Advances in Bioscience and Biotechnology
Vol.4 No.1(2013), Article ID:26922,7 pages DOI:10.4236/abb.2013.41004
Biological implications of 2-chlorocyclohexa-2,5-diene-1,4-dione toward ribonuclease A
![]()
Department of Chemistry, University of Tennessee at Chattanooga, Chattanooga, USA
Email: jisook-kim@utc.edu
Received 22 October 2012; revised 24 November 2012; accepted 9 January 2013
Keywords: Chlorobenzoquinone; PAH; Protein Modification; Ribonuclease A
ABSTRACT
2-Chlorocyclohexa-2,5-diene-1,4-dione (CBQ) or 2- chloro1,4-benzquinone is one of the common metabolites of polycyclic aromatic hydrocarbons generated through industrial processes. This report describes the biological effects of CBQ toward ribonuclease A (RNase). We also investigated the inhibition of RNase modifications and the reactivity of CBQ toward selected amino acids. The study was carried out by incubating RNase or amino acids with CBQ in a concentrationand a time-dependent manner at 37˚C and pH 7.0. SDS-PAGE results showed oligomerization as well as polymeric aggregation of RNase when incubated with CBQ as early as in 10 min. CBQ-induced RNase modifications were inhibited in the presence of NADH or ascorbic acid. CBQ reactivity toward selected amino acids was also evaluated by determining the second-order rate constants for the reactions of CBQ with selected amino acids. It was found that the reactivity toward CBQ decreased in the order of lysine > threonine > serine >> aspartate > cysteine.
1. INTRODUCTION
Chlorinated benzenes (CBs) and their metabolites have been the focus of biochemical and toxicological studies since CBs are commonly used in industrial processes [1-6]. Chlorobenzene is known for targeting reproductive tissues and the nervous system [3,4]. p-Dichlorobenzene, a component for pesticides and mothballs, was found to be associated with neurotoxic effects and carcinogenicity [5,6]. The degradation pathways of CBs were found to be similar to that of polycyclic aromatic hydrocarbons (PAHs) leading to the formation of quinone metabolites [7-11]. Specifically, Li and coworkers carried out an extensive mechanism study on the formation of various halobenzoquinones including chlorobenzoquinones generated as the by-products of drinking water disinfection and the intermediates formed in waste water by oxidants [8,9]. Shen and coworkers revealed that the photocatalytic degradation of chlorophenols involves the formation of chlorobenzoquinones [10,11].
The details on how CBs and chlorobenzoquinones exhibit their toxicity remains to be studied, however one can postulate a mechanism similar to the well-studied case for PAHs [12-16]. The toxicity of PAHs is thought to arise from their metabolites, PAH quinones [13]. PAH quinones are expected to react with cellular proteins and nucleic acids, thus leading to cell toxicity [14-16]. Many studies concentrated primarily on the action of chlorobenzoquinones modifying DNA or the key components of DNA [13,17,18]. Li and coworkers studied the binding interactions of various halobenzoquinones and oligonucleotides [17], while Sturla and coworkers characterized a deoxyguanosine adduct of tetrachlorobenzoquinone [18]. In contrast to chlorobenzoquinones’ effect on DNA, only few studies are available regarding chlorobenzoquinone-induced protein modifications [19-21]. Hanzlik and coworkers pioneered the investigation on protein modifications induced by a quinone-derived bromobenzene, suggesting the mechanism of the quinone’s action as adduct formation [19,20]. Krajewska and coworkers carried out a thorough investigation on the arylation and oxidation of thiol groups in urease through the actions of four quinones including tetrachloro-1,4- benzoquinone [21].
