Advances in Bioscience and Biotechnology
Vol.4 No.5A(2013), Article ID:31996,11 pages DOI:10.4236/abb.2013.45A004
Role of long non-coding RNA in cells: Example of the H19/IGF2 locus
![]()
UMR 8161 CNRS, Institut Biologie de Lille, University of Lille1, Lille, France;
Email: *eric.adriaenssens@ibl.fr
Copyright © 2013 Constance Vennin et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 28 March 2013; revised 1 May 2013; accepted 18 May 2013
Keywords: H19 Gene; Genomic Imprinting; Non-Coding RNA; Cell Cycle
ABSTRACT
In the past decade, studies of non-coding RNAs increase. Non-coding RNAs are divided in two classes: small and long non-coding RNA. It was shown that long non-coding RNAs regulate expression of 70% of genes. Long non-coding RNAs are involved in several cellular processes like epigenetic regulation, dosage compensation, alternative splicing and stem cells maintenance for example. Misregulations of their expression induce diseases such as developmental syndrome or cancer. In this review, we describe some functions of long non-coding RNA in cells. Furthermore, we study the H19/IGF2 cluster: an imprinted genomic locus located on chromosome 11p15.5. Genomic imprinting allows gene expression from a single allele in a parent-origin-dependent manner. This cluster encode for the first long non-coding RNA identified: H19. In 1990, it was established that H19 functions as a riboregulator. Recently, it was shown that H19 is a precursor of microRNA (hsa-miR-675), and several news transcripts were identified at the H19/IGF2 locus. So, the complexity of this locus increasing, in this review, we summarize our current understanding about the H19/IGF2 cluster both in terms of transcription as well as in terms of functions in cells. We highlight the involvement of H19, its new antisense transcript 91H and its microRNA, in the regulation of IGF receptor function and in cell cycle progression.
1. INTRODUCTION
Recently, the ENCODE project have proved that approximately 90% of the genome is transcribed. In cells, about 2% of sequences coding for protein, the others transcripts act as introns or non-coding RNA. These non-coding transcripts are composed of non-protein coding gene and background of transcription. This transcriptional noise would allow maintaining chromatin in a conformation favourable to the transcription of protein-coding gene. In the past decade, studies of non-protein coding gene, also called non-coding RNAs (ncRNAs), have increased and revealed that they are important in cells. NcRNAs are highly regulated and have several functions for normal development of tissues to tumorigenesis. Non-coding RNAs are divided in two classes of RNAs: the small and the long non-coding RNAs.
Small non-coding RNA (<200 nts), notably micro-RNAs (miRNAs), are involved in post-transcriptional regulation of RNA. MiRNAs are the most studied of the small ncRNAs because they regulate 60% of protein-coding gene. Mostly, they are encoded by the genome, transcribed by RNA polymerase II, cleaved by Drosha in the nucleus and by Dicer in the cytosol. Then, they interact with Argonaut protein family to form the RISC complex (RNA-induced silencing complex). This complex allows the function of miRNA [1]. They induce silencing of gene by degrading or inhibiting the translation of mRNA into protein. They are involved in all process of the cell: proliferation, cell cycle progression, differentiation, and apoptosis. Two others small non-coding RNAs are described: piRNA (PIWI-interacting RNA) and snoRNA (small nucleolar RNAs). piRNAs have recently been discovered. They are important for germline development and for the suppression of transposon activity in germline cells. SnoRNA are components of ribonucleoproteins (snoRNPs). They are responsible for posttranscriptional modifications of rRNA that take place in the nucleolus (the nucleus compartment where ribosomes are formed) [1].
Long non-coding RNAs (LncRNAs (>200 nts)) can have different origins: mRNA, antisense transcripts or long-intergenic non-coding RNA (lincRNA). LincRNAs are produced by sequence between two genes or set of genes. In Homo sapiens, 4500 lincRNAs have discovered but only 108 lncRNAs are studied and published [2]. LncRNAs have an important place in cell as they regulate expression of 70% of genes. They are involved, for example, in epigenetic regulation, alternative splice or stem cells maintenance. Misregulations of their expression induce diseases such as cancer.
The first imprinting lncRNA identified is the H19 RNA. H19 is transcript from the H19/IGF2 cluster located on the chromosome 11p15.5. Recently, the complexity of this locus has increased since several new transcripts were identified.
In this review, firstly, we describe some functions of long non-coding RNA in cells. Then, to illustrate our point, we summarize our current understanding about the H19/IGF2 cluster, its complexity and its function in cells.
2. LONG NON-CODING RNA FUNCTIONS
2.1. Dosage Compensation
The lncRNA Xist (X-inactive-specific-transcript), an lncRNA of 17.000 nucleotides, is the most studied. This lncRNA is implicated in the X chromosome inactivation in dosage compensation [3]. Xist is encoded by the X chromosome and acts in cis. Thanks to its conserved repeat motif RepA, Xist interacts with the Polycomb repressive complex (PRC2), the complex responsible for trimethyaltion of histone H3 at Lys27, and targets this complex to the XIC (X-inactivation centre). PRC2 complex induces histone modifications, heterochromatin formation and silencing of the targeted X chromosome. Xist is regulated by two others lncRNAs, one acting negatively Tsix, and the other positively Jpg. Tsix, antisense RNA to Xist, is expressed from the X-active chromosome and inhibit Xist expression in cis. When expressed, Tsix recruits DNA methyltransferases (Dnmt3a) to repress the expression of Xist, and blocks the interaction between Xist and the PRC2 complex.
2.2. Epigenetic Regulation
The lncRNA Air is submitting to the genomic imprinting. It consists of a 108 kb-long transcript. Air promoter is localized in the imprinting centre within the IGF2r gene and it is necessary for the paternal repression of the gene of the locus [4,5]. However, the molecular mechanism remains unclear and authors propose hypothesis of methylation propagation from the IGF2r gene or of repressive ARN/protein complexes formation.
The third lncRNA well studied is located in the cluster Kcnq1/Kcnq1ot1 on the chromosome 11 in position 15.5. Kcnq1ot1 RNA is a 91 kb transcript which is expressed in antisense orientation from a highly conserved and differentially methylated region Kcnq1 ICR or ICR2 present in intron 10 of Kcnq1 gene. Expression of this transcript is exclusively paternal. Indeed, the Kcnq1ot1 promoter shows a maternal specific methylation. This differential epigenetic mark is lost in patients affected by Beckwith-Wiedemann syndrome with RNA biallelic expression [6-8]. More recently, Pandey and colleagues (2004) have documented that the Kcnq1ot transcript has a key role in silencing of genes contained in the Kcnq1 gene imprinted region and that it participates directly or indirectly to the methylation but without RNA interfereence mechanisms [9]. Furthermore, interruption of Kcnq1ot1 RNA production by the insertion of a polyadenylation sequence downstream of the promoter also caused a loss of both silencing activity and methylation spreading. Thus, the antisense RNA plays a key role in the silencing function of the ICR [10].
