSO control cells whereas x63 magnification was used to capture the 5-aza-CdR treated and HMEC control.
2.3. Telomere Length Measurement by Telomere Restriction Fragment Length (TRF)
Genomic DNA was extracted using the WizardTM Genomic DNA isolation Kit (Promega) according to the manufacturer’s protocol. TRF length measurement was performed using the telomere length assay (Roche Diagnosis) and as previously described [17] . Approximately 3 µg of DNA was digested for 2 hours at 37˚C using a mixture of restriction enzymes Hinf I and Rsa I (20 Units for each) and was proceed with electrophoresis on an 8% agarose gel followed by Southern blotting and hybridization according to the manufacturer guideline. The average TRF value was calculated by evaluating the telomeric signal (smear) relative to molecular weight standard as described in the Telomere Length Assay kit (Roche Diagnostics, Risch-Rotkreuz, Switzerland). The standard curve generated from the molecular weight is shown in Figure 1 and Figure 2. The exposed X-ray film was scanned with (Desk Scanner G2710, HP) and analyzed using densitometry software ImageQuant 5.2 (Amersham Biosciences, USA). The mean telomere length was calculated as described in detail elsewhere [18] [19] and the OD
Figure 1. A typical example of a TRF blot. Genomic samples were digested using restriction enzymes and hybridized with specific telomeric probes (see material and methods for details). The overall intensities of hybridization smears (separated by red dotted lines) were quantified as described in methods section. The control samples from the primary HMECs (p5 and p20) as well as the internal control of known telomere lengths is shown on the right. Samples loaded from left to right were as follows: 1) 21NT-untreated, 2) DMSO 72 hrs, 3) 5-aza-CdR 72 hrs, 4) 5-aza-CdR/TSA 72 hrs, 5) 5-aza-CdR 3 weeks, 6) 5-aza-CdR/TSA 3 weeks, 7) 5-aza-CdR 6 weeks, 8) 5-aza-CdR/TSA 6 weeks, 9) DMSO, 10) 5-aza-CdR 2 months, 11) 5-aza-CdR/TSA 2 months, 12) HMEC1 p5, 13) HMEC1 P20, Lane 14) positive control with known telomere length.
Figure 2. Example of the enzymatic digestion of gDNA showing smears of DNA after eletrophoresis. 3 µg of DNA was digested using Hinf I and Rsa I. Samples loaded from left to right were as follows: 1) 21NT-untreated, 2) DMSO 72 hrs, 3) 5-aza-CdR 72 hrs, 4) 5-aza-CdR/TSA 72 hrs, 5) 5-aza-CdR 3 weeks, 6) 5-aza-CdR/TSA 3 weeks, 7) 5-aza-CdR 6 weeks, 8) 5-aza-CdR/TSA 6 weeks, 9) DMSO, 10) 5-aza-CdR 2 months, 11) 5-aza-CdR/TSA 2 months, 12) HMEC1 p5, 13) HMEC1 P20, Lane 14) positive control with known telomere length.
values from the ImageQuant was used in the following formula to calculate the average
telomere length from each sample: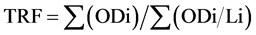 , where ODi is the chemiluminescent signal and ODi/Li is the length of the TRF at position i. We drew a rectangular box containing 30 grids covering the whole telomere fragment smears. The average pixel from 30 grids were calculated and used in the above formula.
, where ODi is the chemiluminescent signal and ODi/Li is the length of the TRF at position i. We drew a rectangular box containing 30 grids covering the whole telomere fragment smears. The average pixel from 30 grids were calculated and used in the above formula.