Accordingly, our laboratory has investigated the nature of protein modifications induced by 2-chlorocyclohexa-2,5-diene-1,4-dione (CBQ) known as 2-chloro-1, 4-benzoquinone using ribonuclease A (RNase). As shown in Figure 1, CBQ can modify a protein in three different ways. First, CBQ can induce cross-linking to form protein oligomers. This involves the oxidation of a lysine to an allysine (i.e., aldehyde containing lysine) followed by the condensation of the formed allysine with
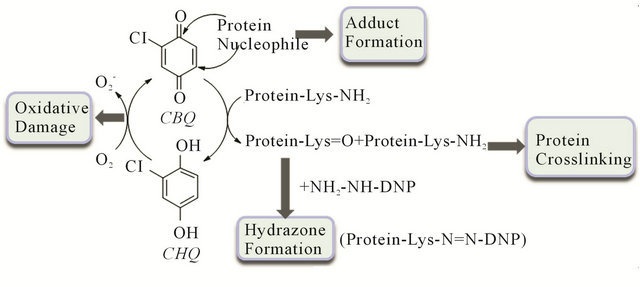
Figure 1. Proposed scheme for the action of CBQ. DNP, dinitrophenylhydrazine.
an intact lysine. Second, redox-cycling between CBQ and 2-chloro-hydroquinone (CHQ) may lead to the formation of reactive oxygen species (ROS) which can be fatal to biological molecules. Third, CBQ can attack a protein and form an adduct with a nucleophilic amino acid in the protein. For the present study, RNase was chosen as a model protein since it is a commonly used protein for studying protein modifications [22,23]. We utilized simple and non-invasive approaches such as gel electrophoresis (i.e., SDS-PAGE) and UV-Vis spectroscopy coupled with a hydrazone trapping method to visualize changes in the molecular weight (MW) of RNase and to monitor the adduct formation between RNase and CBQ. In particular, hydrazone trapping experiments were carried out to monitor the level of oxidized lysine residues [24], and inhibition studies were carried out to see the interplay of reducing agents and redox-cycling of CBQ/CHQ in the presence of NADH and ascorbic acid (AS). In addition, the reactivity of CBQ toward selected amino acids was evaluated kinetically by determining the second-order rate constants with lysine (Lys), serine (Ser), threonine (Thr), aspartate (Asp), and cysteine (Cys).
2. MATERIALS AND METHODS
All chemicals were purchased from Fisher and of reagent grade unless specified otherwise. The water used in the study was Milli-Q water purified by a Millipore system (Milli-Q water). Ribonuclease A (from bovine pancreas) was purchased from Sigma. Electrophoresis units for minigels were purchased from Fisher. Dialysis was carried out using a 3-mL Float-A-Lyzer with a molecular weight (MW) cutoff of 3.5 kDa, which was purchased from Spectrum Laboratories. Spin column was purchased from Millipore (Millipore Microcon-YM3) with a MW cutoff of 3 kDa.
2.1. RNase Modifications Detected by 1D-SDS-PAGE
Aliquots of RNase stock solution were diluted to have a final concentration of 0.145 mM RNase and treated with CBQ (0.50, 1.0, and 5.0 mM) in phosphate buffer (pH 7.0, 50 mM). Each solution was then incubated at 37˚C for various time periods, followed by immediate cooling at 0˚C. The samples above were submitted to trichloroacetic acid (TCA) precipitation according to the published procedure [23]. Then, the TCA-treated samples were submitted to SDS-PAGE analysis. For the SDS-PAGE analysis, all proteins were separated on a 10% SDSPAGE gel according to the method of Laemmli [25]. Pierce Blue Prestained Protein Molecular Weight Marker Mix (Cat. # 26681) was used as a reference to determine MW of protein bands [26]. Protein bands were visualized by staining the gels with 0.1% Coomassie brilliant blue R-250 blue.
2.2. Inhibition of RNase Modifications Detected by SDS-PAGE
RNase (0.145 mM) was incubated with CBQ (5.0 mM) in the presence of NADH or (L)-ascorbic acid (AS) at 0, 1.0, 5.0, 10, and 50 mM, respectively, in phosphate buffer (pH 7.0, 50 mM) at 37˚C for 60 min. Then, each reaction mixture was submitted to SDS-PAGE analysis according to the procedure described above.