2.3. Regulation of Alternative Splicing
The lncRNA Malat1 (Metastasis-associated lung adenocarcinoma transcript 1), also known as NEAT2 (Nuclear-enriched abundant transcript 2), is polyadenylated and overexpressed in various cancers. It is a conserved transcript among mammals of 6 - 7 kb, localized in nuclear. RNA-fish studies have shown that Malat1 is localized in sub-compartment of nuclear: nuclear speckles [11, 12]. Contrary to NEAT1 (a lncRNA essential for nuclear paraspeckle formation), Malat1 is not essential to nuclear speckle integrity. This compartment is composed in majority of factor involved in pre-mRNA splicing, like SR family protein and protein implicated in RNA transport for example. Bernard et al., have shown that Malat1 controls of SR family protein (SF2/ASF) of splicing factor to transcription site. Tripathi and co-workers have established that Malat1 regulates expression levels, localization and activity of SR protein. Targets genes of Malat1 are tissue-dependant. In neuronal cells, Malat1 regulates preferentially splicing of genes involved in synaptogenesis like Neuroligin gene (Nlgn1) and synaptic cell adhesion molecule 1 (SynCAM1).
2.4. Stem Cells Maintenance
Recently, it was observed that 133 lincRNAs were overexpressed and 104 down-regulated in ESC (Embryonic Stem Cell) or iPS (induced Pluripotent Stem Cells) compared with fibroblast [13]. They have shown that twenty-eight lincRNAs upregulated in iPSC, notably lincRNA-RoR, could be regulated by pluripotency transcription factors OCT4, SOX2 or NANOG. Depletion of lincRNA-RoR inhibits iPSC colony formation. They have proved that lincRNA-RoR promotes survival of iPSC and ESC by preventing the activation of stress pathways like p53 response. This RNA is important to reprogramming stem cells whence its name “Regulator of Reprogramming”. So, they identified the first functional lincRNA in establishing iPSC.
Furthermore, a study has identified several ncRNAs implicated in stem cells differentiation [14]. They have demonstrated that lncRNAs are associated with trimethylated H3K4 histones and histone methyltransferase MLL1. These suggest that lncRNAs have a role in epigenetic regulation during ES cell differentiation.
2.5. LncRNA in Cancer
In human, HOX transcription factor are encoded by four HOX cluster on four different chromosomes: HOXA to D. From HOX cluster only 39 transcription factors are expressed but 231 ncRNAs are transcript [15,16]. The well HOX ncRNA studied is HOTAIR (Hox antisense transcript RNA). It is antisense RNA of 2.2 kb, transcript from the HOXC cluster. Studies have shown that HOTAIR regulates expression of genes on HOXD cluster, so acts in trans. Indeed, when expressed, due to its 5’domain, HOTAIR interacts with PRC2 complex, notably Suz12 and EZH2 protein [15]. PRC2 complex induces trimethylation of Histone H3 lysine 27 on HOXD cluster (an inactive methylation). HOTAIR can also interact with the LSD1/CoREST/REST complex: a complex involved in trimethylation of Histone H3 lysine 4 (active chromatin). HOTAIR regulates chromatin conformation from active chromatin to inactive. So, it is scaffold RNA [17]. In several cancers, notably breast cancer, HOTAIR expression is associated to metastasis [18]. Authors have shown that HOTAIR overexpression increases cells invasion and metastasis in mice. They established that HOTAIR invasion is PRC2 complex dependent. So, HOTAIR expression is associated to poor prognosis.
LncRNAs are implicated in several cellular processes (epigenetic regulation, dosage compensation, stem cells self-renewal and differentiation). In some cases, lncRNA expression allows maintains of stem cells pool for example, whereas, sometimes, lncRNA expression is responsible of cancers. So, expression of lncRNA must be well regulated. They can act per se but can also acts as precursor of small ncRNA such as microRNA. The lncRNA H19, the first imprinting ncRNA discover is the precursor of the microRNA: miR-675 [19]. Recently, several groups have identified targets of this miRNA in several cell lines [20,21]. To illustrate our point, we summarize our current understanding about the H19/IGF2 cluster, its transcription complexity and its function in cells.
3. THE H19/IGF2 CLUSTER
3.1. Genomic Imprinting at the H19/IGF2 Cluster
The H19/IGF2 cluster is submitted to genomic imprinting. Genomic imprinting is a form of epigenetic gene regulation that results in expression of a single allele in a parent-of-origin-dependent manner. This form of monoallelic expression is essential for normal development. Despite extensive studies, the molecular mechanisms of genomic imprinting remain unclear. However, some hallmarks of this phenomenon have been identified and we can note that:
• Gene expression is allele-specific and tissue or stagespecific.
• Many of imprinted genes are found in clusters throughout the genome. The clusters contain two or more imprinted genes over a region that can span 1 Mb or more.
• Within each cluster, a common regulating region which are called “imprinting control region” (ICR, also called IC for imprinting Centre or ICE for imprinting control element) controls the imprinting of all genes in the cluster and can act over hundreds of kilobases. ICRs are designed as differentially methylated regions with parental-specific modifications that determine their activity. Deletions of this region lead to the loss of imprinting of multiple genes of the cluster [22,23].
• More recently, it has been reported that non-coding RNA were associated with imprinted clusters and have an essential role in regulating gene expression.
The H19/IGF2 cluster is located on the human chromosome 11 in position p15.5. This 1Mbp domain contains 9 imprinted genes and 2 independent imprinting center. The first imprinting center (ICR) regulates the cluster H19/IGF2 and the second (ICR2), the cluster Kcnq1/Kcnq1ot1.
The H19 gene is one of the first genes proven to be imprinted. This gene is co-regulated negatively with the IGF2 gene located 200kb upstream of the transcription site of the H19 gene. Indeed, H19 is expressed only from the maternal allele whereas IGF2 is expressed from the paternal allele [24]. The paternal allele exhibits several characteristics that explain the silencing of the H19 gene: it is hypermethylated in the promoter region and the promoter shows a compact chromatin structure [25,26]. Moreover, the histone acetylation rate is lower than the one of the maternal allele [27].
Surprisingly, the IGF2 promoter region is not methylated and its chromatin structure is favourable to a biallelic transcription [28]. However, two other differentially methylated regions (DMR) on the expressed paternal allele have been identified within the gene: the DMR1 located 3 kbp upstream the P1 promoter acts as a silencer on the maternal allele when it is unmethylated, and the DMR2, located within exons 5 and 6 is an activator on the paternal allele when it is methylated [29-31].
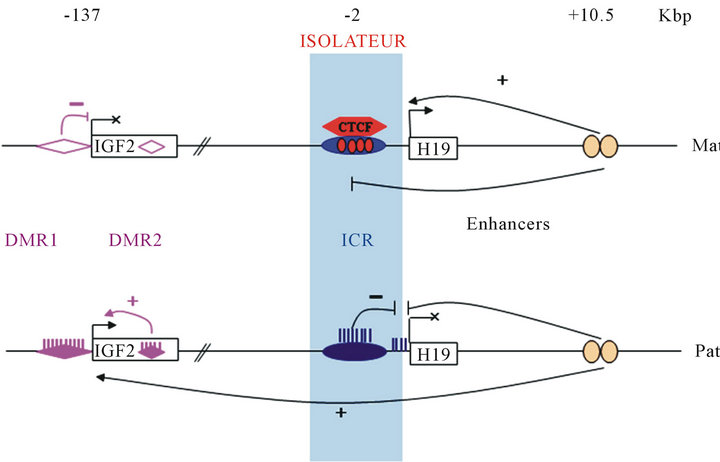
However, DNA methylation is not sufficient to explain the mono-allelic expression. Indeed, the ICR is the key of the genomic imprinting: it controls the chromatin structure and regulates the effect of enhancers located downstream of the H19 gene [32,33]. This region is located 2 to 4 kbp upstream of the transcription site of the H19 gene. In human, it contains seven binding site of zinc-finger protein named CTCF (CCTC-binding factor) but only the sixth is differentially methylated [34]. On the maternal allele, the CTCF protein interacts with non-methylated ICR due to four consensus site (Figure 1) [35]. On the ICR, this protein has a chromatin insulator function as it prevents the action of enhancers on the promoter of IGF2. On the paternal allele, methylation of the ICR represses the H19 expression and prevents the attachment of the CTCF protein [36]. So enhancers can activate the IGF2 expression from this allele. Thus, H19 is expressed from the maternal allele and IGF2 from the paternal allele (Figure 1).