3. Results and discussion
Previously [12] , we reported that the addition of 5-aza-CdR (an epigenetic chemotherapy drug) alone or in combination with TSA to the 21NT breast cancer cells affects shelterin expression. In order to clarify the exact effect of shelterin up-regulation in the breast cancer cell line on telomere length, we set out to measure telomere lengths using three commonly used techniques. TRF is a commonly used telomere length assay that is based on Southern hybridization [18] . It provides the most accurate measure of telomere changes observed in cells in vitro. Alongside TRF we used a cytogenetic method based on the Q-FISH technique [20] [21] known as iQ-FISH [22] . Understanding the mechanism of telomere length stabilization and hence elongation in cancer will have profound implication in the clinical treatment choices. For example, the 5-aza-CdR is currently used clinically, at cytotoxic concentrations to inhibit cancer growth [23] . Resistance to 5-aza-CdR develops rapidly and leads to relapse [24] - [26] ; however, the actual mechanisms leading to resistance are poorly understood [27] . We therefore, have hypothesized that this resistance may be due to the rapid telomere elongation. If treatment of cancer with agents like 5-aza-CdR can lead to increases in telomere length, this in theory may disrupt the genomic stability leading to invasive growth potential and formation of an aggressive cancer phenotype. Therefore, it was imperative to investigate our earlier observation of telomere length elongation in the breast cancer cell lines treated with epigenetic chemotherapy drugs to verify our previous findings.
The treatment of 21NT cell line with 5-aza-CdR and 5-aza-CdR/TSA causes an increase in average telomere length in the 21NT cells as shown in Figures 3(a)-(b). Primary HMEC (p5) was used as the normal control for telomere length measurements with TRF detecting an average length of 7.2 and 9.0 kb respectively, as previously reported [17] .


Figure 3. Telomere length measurement by TRF and iQ-FISH. (a) The mean unmodified Telomere Fluorescence Intensity (uTFI) was measured from untreated 21NT cells and primary HMECs (control) and from various time-points of treated 21NT with 5-aza-CdR and 5-aza- CdR/TSA. The experiment was repeated at least twice with a total number of 100 nuclei quantified for each sample. Error bars represent SEM. (b) Average telomere length measured using Telomere Restriction Fragment (TRF) from the same sample type and time-points as described above. The experiment was performed twice from independent genomic DNA. Error bars represent standard deviation. (c) Scatter plot of telomere lengths measured by the three methods. Each individual average telomere length from treated cells (5-aza-CdR and 5-aza-CdR/TSA) was measured using the three methods previously described. The solid black line represents the median. On average, there is a small but significant increase in the telomere lengths of 21NT cells treated with 5-aza-CdR (p = 0.0033) and 5-aza-CdR/TSA (p = 0.0070).
Telomere elongation in treated 21NT (as compared to untreated controls) peaked at 72 hours when measured with TRF and i-QFISH and continued for three weeks (Figures 3(a)-(b)). This increase in telomere length persisted in the cell populations for two months after treatment, and was detected by the two methods described in this paper and by qPCR as described in [12] . Table 1 summarizes the overall telomere length
elongation in the treated 21NT cells. Over a two-month period an average increase in
telomere lengths of 1.06 kb (TRF) (p = 0.0718), 1.67 kb (qPCR) (p = 0.0570) and 1.83 uTFI (i-QFISH) (p = 0.0232) was observed in the 21NT cells treated with 5-aza-CdR when compared to the untreated control groups. This increase in telomere length was visually noticeable when using the fluorescence microscope (Figure 4).
The TRF method produce results in measurable and comparable units of kb. As it was not possible to convert telomere fluorescence intensity to kb, without having a standard curve from samples of known telomere length, we resorted in using an arbitrary unit of measurements called uTFI. It is interesting to note that the uTFI numerical values were in the same range as the TRF methods, which allowed us to plot the whisker chart as evident in Figure 3(c) and Figure 5(a). This clearly shows that the treatment with 5-aza-CdR and 5-aza-CdR/TSA induces an increase in telomere lengths for individual samples analyzed, with significant overall increases in the two treated groups (p = 0.0070 and p = 0.0033) (Figure 5).
A significant drawback of the TRF method versus the q-PCR analysis is the fact that TRF estimates the length of telomeric sequences that also contain sub-telomeric repeats. These repeats can vary in length based on the last restriction site at a given chromosome arm, thus increasing the heterogeneity of TRFs and influencing the length of sub-telomeric repeats that prevents detection of the true length of telomere repeats [28] . Previous findings by Steinert et al. (2014) showed that sub-telomeres contain 2 - 4kb of sequence that is resistant to enzymatic digestion [29] . TRF analysis provides an average telomere length for the sample (not even a mean length is obtained) and therefore may provide telomere lengths that are shorter than that obtained by other methods.