2.3. RNase Modifications Detected by UV-Vis Spectroscopy
UV-Vis spectra were obtained using Biospec-1601 spectrophotometer by Shimadzu and UV Probe 2.3 Software with a jacketed (temperature controlled) cell compartment to maintain the physiological temperature. The reaction of CBQ (1.0 mM) and RNase (0.012 mM) was initiated by adding an aliquot of a freshly prepared CBQ stock solution to an RNase-containing solution that was equilibrated at 37˚C for 15 min. Then, the reaction mixture was dialyzed against phosphate buffer (pH 7.0, 50 mM, 600 mL × 3 per 3 mL sample) at 4˚C for 4 h. The dialyzed samples in a 1 cm-path length quartz cuvette were submitted to UV-Vis scanning. The unmodified RNase was subjected to the same procedure except the addition of CBQ.
2.4. Reactions of the Modified RNase with 2,4-Dinitrophenylhydrazine
2,4-dinitrophenylhydrazine (DNP) stock solution was prepared according to the published procedure [24]. An aliquot of the post-dialyzed RNase was incubated with an equal volume (0.50 mL) of 10 mM DNP solution at room temperature for 30 min. Then, an aliquot (0.25 mL) of the DNP-treated sample above was loaded into a Millipore spin column and submitted to centrifugation at 12,000 rpm for 2 h at 4˚C to remove liquid and excess DNP. The residue was recovered from the spin column and re-suspended in 3 mL of guanidine (6 M) solution prior to the submission to UV-Vis scanning.
2.5. Reactions of CBQ and Amino Acids Detected by UV-Vis Spectroscopy
The reactions of CBQ (0.010 mM) and selected amino acids (20 mM) were monitored by UV-Vis spectroscopy under pseudo-first-order conditions with excess amino acids in phosphate buffer (pH 7.0, 50 mM) in a 1-cm path-length quartz cuvette. The reaction was initiated by adding an aliquot of a freshly prepared CBQ stock solution to the amino acid-containing solution which was equilibrated at 37˚C for 10 min, and the reaction mixture was submitted to UV-Vis scanning.
2.6. Kinetic Analysis for the Reactions of CBQ with Amino Acids
The pseudo-first-order rate constants (kobs) of the reactions of CBQ and selected amino acids were calculated from the rate of a decrease in the intensity of the absorption due to CBQ using the first-order rate equation:
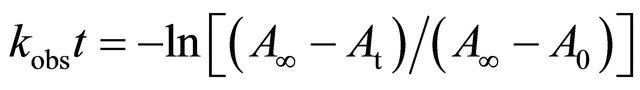 where t is time (min), A∞ is absorbance at infinity which was obtained at 8× half-life or longer, At is absorbance at time t, and A0 is absorbance at the initial time point. The disappearance of CBQ in the reaction was monitored by following the change in absorbance at 255 nm (A255) recorded at Shimadzu UV-Vis spectrophotometer. The pseudo-first-order rate constants were normalized to the second-order rate constants (in units of M−1·s−1) by dividing by the used amino acid concentration.
where t is time (min), A∞ is absorbance at infinity which was obtained at 8× half-life or longer, At is absorbance at time t, and A0 is absorbance at the initial time point. The disappearance of CBQ in the reaction was monitored by following the change in absorbance at 255 nm (A255) recorded at Shimadzu UV-Vis spectrophotometer. The pseudo-first-order rate constants were normalized to the second-order rate constants (in units of M−1·s−1) by dividing by the used amino acid concentration.
3. RESULTS
3.1. RNase Modifications Detected by 1D-SDS-PAGE
To evaluate the efficiency of CBQ in terms of protein cross-linking, RNase (0.145 mM) was incubated with CBQ (0.50, 1.0, 5.0 mM) at various time periods, and the reaction mixtures were visualized by SDS-PAGE. For the 0.50 mM CBQ-treated RNase, three protein bands appeared with the determined MWs of 33 kDa, 53 kDa, and 70 kDa in addition to the intact RNase band at 14 kDa (Figure 2(a)). As CBQ concentration increased to 1.0 mM, a smearing band appeared in the region of high MW. The formation of the smearing band became evident after 120 min (Figure 2(b)). For the 5.0 mM CBQtreated RNase, the multiple bands at the lower MW range and the smearing band at the higher MW range appeared as early as in 10 min of incubation (Figures 2(c) and (d)).