Chromosome conformation capture (3C) analysis shows interaction between different chromosomal regions and suggests that the CTCF protein has a critical role in the epigenetic regulation of the cluster H19/IGF2. Kurukuti and al. 2006 demonstrated that on the maternal allele, CTCF interact with the DMR1 and the Matrix Attachment Region (MAR3) at the IGF2 locus to generate a tight loop around the IGF2 gene [37,38]. This interacttion creates an inactive domain where IGF2 is far away
![]()
Figure 1. Genomic imprinting at the H19/IGF2 locus. Activation of gene transcription is represented by (+), repression by (-) and inhibition of enhancers function by ( ). Relative positions are expressed in kilobase pairs relatively to the H19 transcription start site. Gene expression is regulated by three mechanisms: methylation, enhancers activity and insulator activity. Three DNA region are differentially methylated : DMR 1 and 2 of the IGF2 gene (violet diamond) and ICR (blue oval). On the maternal allele, the CTCF protein interacts with the non-methylated ICR (blue oval). This interaction prohibits enhancers access to the IGF2 gene. Furthermore, the DMR1 non-methylated of IGF2 gene acts as a silencer. On the paternal allele, the methylated ICR repress H19 and inhibits CTCF interaction. So, enhancers can activate IGF2 transcription. Moreover, the methylated DMR2 of IGF2 gene activate also IGF2 transcription.
from the enhancers. Therefore, this gene is in inactive domain so it cannot be expressed from this allele (Figure 2).
On the paternal allele, the methylated ICR interacts with methylated IGF2 DMR2 moving IGF2 into the active chromatin domain [39].
So genomic imprinting of the H19/IGF2 cluster is allowed by DNA methylation, chromatin composition, organization and conformation.
3.2. News Transcripts at the H19/IGF2 Locus
In 1991, an antisense transcript of the IGF2 gene in chicken was identified [40]. Others studies have identified antisense IGF2 transcripts of 3-4 kb in mouse and human (Figure 3) [41,42]. This transcript is expressed only from paternal allele and no open reading frame (ORF) was identified. Its function remains unclear, but it is a good marker for Wilm’s tumor where it is overexpressed [42]. Recently, it was shown that IGF2as is exported in the cytoplasm and associated with polysomes [43]. So, it is not impossible that IGF2as is a protein coding transcript.
We have identified a non-coding transcript, antisense to H19, that we named 91H (Figure 3) [44]. This tran-
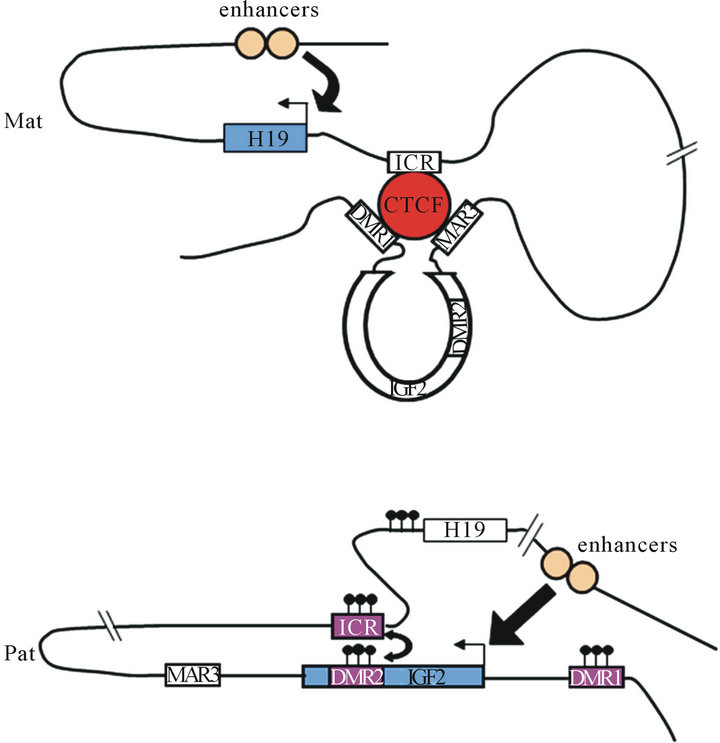
Figure 2. Chromatin loop structure at the H19/IGF2 locus. Chromosome conformation capture revealed that the CTCF protein orchestrate chromatin structure. On the maternal allele, CTCF interact with ICR but also with DMR1 of IGF2 and matrix attachment region (MAR3). These interactions create a loop around the IGF2 gene. Then, enhancers can interact only with H19 promoter and activate its expression. On the paternal allele, the methylated DMR2 of IGF2 interact with the methylated ICR allowing thus moving of IGF2 into the active chromatin domain.
![]()
![]()
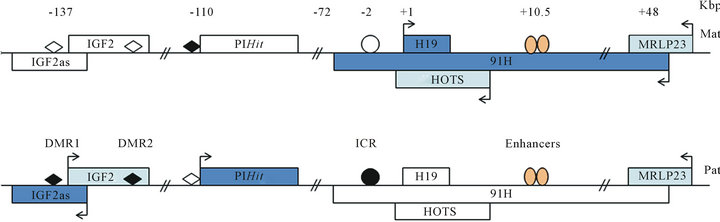
Figure 3. News transcripts at the H19/IGF2 locus. Gene expressed are in blue rectangle with arrow whereas inactive gene are in white rectangle. Protein coding gene are represented by ( ) and non-protein coding gene by ( ). Direction of arrows indicates direction of gene transcription. Relative positions are expressed in kilobase pairs relatively to the H19 transcription start site. ICR is represented by circle and DMR by diamond. Methylation of ICR and DMR is in black. Enhancers are represented by orange oval. The first antisense transcript described is IGF2as. It is ncRNA overexpressed in Wilm’s tumour. Its function remains unclear. Recently, news transcripts have been discovered at the H19/IGF2 locus. On maternal allele, a long antisense transcript to H19 was identified: it is 91H. This ncRNA and its function are conserved in mice. Neither protein was identified, so it is supposed that 91H acts as lncRNA. Recently, from the same allele, a protein coding gene imprinting has discovered: HOTS. This transcript is antisense to H19. It is possible that HOTS protein is coding by 91H RNA. The last transcript discovered from H19/IGF2 locus is PIHit. It is ncRNA expressed after birth from paternal allele.
script is a lncRNA of 120 kb expressed only in human from the maternal allele. It is known that lncRNAs can mediate epigenetic regulation transcription. Indeed, the lncRNA Xist induce X-chromosome inactivation in dosage compensation. So, we have studied effects of 91H expression at the H19/IGF2 locus. By invalidation of 91H with si-RNA, a reduction of IGF2 expression was observed. However, today, the molecular mechanism remains unclear. Recently, it was shown that 91H RNA and its function are conserved among mammals, notably in mice [45]. By 91H overexpression, they have shown that 91H regulates positively IGF2 translation from a novel promoter. More recently, a group identified antisense transcript of H19, expressed from the maternal allele, encoding for a protein named HOTS (H19 opposite tumour suppressor) [46]. But it is not excluded that this protein is encoded by the 91H transcript. Thus, today the function of the 91H transcript remains unclear.