Table 1.Telomere length comparison of three methods: Telomere Restriction Fragment (TRF), quantitative PCR (data from [12] ) and interphase-quantitative FISH (iQ-FISH). The average of untreated and DMSO-treated 21NT cell is compared to the average of the 5-aza-CdR-treated (all time points) and the 5-aza-CdR plus TSA-treated (all time points).
†p = 0.0718 (90% CI), ††p = 0.0570 (90% CI), ‡p = 0.0026 (95% CI), ‡‡p = 0.0035 (95% CI), *p = 0.0232 (95% CI), **p = 0.0053 (95%CI). *Data from [12] .
In this article, we confirm telomere length increase in the breast cancer cell line 21NT within 3 days of exposure to 5-aza-CdR. In some cases, the increase was as much as two fold. 5-aza-CdR is a global demethylatingagent, it will have the effect of altering the methylation pattern of many genes and pathways. It is known that 85% of cancers upregulate telomerase to maintain telomere lengths, the other 15% use the ALT pathway to achieve the same goals [30] . In this paper, we have shown that both pre-spliced and fully spliced hTERT was upregulated 72 hours after 5-aza-CdR treatment (Figure 6). The level of pre-spliced hTERT expression has been shown to correlate with telomerase activity [31] . Also [32] found that when human lymphoid and colon carcinoma cell lines were treated with 5-aza-CdR for 72 hours, a rapid increase in the telomeric recombination (shown by T-SCE) occurred. T-SCE is the mechanism ALT uses to in-
Figure 4. Typical images from an iQ-FISH method. The cell nuclei were hybridized with telomeric PNA probe (Telo-PNA) tagged with green fluorescence signal (FITC) and DNA stained with DAPI (Blue). The total fluorescent intensities of each telomeric signal were converted digitally and the mean average were analyzed. Telomere signals from primary HMECs (with known average telomere length of 9 - 12 kb) are shown for comparison purposes. The scale bar = 10 µm. ×100 lens was used to capture DMSO control cells and ×63 lens was used for 5-aza-CdR treated and HMEC control cells. All backgrounds were taken into account when software were used to measure the telomeric signal using the iQ-FISH protocol. This is only a true representation of more than hundreds of images taken and the overall background is equal in all samples.

Figure 5. Scatter plot of individual telomere lengths measured using the described methods. Determination of telomere lengths usng qPCR technique has been reported in detail previously. The conversion of T/S ratio into telomere lengths in kilobases was done using a standard curve generated to measure absolute telomere lengths of a sample with known telomere lengths as described previously. (a) Serial dilution of the telomere standard starting with 1,018,400 kb to 10,184 kb was used to measure the content of telomeric sequences per sample in kb. (b) Single gene copy gene (36B4) was used as control for every reaction and to determine the genome copies of each sample. The values generated from the qPCR reaction and the standard curves (Figure 5(b)) were used to calculate the total telomere length in kb per human diploid genome. The telomere kb length per reaction value was then divided by diploid genome copy number to give a total telomeric length in kb per human diploid genome.
Figure 6. Gene expression analysis of pre- and fully-spliced hTERT mRNA. 21NT cells were treated with 5-aza-CdR or DMSO for 72 hours before RNA extraction and analysis. Relative qPCR was then performed for both pre-spliced and spliced hTERT using primers from [31] .
crease telomere lengths. Therefore it is possible that a combination of both hTERT (hence telomerase) up-regulation and an increase in T-SCE may have contributed to
the rapid increase in telomere length we observed. We have previously shown [12] that 5-aza-CdR upregulates several members of the Shelterin complex simultaneously. This may also play a role in the rapid increase in telomere length observed as POT1 and TPP1 have been shown to be involved in telomerase regulation [33] .
