Advances in Bioscience and Biotechnology
Vol.06 No.02(2015), Article ID:53960,6 pages
10.4236/abb.2015.62009
Molecular Characterization of Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) Gene of Field Isolate of Edwardsiella tarda
Megha K. Bedekar, Praveena K. Soman, K. V. Rajendran, Gayatri Tripathi, M. Makesh
Aquatic Animal Health, Central Institute of Fisheries Education (ICAR), Mumbai, India
Email: meghakadambedekar@gmail.com
Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 24 January 2015; accepted 8 February 2015; published 11 February 2015
ABSTRACT
The E. tarda bacterial culture isolated from infected fish, was confirmed by morphological and biochemical characterization. GAPDH gene of 996 bp was amplified from bacterial genomic DNA. GAPDH gene was cloned in pTZ57R/T cloning vector. The positive clone was sequenced. The sequencing result showed very homology with published sequence from pathogenic E. tarda. The sequence (nucleotide and amino acid) divergence values were very low between E. tarda isolates from India (KF142190) and China (HQ697337-38) (Figure 4 and Figure 5). However, the divergence value was high when compared with E. tarda isolates from Japan (AB198939) and this value was higher than the inter-specific divergence value (E. tarda-E. ictaluri, E. tarda-E. coli & E. tarda- Klebsiella species).
Keywords:
E. tarda, GAPDH Gene, Vaccine, Cloning

1. Introduction
Aquaculture is the fastest growing sector of agriculture in most part of the world. However, aquaculture growth has been hampered by economic loss due to fish diseases [1] . Ten percent of cultured aquatic animals are lost because of various infectious diseases [2] . Therefore, controlling disease problems in aquaculture industry are a major concern.
Out of various infectious bacterial diseases, Edwardsiellosis caused by Edwardsiella tarda (E. tarda) is a dreadful problem. Lack of effective means of control and prevention, especially the lack of effective vaccines, causes heavy economic losses in aquaculture industries worldwide due to E. tarda infection. E. tarda is a member of family Enterobacteriaceae. It is a Gram-negative, motile and rod shaped bacterium which can affect warm and coldwater species of fish. The bacterium is a pathogen with a wide range of hosts, such as fishes, reptiles, birds and mammals including human. E. tarda is a problem in eel ponds, tilapia culture and channel catfish culture systems. It causes rapidly progressive necrotizing soft tissue infection in humans. Geographic range of E. tarda is worldwide and it is a versatile pathogen with a wide ecological niche.
The DNA based vaccination or genetic immunization is a very promising research field since long years ago. DNA vaccines represent a major advance in fight against infectious diseases. DNA vaccines have several practical and immunological advantages than traditional antigen vaccines. Many virulence genes of E. tarda and genes encoding antigenic proteins have been explored for development of a vaccine and immunodiagnostics.
GAPDH protein of Edwardsiella is reported to be highly antigenic, and is considered to be a strong vaccine candidate to counter both Gram-negative and Gram-positive bacterial infections [3] [4] . The aim of the present study was to clone and sequence the GAPDH gene of E. tarda bacteria isolated from infected fish, and to explore the potential of it as a vaccine candidate against Edwardsiellosis in future.
2. Materials and Methods
2.1. Morphological and Biochemical Characterization of Bacteria
The bacterium E. tarda used in the study was obtained from a field isolate, collected from central India. The bacterium was inoculated in Brain Heart Infusion Broth (BHI broth) (Himedia, India). The broth was incubated for 18 - 24 h at 37˚C. One loop of bacteria from the incubated broth was streaked on Nutrient Agar and Salmonella Shigella Agar (SS agar) media.
Based on the principle of pH change and substrate utilisation biochemical tests were carried out. For biochemical analysis of bacteria Hi 25 Enterobacteriaceae identification kit (Himedia, India) was used. Briefly, a single isolated colony of E. tarda was picked and inoculated in 5 mL of BHI broth and incubated at 37˚C for 4 - 6 hours. Kit was opened aseptically and each well was inoculated with 50 μL of inoculum by surface inoculation method. The kit was incubated at 37˚C for 18 h. Results were interpreted using the chart supplied along with kit.
2.2. DNA Isolation from E. tarda
The genomic DNA from E. tarda was isolated by using GenElute bacterial genomic DNA kit (Sigma USA), according to manufacturer’s protocol.