A new paternal transcript was identified in mice. This transcript, PIHit (Paternally-expressed IGF2/H19 intergenic transcript), is coding by intergenic sequence, between IGF2 and H19, and expressed, in mice, principally 8 days after birth (Figure 3) [47]. Then, its expression decreases rapidly during the third post-natal week. It is expressed at similar level to mRNA (IGF2), capped but no polyadenylated. Neither ORF was identified, so it is supposed that it is a lncRNA. Authors have identified transcription start site but not the 3’end, which is why it is a transcript of 5 to 6 kb. Neither function has been associated to PIHit RNA. By 3C, they observed two chromatin conformation of paternal allele. They supposed that there is a dynamic system permitting IGF2 or PIHit expression. However, it cannot exclude that there is chromatin conformation cell lines specific.
4. THE H19 RNA AND ITS FUNCTION IN CELLS
4.1. The H19 RNA
The H19 gene was discovered in the mouse in 1984 and in the human in 1992 [48-50]. This gene is composed of five exons and encoded an mRNA of 2.3 kb. This RNA is transcribed by the RNA polymerase II, polyadenylated, capped and spliced with conserved secondary RNA structure. But, no conserved open reading frame was identified. Even if deletion and/or mutation produce a 26 kDa protein, no endogenous translation has so far been identified [51]. So, in 1990, Brannan et al. have proposed that H19 RNA functions as a riboregulateur of which expression is developmentally regulated [52].
It is well established that a ncRNA can be precursor of microRNA. There are different biogenesis pathways of microRNA, but generally stem-loop structure RNA are recognized by protein like DGCR8, cleaved by Drosha and Dicer to generate the duplex miR-5p/miR-3p. Then, the duplex interacts with Argonaut protein family and is incorporated in the RISC complex. MicroRNA can also be generated in Drosha or Dicer-independent pathways [53]. Introns from the splicing or tRNA (tRNA-Ile for example) can be directly recognized by Dicer, cleaved by this enzyme and incorporated in the RISC. There is a microRNA (miR-451) cleaved by Drosha which is directly recognized by Ago and incorporated in the RISC.
In 2007, Cai and Cullen have demonstrated that H19 is precursor of microRNAs: miR-675-5p and miR-675-3p [19]. They are generated by the exon1 of the gene. Today, few targets of the miR-675 have identified. Due to its microRNA, it was shown that H19 can regulate placental growth and cell cycle.
4.2. H19/91H Regulate IGF Associated Phenotypes
We and others groups have established that H19 regulates IGF2 ligand and receptor expression. Expression of H19 and IGF2 are regulated by enhancers located downstream of H19. Actions of enhancers are regulated by imprinting control region (ICR), located between H19 and IGF2. Expression of H19 and IGF2 are allele-dependent. On the maternal allele, the CTCF protein interacts with ICR non-methylated, DMR1 of IGF2 and the MAR3 domain [35,37,38]. This interaction creates a loop containing IGF2 gene. Enhancers cannot active transcription of IGF2 gene when chromatin is in this conformation. So, only H19 is expressed from the maternal allele. On the paternal allele, ICR is methylated, so the protein CTCF is absent on this allele. ICR methylated interacts with IGF2 DMR2 methylated too. This interaction allows action of enhancers on IGF2 promoter and then IGF2 expression [30]. H19 and IGF2 are in competition for enhancers. Furthermore, it was shown that deletion of H19 and its flanking region affect expression of IGF2. So, H19 and region flanking regulate IGF2 expression in cis.
Moreover, H19 is a RNA polyadenylated, spliced and exported in the cytosol. In cytosol, a group have shown that H19 is associated to polysomes [54]. These polysomes have similar size to those associated to IGF2 mRNA. Then, they have found an inverse co-regulation between H19 expression and IGF2 translation in cytosol. In Wilm’s tumor, H19-negative cells show overexpression of IGF2 3 fold higher then control. Inversely, in H19-positive cells, IGF2 expression protein was reduced. So, they hypothesized, that H19 regulates translation of IGF2 mRNA in trans. Moreover, it was shown a co-regulation between H19 and IGF2 transcription. In breast cancer cells, when H19 is overexpressed, IGF2 expression decreases severely [55,56]. H19 regulates negatively transcription of IGF2 in trans. So, it was supposed that H19 acts as a trans-riboregulateur.
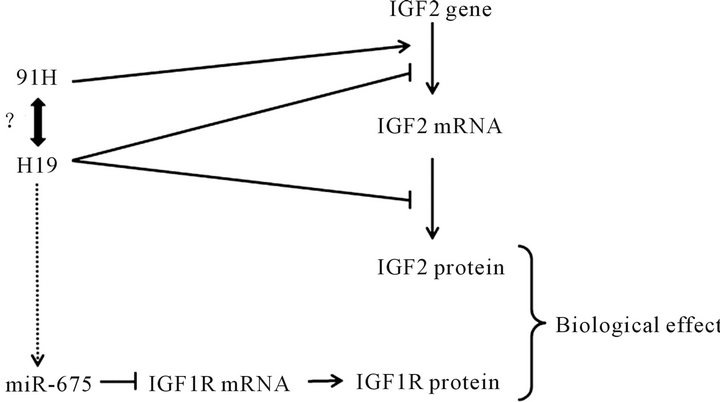
In mouse placental cells, the expression of miR-675, from H19 gene, is regulated negatively by HuR protein [21]. They observed a relation between miR-675 expression and size of placenta. Indeed, when miR-675-3p is expressed (from E11.5 until term), a reduction size of placenta is observed. This reduction is due to a decrease of cells proliferation but not an increase of apoptosis. They established that miR-675-3p interacts with two seed on 3’UTR IGF1r and inhibits its translation. So, H19 is a key regulator of IGF ligand and receptor expression (Figure 4).
Intriguingly, we have shown that the lncRNA 91H, transcript antisense to H19, affects a little H19 expression but regulates positively IGF2 [44]. However, the mechanism remains unclear. We supposed that 91H interacts with proteins that modulate expression of genes. More recently, a group have shown that 91H overexpression, in mouse, upregulates IGF2 expression [45]. They supposed that 91H activate a novel promoter of IGF2.
4.3. Cell Cycle Regulation by H19 Promotes Cancer
H19 is implicated in embryonic development. It is expressed in blastocyst stage of development and accumulated at high level in tissues of endodermal and mesodermal origins as well as ectodermal origin [57-60]. After birth, the gene is repressed in all tissues except skeletal muscle [61]. Misregulations of H19 expression during development induce developmental syndrome like Silver-Russel syndrome or Beckwith-Wiedemann syndrome [62,63]. In adulthood, function of H19 is controversial: it was supposed that H19 act as tumor suppressor or oncogene. Nevertheless, several data show that H19 act as oncogene in various cancer tissues: breast [61,64,65], uterus [66], bladder [67,68] and gastric [69]. Indeed, today, it was clearly established that H19, per se or through its microRNA, regulates different check-point of the cell cycle.