3.2. Inhibition of RNase Modifications Detected by 1D-SDS-PAGE
In order to examine the effect of common antioxidants on the redox-cycling of CBQ/CHQ, RNase and CBQ were reacted in the absence and presence of NADH or AS at pH 7.0 and 37˚C for 60 min. For the unmodified RNase, a single band appeared at the bottom of the gel (Figures 3(a) and (b), first lane). At 5.0 mM of CBQ and various amounts of NADH, multiple bands including a smearing band appeared except at the higher amount of NADH. A similar feature was observed when AS was present instead of NADH (Figure 3(b)). Figure 3(c) shows the solutions of the same incubation reactions as the series from Figure 3(b). As the concentrations of NADH or AS increased from 5.0 to 50 mM, there was a reduction in the formation of the multiple bands as well as a decrease in the intensity of the smearing band. The disappearance of the smearing band was almost complete when 10-fold NADH or 2-fold AS was present in the system. These results showed that both NADH and AS were able to suppress the formation of the multiple protein bands, and that AS was found to be more effective, exhibiting the same level of inhibition with 5-fold lower concentration than when using NADH.
3.3. RNase Modifications Detected by UV-Vis Spectroscopy after Dialysis
In order to examine adduct formation as well as oxidation of RNase induced by CBQ, UV-Vis spectroscopic experiments were coupled with a hydrazone trapping method. Reactions were carried out by incubating RNase with CBQ at pH = 7.0 and 37˚C for 1, 24, and 48 h, respectively. Figures 4(a) and (b) show the representative UV-Vis spectra of the unmodified RNase and the modified RNase, respectively, after dialysis. The spectra of the unmodified RNase were recorded up to 24 h, revealing λmax at 277 nm, and showed no feature change over time (Figures 4(a)). However, the CBQ-treated/
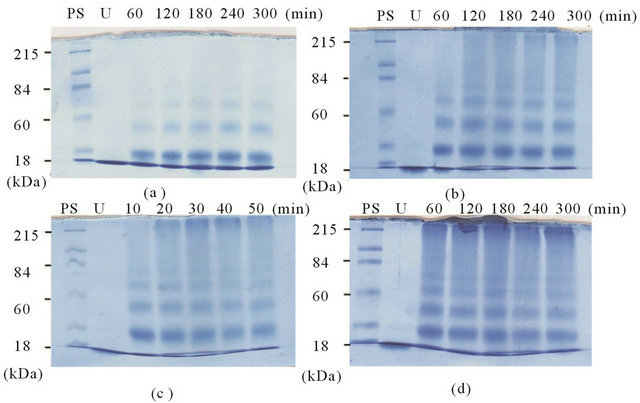
Figure 2. RNase (0.145 mM) + CBQ (0.50, 1.0, 5.0 mM, respectively) in phosphate buffer at 37˚C. PS, protein standard molecular marker; U, unmodified RNase. (a) 0.50 mM CBQ for 10 to 50 min; (b) 1.0 mM CBQ for 10 to 50 min; (c) 5.0 mM CBQ for 10 to 50 min; (d) 5.0 mM CBQ for 60 to 300 min.
 (a) (b) (c)
(a) (b) (c)
Figure 3. Inhibition of RNase modifications at 37˚C. CBQ at 0 and 5.0 mM. NADH and AS at 0, 1.0, 5.0, 10, 50 mM. (a) RNase + CBQ + NADH; (b) RNase + CBQ + AS; (c) Visual observation of AS inhibition. Same reaction conditions as (b).