2.3. PCR Amplification of GAPDH Gene of E. tarda
Specific primer set containing forward primer “CTC GAG ATG ACT ATC AAA GTA GGT ATC A” and reverse primer ACG CGT TTA CTT AGA GAT GTG TGC GA was used to amplify the gene GAPDH. PCR was performed using 25 μL reaction volume, each containing 2 μL (100 μg) of template DNA, 2.5 μL of 10× buffer, 2 μL of 25 mM MgCl2, 0.5 μL of 10 mM dNTP mix, 0.25 μL of Taq DNA polymerase (5U), 0.5 μL each of forward and reverse primers (25 pmol) and rest nuclease-free water. Amplification was carried out at the following cyclic conditions in the thermocycler (Quanta Biotech, England) 95˚C for 5 min, 30 cycles of 95˚C for 1 min, 56˚C for 1 min and 72˚C for 1 min, then a single step of final extension at 72˚C for 10 min. Size of the amplified product was then determined on 1.2% agarose gel electrophoresis.
The 996 bp DNA fragment of GAPDH was eluted from gel using Fermentas GeneJET Gel Extraction Kit. The PCR product was eluted in 25 μL elution buffer. Concentration of elute was measured using Nano Drop (Thermo Scientific, USA).
2.4. Cloning of GAPDH Gene in pTZ57R/T Cloning Vector
InsTA cloning vector pTZ57R/T was supplied by Thermo Scientific, India. The host strain DH5α of E. coli was available in the Aquatic Animal Health Management Laboratory, CIFE.
Four μL of the gene eluted PCR product and 3 μL of the pTZ57R/T vector were ligated in 30 μL ligation reaction, with 5× ligation buffer, T4 DNA ligase and nuclease-free water. This mixture was incubated at room temperature for 1 h.
2.5. Preparation of Competent E. coli Cells and Transformation
One loop from fresh colony of DH5α cells was inoculated in 2 mL of C medium which was supplied with Tranform Aid bacterial transformation kit (Fermentas, India) and incubated at 37˚C overnight. One fifty μL of overnight bacterial culture was added to 1.5 mL of prewarmed C medium and incubated for 20 min at 37˚C in a shaker. Pelleted cells were resuspended in 300 μL of T-solution and incubated for 5 min and centrifuged. The pellet was resuspended in 120 μL of T-solution and to this 5 μL of ligation mixture was added. This mix was incubated on ice for 5 min. The transformed cells were spreaded immediately on LB agar plates containing ampicillin (100 μg/mL), IPTG (10 mM) and X GAL (2% W/V), under sterile conditions. Appropriate positive and negative controls were processed simultaneously. Plate was incubated at 37˚C for 18 h.
2.6. Screening of Recombinant InsT/A Clones
Screening of the colonies was done by sub culturing white colonies in LB broth containing Ampicillin (100 µg/mL) and incubated at 37˚C overnight in a shaker incubator. The plasmid DNA was extracted by GeneJET Plasmid Miniprep Kit (Fermentas, USA). For plasmid extraction single colony from freshly streaked selective plate was picked and inoculated in 3 mL LB-ampicillin broth and incubated at 37˚C in shaking incubator for 16 h. Bacterial cells were harvested by centrifugation at 8000× for 2 min. Pelleted cells were resuspended in 250 μL of re-suspension solution lysed by adding 250 μL of lysis solution. Then 350 μL of neutralisation solution was added. The supernatant was transferred to the supplied GeneJET spin column. Plasmid was eluted in 25 μL of nuclease free water, after washing with washing buffer.
2.7. Restriction Enzyme Analysis of Recombinant Clones
To confirm the cloning of GAPDH gene in T/A vector double enzyme digestion reaction was carried out. Plasmid DNA (1 μL), 10× FD buffer (2 μL), Xho I (1 μL), Mlu I (1 μL) and nuclease-free water (15 μL) were mixed thoroughly to make a 20 μL reaction volume. The mixture was incubated at 37˚C for 5 min and 80˚C for 10 min. The digested samples were analysed on 1.2% agarose gel in 0.5× TAE buffer.
2.8. Sequence Analysis of GAPDH Gene
The pTZ/GPD plasmid, 15 µL was sent for sequencing to Bioinnovations, India. The nucleotide sequence was screened using VecScreenprogramme on the internet (http://www.ncbi.nlm.nih.gov/VecScreen/). The sequence was designated as E tardaGAPDH-MP-INDIA. This sequence was aligned with few available GAPDH sequences from E tarda and other bacteria (Table 1). To find homologous sequence single nucleotide-nucleotide alignment was done using the programme Basic Local Alignment Search Tool (BLAST) on internet using web site http://www.ncbi.nlm.nih.gov/blast. Nucleotide and aminoacid based Neighbour-Joining (NJ) trees were constructed by implying Kimura 2 Parameter and Jones-Taylor-Thornton (JTT) substitution model, respectively using MEGA software v5.0 [5] .