H19 mRNA generates two microRNAs: miR-675-5p and miR-675-3p [19]. The mir-675-5p is most studied but few targets have been identified. The first target identified is 3’UTR of Retinoblastoma (RB) mRNA in colon cancer cells [20]. Authors have shown a negative co-regulation between H19 and RB in human colorectal
![]()
![]()
Figure 4. Regulation of IGF by H19 and its antisense transcript 91H. Arrows represent positive regulation ( ) and vertical bar negative regulation ( ). H19 overexpression regulates negatively IGF2 transcription and translation. The lncRNA 91H regulates positively transcription of H19 and IGF2 but the mechanism is unclear. H19 is precursor of microRNA: miR- 675. MiR-675 inhibits translation of IGF1r mRNA. So, H19 is a key regulator of IGF ligand and receptor.
tumour. They demonstrated a reduced expression of RB in tumour whereas H19 and miR-675-5p are overexpressed (Figure 5). Owing to a reporter luciferase vector, they established the interaction and the negative effect of miR-675-5p on 3’UTR of RB. They have shown that miR-675-5p increases clonogenicity in soft agar of human colon cancer cells. So, in colon cancer cells lines, H19 and miR-675-5p increase proliferation of cells.
In 1998, it was shown that the H19 expression is regulated by p53 protein [70]. Indeed, the H19 promoter contains consensus site to interaction with p53. A negative regulation of p53 on this promoter was observed. In parallel, in human breast tumour, H19 is overexpressed in 70% of tumour independent of p53 expression [71]. However, it was shown that H19 is located in stromal cells whereas p53 is located in epithelial cells.
Recently, a group studying gastric cancer shows that H19 expression is increased in this disease [69]. They observed an increase cell proliferation and a reduction of apoptosis when H19 is overexpressed. So, they studied the effect of H19 overexpression on a protein inhibiting cell cycle proliferation and inducing apoptosis: the p53
![]()
![]()
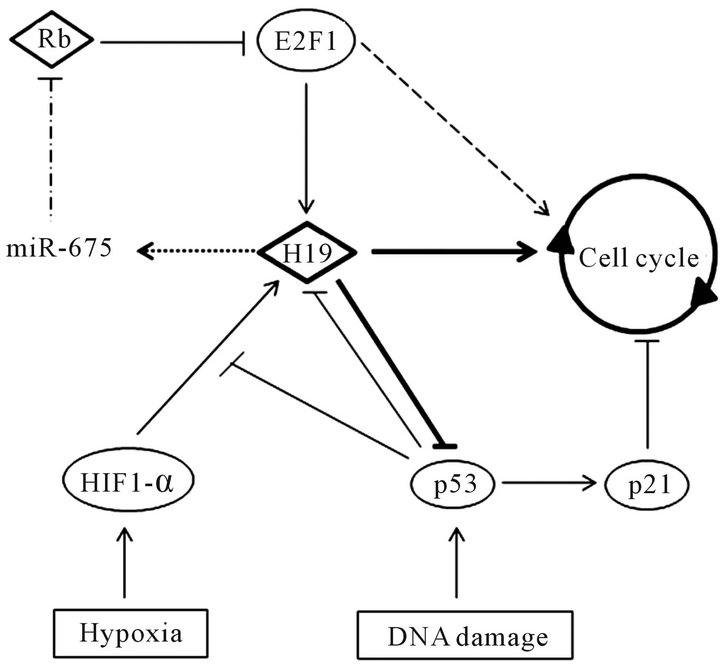
Figure 5. Regulation of cell cycle by H19. Proteins are represented by circle, RNA by diamond, cellular stress by rectangle and cell cycle by circle with arrow. Activation of gene expression is represented by arrow ( ) and repression by vertical bar ( ). Mir-675 is processed from H19 transcript and represses translation of an inhibitor cell cycle progression mRNA: Rb (Retinoblastoma). So, E2F1 protein is activated and promotes H19 overexpression and expression of genes involved in cell cycle progression. H19 overexpression facilitates G1/S transition. Hypoxia, by activating HIF1-α, induces H19 expression in p53-dependent manner. DNA damage activates the p53 protein. P53 repress H19 expression and inhibits cell cycle progression by activating p21. Recently, it was shown that H19 overexpression inhibits p53 activity. So, H19 is a key regulator of cell cycle progression.
protein. Thanks to a RNA-immunoprecipitation (RIP), they have shown that H19 RNA can interact physically with the p53 protein. By a luciferase reporter system, they demonstrated that H19 RNA regulates negatively the p53 protein may be by blocking this phosphorylation. So, the H19 overexpression in gastric cancer cells contributes to tumorigenesis by regulating p53 activation.
In tumour, some cells are in hypoxic condition. So, a team has studied effect of hypoxia on H19 expression. Upon hypoxia, they observed an increase rate of H19 RNA [72,73]. So, they have verified that the activation is due to the activation of the HIF1-α pathway (pathway activated during hypoxia). Invalidation of HIF1-α by RNA interference induces a diminution of H19 overexpression upon hypoxia. Furthermore, they have observed that H19 is overexpressed only when p53 is mutated or absent. If, p53 is not mutated, they have observed a decrease H19 expression. It has previously been reported that p53 inhibits action of HIF1-α by increasing its ubiquitination and degradation [74]. So, upon hypoxia, H19 is overexpressed by activation of HIF1-α pathway and p53-dependent manner.
Surprisingly, in breast cancer cells (MCF-7), they have observed an overexpression of H19 upon hypoxia although p53 is present. In this cell, the p53 protein is principally in the cytoplasm. So, to repress activation of transcription by HIF1-α, p53 must be in the nucleus. Taken these results together, we can hypothesize that, in MCF-7 cells, H19 interacts with the p53 protein and inhibits its activation by sequestering p53 protein in the cytoplasm.
Furthermore, it was shown that H19 facilitates cell cycle transition G1/S [75]. This check-point is regulated particularly by the E2F1 protein. The H19 promoter contains two consensus sites for this protein. It was studied the potential role of E2F1 on the H19 promoter. Using luciferase system, it was reported that E2F1 induced H19 expression through theses two sites. Moreover, the RB protein and E2F6 factor inhibit the activation of E2F1. So, E2F1 is negatively regulated by RB and E2F6. Recently, it was shown that H19, thanks to its microRNA, regulates negatively RB expression [20]. So, theses studies have demonstrated a positive feedback loop between H19 and E2F1 (Figure 5). Then, in breast cancer cells lines as BT20, T47D and MCF-7, E2F1 and H19 are overexpressed. It was shown that H19 overexpression conferred a growth advantage on cells. Indeed, an increase S-phase entry was observed when cells overexpress this gene [75]. So, H19, through a positive regulation by E2F1, active cell cycle progression and promotes growth of breast cancer cells.
To resume, it was shown that, in cancer cell lines, H19 control p53 activity, reduced translation of the RB mRNA and promotes the G1/S cell cycle transition (Figure 5). So, H19 have a key role in the regulation of cell cycle and could be implicated in cancer progression.
6. CONCLUSION
Recently, the ENCODE project have proved that 90% of the genome is transcribed, but protein-coding genes represent only 2% of transcripts. So, the genome encode for ncRNAs which have an important place in function of cells. Today, small non-coding RNAs are most studied than lncRNAs. However, lncRNAs regulate the expression of 70% of genes. So general, it is accepted that lncRNAs regulate gene expression by interacting with PRC complex chromatin modifications, including PCR2. In this review, we show that lncRNA, notably H19 RNA, may have several functions in cell without involving the complex. So, we show that lncRNAs can act per se and/or as precursor of microRNA. Furthermore, we show that lncRNAs can be involved in different cellular process. For example, the H19 RNA and its microRNA regulate negatively expression of IGF2 ligand and IGF1 receptor; and control positively cell cycle progression. So, today, it is clear that lncRNAs have function as important as protein within cellular processes.