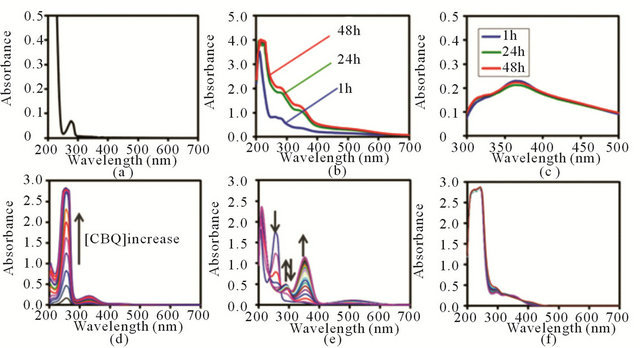
Figure 4. UV-Vis spectra of (a) Unmodified control Rnase; (b) Post-dialyzed RNase after CBQ (1.0 mM) treatment for 1, 24, and 48 h; (c) Post-dialyzed and DNP-treated samples of the modified Rnase; (d) CBQ (0.010 to 0.325 mM); (e) Reaction progress of CBQ (0.010 mM) + Lys (20 mM). Arrows represent the peak movement direction in time. (f) Reaction progress of CBQ (0.010 mM) + Cys (20 mM).
post-dialyzed RNase samples exhibited a significantly different spectral feature compared to the unmodified RNase, accompanying two chromophores developed at 280 nm and 350 nm as time progressed (Figures 4(b)). There was 2-fold increase in A280 (from 0.77 to 1.82) and approximately 3-fold increase in A350 (from 0.36 to 1.04) as the incubation time increased from 1 h to 24 h. However, this increasing trend in A280 and A350 slowed down and reached a plateau after 24 h.
3.4. Reactions of CBQ and Amino Acids Detected by UV-Vis Spectroscopy
In order to get insight on how fast CBQ reacts with nucleophilic amino acids, CBQ were reacted with selected nucleophilic amino acids (Lys, Ser, Thr, Asp, and Cys, respectively) in phosphate buffer at 37˚C. The reactions were monitored by UV-Vis spectroscopy in a timely manner under aerobic and pseudo-first-order conditions using excess amino acids (200-fold more).
For the control, the spectra of CBQ in phosphate buffer at 37˚C were obtained, where CBQ concentrations were varied from 0.010 mM to 0.325 mM in a 0.025 increment for each scan, respectively (Figure 4(d)). The CBQ spectra display two distinctive chromophores at 255 nm (λ255) and 330 nm (λ330) over various CBQ concentration ranges. Figure 4(e) illustrates the time-progressive UV-Vis spectra of a representative reaction of CBQ (0.10 mM) and Lys (20 mM) in phosphate buffer at 37˚C, which was monitored every 2.5 min up to 92.5 min. As CBQ was reacted with Lys, λ330 shifted to λ354, and a new chromophore developed at 290 nm which was absent in the control CBQ spectra.
The time-progressive spectral feature of CBQ + Lys included the decrease of A255, the increase followed by the decrease of A290, and the increase of A354. The timedependent spectra of CBQ + other amino acids, except for CBQ + Cys, presented similar features, showing absorption bands at 255, 290, and 354 nm. In contrast to the reactions of CBQ + Lys, the reaction progress of CBQ + Cys appeared to be slower (Figure 4(f)), as shown in the UV-Vis spectra monitored every 30 min up to 300 min. In order to evaluate the reactivity of CBQ toward various amino acids, pseudo-first-order rate constants were normalized to obtain second-order rate constants, by following the time-dependent change in A255 corresponding to the consumption of CBQ. The calculated second-order rate constants were 0.20, 0.11, 0.14, and 0.006 M−1·s−1 for the reactions of CBQ with Lys, Ser, Thr, and Asp, respectively. Kinetic analysis of CBQ + Cys was not possible due to the slow progress of the reaction and the overlap of A255 with other chromophores.
4. DISCUSSION
Based on the observed protein bands at 33, 53, and 70 kDa in SDS-PAGE (Figure 2), the presence of CBQ was found to result in RNase oligomerization. The smearing band at the higher MW regions indicates that CBQ caused also extensive polymeric aggregation, in addition to oligomerization. Figure 2(c) illustrates the polymerization occurred as early as in 10 min with a higher concentration of CBQ.