Our previous study has shown similar telomere length elongation in two other breast cancer cell lines, BT474 and MCF7 upon treatment with 5-aza-CdR and TSA assessed with q-PCR [12] . Therefore, the current observation reported here is consistent with our previous study, marking the epigenetic regulation of telomere length maintenance an important area to be considered in clinical therapeutic approaches.
Increase in telomere length may influence the clinical outcome of patients treated with drugs such as 5-aza-CdR and TSA. For instance, prostate and breast cancer cell lines that have elongated telomeres through hTERT overexpression show signs of differentiation in vitro [34] compared to their parental cell type. Whereas, hepatocellular carcinoma cells (HCC) with long telomeres tend to develop greater invasive ability compared to similar HCC with short telomeres [35] . Therefore, the mechanism of telomere length elongation through epigenetic regulating drug needs to be further investigated.
Acknowledgements
We thank Professor Robert Newbold for his support and for providing the opportunity to carry out this work within the Institute of Cancer Genetics and Pharmacogenomics, Brunel University London. HY was supported by a triennial project grant (Strategic Award) from the National Centre for Replacement, Refinement, and Reduction (NC3Rs) of animals in research (NC. K500045.1 and G0800697).
Cite this paper
Motevalli, A., Yasaei, H., Virmouni, S.A., Mirabdulhagh, M., Slijepcevic, P. and Roberts, T. (2016) Telomere Elongation in the Breast Cancer Cell Line 21NT after Treatment with an Epigenetic Modifying Drug. Journal of Cancer Therapy, 7, 700-711. http://dx.doi.org/10.4236/jct.2016.710072
References
- 1. Jacobs, J.J. (2013) Loss of Telomere Protection: Consequences and Opportunities. Frontiers in Oncology, 3, 88.
http://dx.doi.org/10.3389/fonc.2013.00088 - 2. Mather, K.A., Jorm, A.F., Parslow, R.A. and Christensen, H. (2011) Is Telomere Length a Biomarker of Aging? A Review. The Journals of Gerontology. Series A, Biological Sciences and Medical Sciences, 66, 202-213.
http://dx.doi.org/10.1093/gerona/glq180 - 3. von Zglinicki, T. and Martin-Ruiz, C.M. (2005) Telomeres as Biomarkers for Ageing and Age-Related Diseases. Current Molecular Medicine, 5, 197-203.
http://dx.doi.org/10.2174/1566524053586545 - 4. Collado, M., Blasco, M.A. and Serrano, M. (2007) Cellular Senescence in Cancer and Aging. Cell, 130, 223-233.
http://dx.doi.org/10.1016/j.cell.2007.07.003 - 5. Sfeir, A. and de Lange, T. (2012) Removal of Shelterin Reveals the Telomere End-Protection Problem. Science, 336, 593-597.
http://dx.doi.org/10.1126/science.1218498 - 6. Hanahan, D. and Weinberg, R.A. (2000) The Hallmarks of Cancer. Cell, 100, 57-70.
http://dx.doi.org/10.1016/S0092-8674(00)81683-9 - 7. Hiyama, K. (2009) Telomeres and Telomerase in Cancer. Humana, Springer, London, Totowa.
http://dx.doi.org/10.1007/978-1-60327-879-9 - 8. Kim, N.W., Piatyszek, M.A., Prowse, K.R., Harley, C.B., West, M.D., Ho, P.L., Coviello, G.M., Wright, W.E., Weinrich, S.L. and Shay, J.W. (1994) Specific Association of Human Telomerase Activity with Immortal Cells and Cancer. Science, 266, 2011-2015.
http://dx.doi.org/10.1126/science.7605428 - 9. Donate, L.E. and Blasco, M.A. (2011) Telomeres in Cancer and Ageing. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 366, 76-84.
http://dx.doi.org/10.1098/rstb.2010.0291 - 10. Salhab, M., Jiang, W.G., Newbold, R.F. and Mokbel, K. (2008) The Expression of Gene Transcripts of Telo-Mere-Associated Genes in Human Breast Cancer: Correlation with Clinico-Pathological Parameters and Clinical Outcome. Breast Cancer Research and Treatment, 109, 35-46.