REFERENCES
- Esteller, M. (2011) Non-coding RNAs in human disease. Nature Reviews. Genetics, 12, 861-874. doi:10.10398/nrg3074
- Knowling, S. and Morris, K.V. (2011) Non-coding RNA and antisense RNA. Nature’s trash or treasure? Biochimie, 93, 1922-1927. doi:10.1016/j.biochi.2011.07.031
- Lee, J.T. (2012) Epigenetic regulation by long non-coding RNAs. Science, 338, 1435-1439. doi:10.1126/science.1231776
- Rougeulle, C. and Heard, E. (2002) Antisense RNA in imprinting: spreading silence through Air. Trends in Genetics, 18, 434-437. doi:10.1016/S0168-9525(02)0274-X
- Sleutels, F., Zwart, R. and Barlow, D.P. (2002) The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature, 415, 810-813. doi:10.1038/415810a
- Mitsuya, K., Meguro, M., Lee, M.P., Katoh, M., Schulz, T.C., Kugoh, H., Yoshida, M.A., Niikawa, N., Feinberg, A.P. and Oshimura, M. (1999) LIT1, an imprinted antisense RNA in the human KvLQT1 locus identified by screening for differentially expressed transcripts using monochromosomal hybrids. Human Molecular Genetics, 8, 1209-1217. doi:10.1093/hmg/8.7.1209
- Lee, M.P., DeBaun, M.R., Mitsuya, K., Galonek, H.L., Brandenburg, S., Oshimura, M. and Feinberg, A.P. (1999) Loss of imprinting of a paternally expressed transcript, with antisense orientation to KVLQT1, occurs frequently in Beckwith-Wiedemann syndrome and is independent of insulin-like growth factor II imprinting. Proceedings of the National Academy of Sciences of the USA, 96, 5203- 5208. doi:10.1073/pnas.96.9.5203
- Du, M., Zhou, W., Beatty, L.G., Weksberg, R. and Sadowski, P.D. (2004) The KCNQ1OT1 promoter, a key regulator of genomic imprinting in human chromosome 11p15.5. Genomics, 84, 288-300. doi:10.1016/j.ygeno.2004.03.008
- Pandey, R.R., Ceribelli, M., Singh, P.B., Ericsson, J., Mantovani, R. and Kanduri, C. (2004) NF-Y regulates the antisense promoter, bidirectional silencing, and differential epigenetic marks of the Kcnq1 imprinting control region. Journal of Biological Chemistry, 279, 52685-52693. doi:10.1074/jbc.M408084200
- Thakur, N., Tiwari, V.K., Thomassin, H., Pandey, R.R., Kanduri, M., Göndör, A., Grange, T., Ohlsson, R. and Kanduri, C. (2004) An antisense RNA regulates the bidirectional silencing property of the Kcnq1 imprinting control region. Molecular and Cellular Biology, 24, 7855- 7862. doi:10.1128/MCB.24.18.7855-7862.2004
- Bernard, D., Prasanth, K.V., Tripathi, V., Colasse, S., Nakamura, T., Xuan, Z., Zhang, M.Q., Sedel, F., Jourdren, L., Coulpier, F., et al. (2010) A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO Journal, 29, 3082-3093. doi:10.1038/emboj.2010.199
- Tripathi, V., Ellis, J.D., Shen, Z., Song, D.Y., Pan, Q., Watt, A.T., Freier, S.M., Bennett, C.F., Sharma, A., Bubulya, P.A., et al. (2010) The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Molecular Cell, 39, 925-938. doi:10.1016/j.molcel.2010.08.011
- Loewer, S., Cabili, M.N., Guttman, M., Loh, Y.-H., Thomas, K., Park, I.H., Garber, M., Curran, M., Onder, T., Agarwal, S., et al. (2010) Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nature Genetics, 42, 1113-1117. doi:10.1038/ng.710
- Dinger, M.E., Amaral, P.P., Mercer, T.R., Pang, K.C., Bruce, S.J., Gardiner, B.B., Askarian-Amiri, M.E., Ru, K., Soldà, G., Simons, C., et al. (2008) Long non-coding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Research, 18, 1433-1445. doi:10.1101/gr.078378.108
- Rinn, J.L., Kertesz, M., Wang, J.K., Squazzo, S.L., Xu, X., Brugmann, S.A., Goodnough, L.H., Helms, J.A., Farnham, P.J., Segal, E., et al. (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by non-coding RNAs. Cell, 129, 1311-1323. doi:10.1016/j.cell.2007.05.022
- Tano, K. and Akimitsu, N. (2012) Long non-coding RNAs in cancer progression. Frontiers in Genetics, 3, 219. doi:10.3389/fgene.2012.00219
- Shore, A.N., Herschkowitz, J.I. and Rosen, J.M. (2012) Non-coding RNAs Involved in Mammary Gland Development and Tumorigenesis: There’s a Long Way to Go. Journal of Mammary Gland Biology and Neoplasia, 17, 43-58. doi:10.1007/s10911-012-9247-3
- Gupta, R.A., Shah, N., Wang, K.C., Kim, J., Horlings, H.M., Wong, D.J., Tsai, M.-C., Hung, T., Argani, P., Rinn, J.L., et al. (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature, 464, 1071-1076. doi:10.1038/nature08975
- Cai, X. and Cullen, B.R. (2007) The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA, 13, 313-316. doi:10.1261/rna.351707
- Tsang, W.P., Ng, E.K.O., Ng, S.S.M., Jin, H., Yu, J., Sung, J.J.Y. and Kwok, T.T. (2010) Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis, 31, 350-358. doi:10.1093/carcin/bgp181
- Keniry, A., Oxley, D., Monnier, P., Kyba, M., Dandolo, L., Smits, G. and Reik, W. (2012) The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and IGF1r. Nature Cell Biology, 14, 659-665.
- Leighton, P.A., Ingram, R.S., Eggenschwiler, J., Efstratiadis, A. and Tilghman, S.M. (1995) Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature, 375, 34-39. doi:10.1038/375034a0
- Ripoche, M.A., Kress, C., Poirier, F. and Dandolo, L. (1997) Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes and Development, 11, 1596-1604. doi:10.1101/gad.11.12.1596
- DeChiara, T.M., Robertson, E.J. and Efstratiadis, A. (1991) Parental imprinting of the mouse insulin-like growth factor II gene. Cell, 64, 849-859. doi:10.1016/0092-8674(91)90513-X
- Bartolomei, M.S., Webber, A.L., Brunkow, M.E. and Tilghman, S.M. (1993) Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes and Development, 7, 1663-1673. doi:10.1101/gad.7.9.1663
- Ferguson-Smith, A.C., Sasaki, H., Cattanach, B.M. and Surani, M.A. (1993) Parental-origin-specific epigenetic modification of the mouse H19 gene. Nature, 362, 751- 755. doi:10.1038/362751a0
- Grandjean, V., O’Neill, L., Sado, T., Turner, B. and Ferguson-Smith, A. (2001) Relationship between DNA methylation, histone H4 acetylation and gene expression in the mouse imprinted IGF2-H19 domain. FEBS Letters, 488, 165-169. doi:10.1016/S0014-5793(00)02349-8
- Sasaki, H., Jones, P.A., Chaillet, J.R., Ferguson-Smith, A.C., Barton, S.C., Reik, W. and Surani, M.A. (1992) Parental imprinting: potentially active chromatin of the repressed maternal allele of the mouse insulin-like growth factor II (IGF2) gene. Genes and Development, 6, 1843- 1856. doi:10.1101/gad.6.10.1843
- Feil, R., Walter, J., Allen, N.D. and Reik, W. (1994) Developmental control of allelic methylation in the imprinted mouse IGF2 and H19 genes. Development, 120, 2933-2943.