3. Result & Discussion
In the present study, GAPDH gene of an Edwardsiella tarda isolated from infected fish collected from central India was cloned and sequenced.
3.1. Characterization of E. tarda
The E. tarda was isolated from infected fish, from central India. Colonies of bacteria were identified as E. tarda on the basis of colony morphology, staining characteristics and biochemical properties. The colonies appeared transparent with black centre, after overnight incubation at 37˚C in SS agar medium. On staining with Gram’s stain it appeared Gram-negative small rods to coccobacilli. The length was about 2 - 4.5 times more than the width and was non-sporing and non-capsulated bacteria. Biochemical tests showed positive result, especially Indole, Methyl Red and H2S production which are the clear distinguishing factors of E. tarda from E. ictaluri [6] . Also the bacteria showed positive catalase reaction with H2O2 (Figure 1(a) and Figure 1(b)).
3.2. Cloning and Sequencing of GAPDH Gene of E. tarda
After bacterial genomic DNA extraction, PCR amplification of GAPDH gene was achieved at annealing tem-
perature 56˚C. A 996 base pair PCR-amplified GAPDH gene was obtained. The size of the gene was similar to the earlier reports [3] .
Successful ligation of PCR amplified GAPDH gene was achieved at 22˚C incubation. On transformation, in DH5α cells, white and blue colonies on LB-ampicillin plates containing X-GAL and IPTG were produced. Plasmid from the white colonies were confirmed as positive clones by release of GAPDH fragment from the plasmid by double digestion with MIu I and Xho I enzymes (Figure 2).
3.3. Sequence Analysis of GAPDH Gene
The nucleotide sequences obtained from GAPDH, is submitted to NCBI, Gene Bank (accession no. KF142190). The sequence showed a complete functional open reading frame of 996 bp containing initiation codon of ATG and termination codon TAA (Figure 3).
Table 1. List of GAPDH sequences from bacterial origin used for sequence similarity analysis.

Figure 1. (a) Edwardsiella tarda growth on Salmonella Shigella Agar, after 18 h growth at 37˚C; (b) Edwardsiella tarda culture showing catalase positive reaction.
Figure 2. Agarose gel electrophoresis (1%) showing release of 996 bp GAPDH fragment from pTZ vector after restriction digestion with MIu I and Xho I enzymes. Lane 1: release of 996 bp GAPDH fragment from pTZ vector, confirming the cloning of the gene in vector; Lane 2: PCR amplified 996 bp GAPDH gene fragment; Lane M: 1 Kb plus marker; and Lane 3: 100 bp DNA Molecular weight marker.
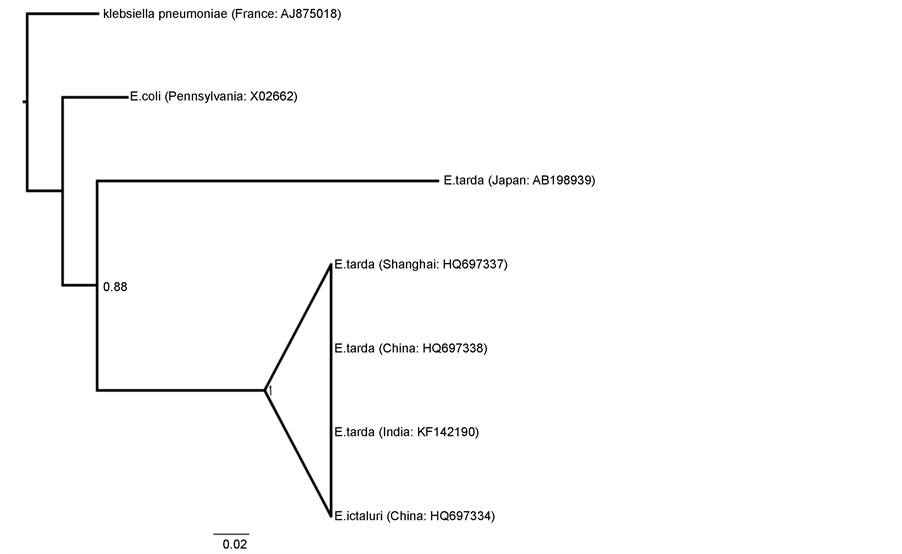
The sequence (nucleotide and amino acid) divergence values were very low between E. tarda isolates from India (KF142190) and China (HQ697337-38) (Figure 4 and Figure 5). However, the divergence value was high when compared with E. tarda isolates from Japan (AB198939) and this value was more than the Inter-specific divergence value (E. tarda-E. ictaluri, E. tarda-E. coli & E. tarda-Klebsiella species). This could be due to accumulation of more synonymous mutations in E. tarda isolates from Japan. As per the NJ tree of GAPDH sequences, Indian isolate of the E. tarda showed close clustering with other Asian countries isolates (Japan and China) (Figure 6 and Figure 7). Although GAPDH sequence of E. ictaluri clustered with E. tarda, this clustering does not have strong bootstrap value.