http://dx.doi.org/10.1007/s10549-007-9622-8 - 11. Butler, K.S., Hines, W.C., Heaphy, C.M. and Griffith, J.K. (2012) Coordinate Regulation between Expression Levels of Telomere-Binding Proteins and Telomere Length in Breast Carcinomas. Cancer Medicine, 1, 165-175.
http://dx.doi.org/10.1002/cam4.14 - 12. Motevalli, A., Yasaei, H., Virmouni, S.A., Slijepcevic, P. and Roberts, T. (2014) The Effect of Chemotherapeutic Agents on Telomere Length Maintenance in Breast Cancer Cell Lines. Breast Cancer Research and Treatment, 145, 581-591.
http://dx.doi.org/10.1007/s10549-014-2975-x - 13. Benitez-Buelga, C., Sanchez-Barroso, L., Gallardo, M., Apellaniz-Ruiz, M., Inglada-Perez, L., Yanowski, K., Car-rillo, J., Garcia-Estevez, L., Calvo, I., Perona, R., Urioste, M., Osorio, A., Blasco, M.A., Rodriguez-Antona, C. and Benitez, J. (2015) Impact of Chemotherapy on Telomere Length in Sporadic and Familial Breast Cancer Patients. Breast Cancer Research and Treatment, 149, 385-394.
http://dx.doi.org/10.1007/s10549-014-3246-6 - 14. Band, V., Zajchowski, D., Swisshelm, K., Trask, D., Kulesa, V., Cohen, C., Connolly, J. and Sager, R. (1990) Tumor Progression in Four Mammary Epithelial Cell Lines Derived from the Same Patient. Cancer Research, 50, 7351-7357.
- 15. McIlrath, J., Bouffler, S.D., Samper, E., Cuthbert, A., Wojcik, A., Szumiel, I., Bryant, P.E., Riches, A.C., Thompson, A., Blasco, M.A., Newbold, R.F. and Slijepcevic, P. (2001) Telomere Length Abnormalities in Mammalian Radiosensitive Cells. Cancer Research, 61, 912-915.
- 16. Yasaei, H. and Slijepcevic, P. (2010) Defective Artemis Causes Mild Telomere Dysfunction. Genome Integrity, 1, 3.
http://dx.doi.org/10.1186/2041-9414-1-3 - 17. Garbe, J.C., Bhattacharya, S., Merchant, B., Bassett, E., Swisshelm, K., Feiler, H.S., Wyrobek, A.J. and Stampfer, M.R. (2009) Molecular Distinctions between Stasis and Telomere Attrition Senescence Barriers Shown by Long-Term Culture of Normal Human Mammary Epithelial Cells. Cancer Research, 69, 7557-7568.
http://dx.doi.org/10.1158/0008-5472.CAN-09-0270 - 18. Kimura, M., Stone, R.C., Hunt, S.C., Skurnick, J., Lu, X., Cao, X., Harley, C.B. and Aviv, A. (2010) Measurement of Telomere Length by the Southern Blot Analysis of Terminal Restriction Fragment Lengths. Nature Protocols, 5, 1596-1607.
http://dx.doi.org/10.1038/nprot.2010.124 - 19. Wang, F., Podell, E.R., Zaug, A.J., Yang, Y., Baciu, P., Cech, T.R. and Lei, M. (2007) The POT1-TPP1 Telomere Complex Is a Telomerase Processivity Factor. Nature, 445, 506-510.
http://dx.doi.org/10.1038/nature05454 - 20. Hultdin, M., Gronlund, E., Norrback, K., Eriksson-Lindstrom, E., Just, T. and Roos, G. (1998) Telomere Analysis by Fluorescence in Situ Hybridization and Flow Cytometry. Nucleic Acids Research, 26, 3651-3656.
http://dx.doi.org/10.1093/nar/26.16.3651 - 21. Slijepcevic, P. (2001) Telomere Length Measurement by Q-FISH. Methods in Cell Science, 23, 17-22.
http://dx.doi.org/10.1023/A:1013177128297 - 22. Ojani, M. (2012) Relationship between DNA Damage Response and Telomere Maintenance. Brunel University, London.