- Murrell, A., Heeson, S., Bowden, L., Constância, M., Dean, W., Kelsey, G. and Reik, W. (2001) An intragenic methylated region in the imprinted IGF2 gene augments transcription. EMBO Reports, 2, 1101-1106. doi:10.1093/embo-reports/kve248
- Constância, M., Dean, W., Lopes, S., Moore, T., Kelsey, G. and Reik, W. (2000) Deletion of a silencer element in IGF2 results in loss of imprinting independent of H19. Nature Genetics, 26, 203-206. doi:10.1038/79930
- Tremblay, K.D., Duran, K.L. and Bartolomei, M.S. (1997) A 5’ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Molecular and Cellular Biology, 17, 4322- 4329.
- Drewell, R.A., Arney, K.L., Arima, T., Barton, S.C., Brenton, J.D. and Surani, M.A. (2002a) Novel conserved elements upstream of the H19 gene are transcribed and act as mesodermal enhancers. Development, 129, 1205- 1213.
- Takai, D., Gonzales, F.A., Tsai, Y.C., Thayer, M.J. and Jones, P.A. (2001) Large scale mapping of methylcytosines in CTCF-binding sites in the human H19 promoter and aberrant hypomethylation in human bladder cancer. Human Molecular Genetics, 10, 2619-2626. doi:10.1093/hmg/10.23.2619
- Hark, A.T., Schoenherr, C.J., Katz, D.J., Ingram, R.S., Levorse, J.M. and Tilghman, S.M. (2000) CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/IGF2 locus. Nature, 405, 486-489. doi:10.1038/35013106
- Srivastava, M., Hsieh, S., Grinberg, A., Williams-Simons, L., Huang, S.P. and Pfeifer, K. (2000) H19 and IGF2 monoallelic expression is regulated in two distinct ways by a shared cis-acting regulatory region upstream of H19. Genes and Development, 14, 1186-1195.
- Murrell, A., Heeson, S. and Reik, W. (2004) Interaction between differentially methylated regions partitions the imprinted genes IGF2 and H19 into parent-specific chromatin loops. Nature Genetics, 36, 889-893.
- Kurukuti, S., Tiwari, V.K., Tavoosidana, G., Pugacheva, E., Murrell, A., Zhao, Z., Lobanenkov, V., Reik, W. and Ohlsson, R. (2006) CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to IGF2. Proceedings of the National Academy of Sciences of the USA, 103, 10684-10689. doi:10.1073/pnas.0600326103
- Dekker, J., Rippe, K., Dekker, M. and Kleckner, N. (2002) Capturing chromosome conformation. Science, 295, 1306- 1311. doi:10.1126/science.1067799
- Taylor, E.R., Seleiro, E.A. and Brickell, P.M. (1991) Identification of antisense transcripts of the chicken insulin-like growth factor-II gene. Journal of Molecular Endocrinology, 7, 145-154. doi:10.1677/jme.0.0070145
- Moore, T., Constancia, M., Zubair, M., Bailleul, B., Feil, R., Sasaki, H. and Reik, W. (1997) Multiple imprinted sense and antisense transcripts, differential methylation and tandem repeats in a putative imprinting control region upstream of mouse IGF2. Proceedings of the National Academy of Sciences USA, 94, 12509-12514. doi:10.1073/pnas.94.23.12509
- Okutsu, T., Kuroiwa, Y., Kagitani, F., Kai, M., Aisaka, K., Tsutsumi, O., Kaneko, Y., Yokomori, K., Surani, M.A., Kohda, T., et al. (2000) Expression and imprinting status of human PEG8/IGF2as, a paternally expressed antisense transcript from the IGF2 locus, in Wilms’ tumors. Journal of Biochemistry, 127, 475-483. doi:10.1093/oxfordjournals.jbchem.a022630
- Duart-Garcia, C. and Braunschweig, M.H. (2013) The IGF2as transcript is exported into cytoplasm and associated with polysomes. Biochemical Genetics, 51, 119-130. doi:10.1007/s10528-012-9547-8
- Berteaux, N., Aptel, N., Cathala, G., Genton, C., Coll, J., Daccache, A., Spruyt, N., Hondermarck, H., Dugimont, T., Curgy, J.-J., et al. (2008) A novel H19 antisense RNA overexpressed in breast cancer contributes to paternal IGF2 expression. Molecular and Cellular Biology, 28, 6731-6745. doi:10.1128/MCB.02103-07
- Tran, V.G., Court, F., Duputié, A., Antoine, E., Aptel, N., Milligan, L., Carbonell, F., Lelay-Taha, M.-N., Piette, J., Weber, M., et al. (2012) H19 antisense RNA can up-regulate IGF2 transcription by activation of a novel promoter in mouse myoblasts. PLoS ONE, 7, e37923. doi:10.1371/journal.pone.0037923
- Onyango, P. and Feinberg, A.P. (2011) A nucleolar protein, H19 opposite tumor suppressor (HOTS), is a tumor growth inhibitor encoded by a human imprinted H19 antisense transcript. Proceedings of the National Academy of Sciences of the USA, 108, 16759-16764. doi:10.1073/pnas.1110904108
- Court, F., Baniol, M., Hagege, H., Petit, J.S., Lelay-Taha, M.-N., Carbonell, F., Weber, M., Cathala, G. and Forne, T. (2011) Long-range chromatin interactions at the mouse IGF2/H19 locus reveal a novel paternally expressed long non-coding RNA. Nucleic Acids Research, 39, 5893-5906. doi:10.1093/nar/gkr209
- Pachnis, V., Belayew, A. and Tilghman, S.M. (1984) Locus unlinked to alpha-fetoprotein under the control of the murine raf and Rif genes. Proceedings of the National Academy of Sciences of the USA, 81, 5523-5527. doi:10.1073/pnas.81.17.5523
- Pachnis, V., Brannan, C.I. and Tilghman, S.M. (1988) The structure and expression of a novel gene activated in early mouse embryogenesis. The EMBO Journal, 7, 673- 681.
- Zhang, Y. and Tycko, B. (1992) Monoallelic expression of the human H19 gene. Nature Genetics, 1, 40-44. doi:10.1038/ng0492-40
- Joubel, A., Curgy, J.J., Pelczar, H., Begue, A., Lagrou, C., Stehelin, D. and Coll, J. (1996) The 5’ part of the human H19 RNA contains cis-acting elements hampering its translatability. Cellular and Molecular Biology, 42, 1159- 1172.
- Brannan, C.I., Dees, E.C., Ingram, R.S. and Tilghman, S.M. (1990) The product of the H19 gene may function as an RNA. Molecular and Cellular Biology, 10, 28-36.