E. tarda is one of the most important pathogen in the context of dreadful disease in aquaculture. Because of the incapability of antibiotics and other subunit vaccines, researchers are focussing towards DNA vaccine candidates. A 37 kDa outer membrane protein was detected in several E. tarda serotype strains and was designated as an effective vaccine candidate against experimental E. tarda infection in Japanese flounder [3] . A recombinant GAPDH of E. tarda was reported to be an effective vaccine antigen against E. tarda infection in Japanese flounder [7] . The gene GAPDH is conserved in a series of bacteria including Gram-positive and Gram-negative.
Figure 3. Nucleotide sequence of 996 bp GAPDH gene of Edwardsiella tarda isolate collected from infected fish from central India, submitted to NCBI GENBANK (Accession No. KF142190).
Figure 4. Percentage nucleotide similarity and divergence based on the 996 bpnucleotide sequences of GAPDH gene from different Edwardsiella tarda isolates. Sequence in the box is the one which is analysed. The analysis is done by Clustal W algorithm. Table showing sequence distance on the basis of pairwise sequence homology.
Figure 5. Percentage nucleotide similarity and divergence based on the deduced amino acid sequences of GAPDH gene from different Edwardsiella tarda isolates. Table showing sequence distance on the basis of pairwise sequence homology of aminoacid sequence.
Figure 6. Neighbour-Joining tree (cladogram) based on deduced 996 bp nucleotide similarity of GAPDH gene from different Edwardsiella tarda isolates showing close cluster of E. tarda sequences with E. ictaluri.
So the vaccine construct is believed to work against a number of bacteria. In our study, as per the secondary structure prediction, GAPDH sequence showed high antigenic index and surface probability, which make it an ideal candidate for generating immunogenicity. This result is in agreement to the experimental work of Kawai et al. [3] .
Figure 7. Neighbour-Joining tree (cladogram) based on deduced amino acid sequence similarity of GAPDH gene of different Edwardsiella tarda isolates showing close cluster of E. tarda aminoacid sequences with E. ictaluri.
4. Conclusion
Objective of our study was to characterize the GAPDH gene of an Indian isolate of E. tarda. The study confirmed that the bacterial GAPDH gene was highly conserved and could be exploited as the potential candidate as vaccine gene against E. tarda and other Gram-negative bacteria.
Acknowledgements
Authors are thankful to Director and Vice Chancellor of the Central Institute of Fisheries Education, Mumbai Dr. W. S. Lakra, for providing facilities for research work. Authors are also thankful to Dr. Sudhir Jain, VAS, Manpur, for providing the infected samples.
References
- Hanneson, R. (2003) Aquaculture and Fisheries. Marine Policy, 27, 169-178. http://dx.doi.org/10.1016/S0308-597X(02)00083-0
- Leong, J.C. and Fryer, J.L. (1993) Viral Vaccines for Aquaculture. Annual Review of Fish Diseases, 3, 225-240. http://dx.doi.org/10.1016/0959-8030(93)90036-B
- Kawai, K., Liu, Y., Ohnishi, K. and Ohnishi, S. (2004) A Conserved 37 kDa Outer Membrane Protein of Edwardsiella tarda Is an Effective Vaccine Candidate. Vaccine, 22, 3411-3418. http://dx.doi.org/10.1016/j.vaccine.2004.02.026
- Bolton, A., Song, X.M., Willson, P., Fontaine, M.C., Potter, A.A. and Perez-Casal, J. (2004) Use of the Surface Proteins GapC and Mig of Streptococcus dysgalactiae as Potential Protective Antigens against Bovine Mastitis. Canadian Journal of Microbiology, 50, 423-432. http://dx.doi.org/10.1139/w04-016
- Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M. and Kumar, S. (2011) MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution, 28, 2731-2739. http://dx.doi.org/10.1093/molbev/msr121
- Park, S.B., Aoki, T. and Jung, T.S. (2012) Pathogenesis of and Strategies for Preventing Edwardsiella tarda Infection in Fish. Veterinary Research, 43, 67. http://dx.doi.org/10.1186/1297-9716-43-67
- Liu, Y., Oshima, S. and Kurohara, K. (2005) Vaccine Efficacy of Recombinant GAPDH of Edwardsiella tarda against Edwardsiellosis. Microbiology and Immunology, 49, 605-612. http://dx.doi.org/10.1111/j.1348-0421.2005.tb03652.x