- 23. Connolly, R. and Stearns, V. (2012) Epigenetics as a Therapeutic Target in Breast Cancer. Journal of Mammary Gland Biology and Neoplasia, 17, 191-204.
http://dx.doi.org/10.1007/s10911-012-9263-3 - 24. Ishikawa, T. (2014) Novel Therapeutic Strategies Using Hypomethylating Agents in the Treatment of Myelo-Dysplastic Syndrome. International Journal of Clinical Oncology, 19, 10-15.
http://dx.doi.org/10.1007/s10147-013-0651-5 - 25. Ozbalak, M., Cetiner, M., Bekoz, H., Atesoglu, E.B., Ar, C., Salihoglu, A., Tuzuner, N. and Ferhanoglu, B. (2012) Azacitidine Has Limited Activity in “Real Life” Patients with MDS and AML: A Single Centre Experience. Hematological Oncology, 30, 76-81.
http://dx.doi.org/10.1002/hon.986 - 26. Prebet, T., Gore, S.D., Esterni, B., Gardin, C., Itzykson, R., Thepot, S., Dreyfus, F., Rauzy, O.B., Recher, C., Ades, L., Quesnel, B., Beach, C.L., Fenaux, P. and Vey, N. (2011) Outcome of High-Risk Myelodysplastic Syndrome after Azacitidine Treatment Failure. Journal of Clinical Oncology, 29, 3322-3327.
http://dx.doi.org/10.1200/JCO.2011.35.8135 - 27. Zeidan, A.M., Linhares, Y. and Gore, S.D. (2013) Current Therapy of Myelodysplastic Syndromes. Blood Reviews, 27, 243-259.
http://dx.doi.org/10.1016/j.blre.2013.07.003 - 28. Vera, E. and Blasco, M.A. (2012) Beyond Average: Potential for Measurement of Short Telomeres. Aging, 4, 379-392.
http://dx.doi.org/10.18632/aging.100462 - 29. Steinert, S., Shay, J.W. and Wright, W.E. (2004) Modification of Subtelomeric DNA. Molecular and Cellular Biology, 24, 4571-4580.
http://dx.doi.org/10.1128/MCB.24.10.4571-4580.2004 - 30. Newbold, R.F. (2002) The Significance of Telomerase Activation and Cellular Immortalization in Human Cancer. Mutagenesis, 17, 539-550.
http://dx.doi.org/10.1093/mutage/17.6.539 - 31. Ducrest, A.L., Amacker, M., Mathieu, Y.D., Cuthbert, A.P., Trott, D.A., Newbold, R.F., Nabholz, M. and Lingner, J. (2001) Regulation of Human Telomerase Activity: Repression by Normal Chromosome 3 Abolishes Nuclear Telomerase Reverse Transcriptase Transcripts but Does Not Affect c-Myc Activity. Cancer Research, 61, 7594-7602.
- 32. Vera, E., Canela, A., Fraga, M.F., Esteller, M. and Blasco, M.A. (2008) Epigenetic Regulation of Telomeres in Human Cancer. Oncogene, 27, 6817-6833.
http://dx.doi.org/10.1038/onc.2008.289 - 33. Wang, F., Podell, E.R., Zaug, A.J., Yang, Y., Baciu, P., Cech, T.R. and Lei, M. (2007) The POT1-TPP1 Telomere Complex Is a Telomerase Processivity Factor. Nature, 445, 506-510.
http://dx.doi.org/10.1038/nature05454 - 34. Hirashima, K., Migita, T., Sato, S., Muramatsu, Y., Ishikawa, Y. and Seimiya, H. (2013) Telomere Length Influences Cancer Cell Differentiation in Vivo. Molecular and Cellular Biology, 33, 2988-2995.
http://dx.doi.org/10.1128/MCB.00136-13 - 35. Ko, E. and Jung, G. (2014) Positive Association of Long Telomeres with the Invasive Capacity of Hepatocellular Carcinoma Cells. Biochemical and Biophysical Research Communications, 447, 358-363.
http://dx.doi.org/10.1016/j.bbrc.2014.04.022

#Authors contributed equally.