- Yang, J.-S. and Lai, E.C. (2011) Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Molecular Cell, 43, 892-903. doi:10.1016/j.molcel.2011.07.024
- Li, Y.M., Franklin, G., Cui, H.M., Svensson, K., He, X.B., Adam, G., Ohlsson, R. and Pfeifer, S. (1998) The H19 transcript is associated with polysomes and may regulate IGF2 expression intrans. Journal of Biological Chemistry, 273, 28247-28252. doi:10.1074/jbc.273.43.28247
- Wilkin, F., Paquette, J., Ledru, E., Hamelin, C., Pollak, M., Deal, C.L. and Mamelin, C. (2000) H19 sense and antisense transgenes modify insulin-like growth factor-II mRNA levels. European Journal of Biochemistry, 267, 4020-4027. doi:10.1046/j.1432-1327.2000.01438.x
- Lottin, S., Adriaenssens, E., Dupressoir, T., Berteaux, N., Montpellier, C., Coll, J., Dugimont, T. and Curgy, J.J. (2002) Overexpression of an ectopic H19 gene enhances the tumorigenic properties of breast cancer cells. Carcinogenesis, 23, 1885-1895. doi:10.1093/carcin/23.11.1885
- Poirier, F., Chan, C.T., Timmons, P.M., Robertson, E.J., Evans, M.J. and Rigby, P.W. (1991) The murine H19 gene is activated during embryonic stem cell differentiation in vitro and at the time of implantation in the developing embryo. Development, 113, 1105-1114.
- Lustig, O., Ariel, I., Ilan, J., Lev-Lehman, E., De-Groot, N. and Hochberg, A. (1994) Expression of the imprinted gene H19 in the human fetus. Molecular Reproduction and Development, 38, 239-246. doi:10.1002/mrd.1080380302
- Ohlsson, R., Hedborg, F., Holmgren, L., Walsh, C. and Ekström, T.J. (1994) Overlapping patterns of IGF2 and H19 expression during human development: Biallelic IGF2 expression correlates with a lack of H19 expression. Development, 120, 361-368.
- [61] Hemberger, M., Redies, C., Krause, R., Oswald, J., Walter, J. and Fundele, R.H. (1998) H19 and IGF2 are expressed and differentially imprinted in neuroecto-dermderived cells in the mouse brain. Development Genes and Evolution, 208, 393-402.doi:10.1007/s004270050195
- [62] Douc-Rasy, S., Coll, J., Barrois, M., Joubel, A., Prost, S., Dozier, C., Stéhelin, D. and Riou, G. (1993) Expression of the human fetal BAC/H19 gene in invasive cancer. International Journal of Oncology, 2, 753-758.
- [63] Okamoto, K., Morison, I.M., Taniguchi, T. and Reeve, A.E. (1997) Epigenetic changes at the insulin-like growth factor II/H19 locus in developing kidney is an early event in Wilms tumorigenesis. Proceedings of the National Academy of Sciences of the USA, 94, 5367-5371. doi:10.1073/pnas.94.10.5367
- [64] Steenman, M.J., Rainier, S., Dobry, C.J., Grundy, P., Horon, I.L. and Feinberg, A.P. (1994) Loss of imprinting of IGF2 is linked to reduced expression and abnormal methylation of H19 in Wilms’ tumour. Nature Genetics, 7, 433-439. doi:10.1038/ng0794-433
- [65] Dugimont, T., Curgy, J.J., Wernert, N., Delobelle, A., Raes, M.B., Joubel, A., Stehelin, D. and Coll, J. (1995) The H19 gene is expressed within both epithelial and stromal components of human invasive adenocarcinomas. Biology of the Cell, 85, 117-124. doi:10.1016/0248-4900(96)85272-5
- [66] Adriaenssens, E., Lottin, S., Dugimont, T., Fauquette, W., Coll, J., Dupouy, J.P., Boilly, B. and Curgy, J.J. (1999) Steroid hormones modulate H19 gene expression in both mammary gland and uterus. Oncogene, 18, 4460-4473. doi:10.1038/sj.onc.1202819
- [67] Ariel, I., Ayesh, S., Perlman, E.J., Pizov, G., Tanos, V., Schneider, T., Erdmann, V.A., Podeh, D., Komitowski, D., Quasem, A.S., et al. (1997) The product of the imprinted H19 gene is an oncofetal RNA. Molecular Pathology, 50, 34-44. doi:10.1136/mp.50.1.34
- [68] Ariel, I., Lustig, O., Schneider, T., Pizov, G., Sappir, M., De-Groot, N. and Hochberg, A. (1995) The imprinted H19 gene as a tumor marker in bladder carcinoma. Urology, 45, 335-338. doi:10.1016/0090-4295(95)80030-1
- [69] Elkin, M., Shevelev, A., Schulze, E., Tykocinsky, M., Cooper, M., Ariel, I., Pode, D., Kopf, E., De Groot, N. and Hochberg, A. (1995). The expression of the imprinted H19 and IGF2 genes in human bladder carcinoma. FEBS Letters, 374, 57-61. doi:10.1016/0014-5793(95)01074-O
- [70] Yang, F., Bi, J., Xue, X., Zheng, L., Zhi, K., Hua, J. and Fang, G. (2012) Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS Journal, 279, 3159-3165. doi:10.1111/j.1742-4658.2012.08694.x
- [71] Dugimont, T., Montpellier, C., Adriaenssens, E., Lottin, S., Dumont, L., Iotsova, V., Lagrou, C., Stéhelin, D., Coll, J. and Curgy, J.J. (1998) The H19 TATA-less promoter is efficiently repressed by wild-type tumor suppressor gene product p53. Oncogene, 16, 2395-2401. doi:10.1038/sj.onc.1201742
- [72] Adriaenssens, E., Dumont, L., Lottin, S., Bolle, D., Leprêtre, A., Delobelle, A., Bouali, F., Dugimont, T., Coll, J. and Curgy, J.J. (1998) H19 overexpression in breast adenocarcinoma stromal cells is associated with tumor values and steroid receptor status but independent of p53 and Ki-67 expression. American Journal of Pathology, 153, 1597-1607. doi:10.1016/S0002-9440(10)65748-3
- [73] Matouk, I.J., DeGroot, N., Mezan, S., Ayesh, S., Abu-lail, R., Hochberg, A. and Galun, E. (2007) The H19 noncoding RNA is essential for human tumor growth. PLoS ONE, 2, e845. doi:10.1371/journal.pone.0000845
- [74] Matouk, I.J., Mezan, S., Mizrahi, A., Ohana, P., Abu-Lail, R., Fellig, Y., Degroot, N., Galun, E. and Hochberg, A. (2010) The oncofetal H19 RNA connection: Hypoxia, p53 and cancer. Biochimica et Biophysica Acta, 1803, 443-451.
- [75] Ravi, R., Mookerjee, B., Bhujwalla, Z.M., Sutter, C.H., Artemov, D., Zeng, Q., Dillehay, L.E., Madan, A., Semenza, G.L. and Bedi, A. (2000) Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1alpha. Genes and Development, 14, 34-44.
- [76] Berteaux, N., Lottin, S., Monté, D., Pinte, S., Quatannens, B., Coll, J., Hondermarck, H., Curgy, J.-J., Dugimont, T. and Adriaenssens, E. (2005) H19 mRNA-like non-coding RNA promotes breast cancer cell proliferation through positive control by E2F1. Journal of Biological Chemistry, 280, 29625-29636. doi:10.1074/jbc.M504033200
NOTES
*Corresponding author.