Previously, we reported that 1,4-benzoquinone (PBQ) exhibited a similar modification feature in 30 min which is 3-fold longer than the one for CBQ [23]. This suggests that CBQ is more effective than PBQ in terms of RNase polymerization. Without including covalent linkages of RNase to CBQ, the calculated MW values for RNase oligomers are 27, 41, and 55 kDa, respectively. The MW values observed in SDS-PAGE experiment are slightly higher than the calculated MW values for oligomers suggesting that adduct formation of RNase and CBQ occurred simultaneously with RNase crosslinking.
The inhibition study shows that the presence of NADH or AS suppressed RNase modifications, based on the reduced number and intensity of protein bands, with AS being more efficient than NADH (Figure 3). The lack of RNase polymerization observed in SDS-PAGE (Figures 3(a) and (b)) is in agreement with the observed loss of reddish-brown color of the incubation solution as the amount of AS increases (Figure 3(c)). It is known that NADH and AS intervene with the redox cycling of quinones, generating superoxide [27-31]. Our SDSPAGE results show that NADH and AS can influence redox-cycling of CBQ and serve as efficient inhibiting agents for the CBQ-induced RNase modifications.
The full-scan UV-Vis spectra of the modified, postdialyzed RNase (obtained after removal of CBQ and CHQ) show two chromophores at 280 and 350 nm (Figure 4(b)), which are not present in the spectra of the unmodified RNase (Figure 4(a)). These newly developed chromophores for the modified RNase indicate that RNase was covalently linked to CBQ. The time-dependent changes in A280 and A350 occurred within 24 h of incubation and plateaued in time. This plateauing feature is anticipated since RNase has a limited number of nucleophilic amino acids susceptible to alkylation. Figure 4(c) presents the time-dependent UV-Vis spectra for the reaction of the modified RNase and DNP, after dialysis, showing the formation of hydrazone. This suggests the presence of oxidized Lys in the modified RNase. Lys in RNase, whether solvent exposed or buried inside the protein, may react rapidly with CBQ (based on the determined rate constant) and be converted into allysine. Then, the generated allysine and an intact Lys at the protein surface can participate in cross-linking as shown in Figure 1. The allysines that are not involved in crosslinking can be trapped by DNP, forming hydrazones (Figure 1). Figure 4(b) shows the spectral changes of the modified, post-dialyzed RNase upon 1, 24, and 48 h incubations with CBQ. The spectral changes between 1 h and 24 h incubations indicate that alkylation is not completed within 1 h. Considering the similar rate constants for reactions of CBQ with Lys (0.20 M−1·s−1), Thr (0.14 M−1·s−1), and Ser (0.11 M−1·s−1), most of the spectral changes monitored in 1 h should correspond to the alkylation of these three amino acids. The lower rate constant for CBQ + Asp (0.006 M−1·s−1) and the slow spectral change for CBQ + Cys (Figure 4(f)) are consistent with the later spectral development observed for the post-dialyze RNase between 1 - 48 h incubations (Figure 4(b)). The lower rate constant for CBQ + Asp is expected since the α-carboxylate group of Asp does not serve as a good nucleophile. However, the slow reaction of Cys and CBQ is unexpected considering that BQ and Cys of RNase are known to form a well-identified adduct [32,33].
5. CONCLUSION
In conclusion, our study revealed that the nature of CBQ-induced RNase modifications is complex involving cross-linking, adduct formation, and redox cycling. We presented the evidence that CBQ induces massive polymeric aggregation of RNase, which can lead to potential toxicity of CBQ. In addition, we demonstrated that the activity of CBQ was inhibited by antioxidants such as NADH and AS. The reactions of selected amino acids and CBQ showed that CBQ can react readily with nucleophilic amino acids such as Lys, Thr, and Ser.
6. ACKNOWLEDGEMENTS
The study was supported in part by UC Foundation Fund, Provost Student Research Award, and Grote Fund at the University of Tennessee at Chattanooga.
REFERENCES
- Hrudey, S.E. (2009) Chlorination disinfection by-products, public health risk tradeoffs and me. Water Research, 43, 2057-2092. doi:10.1016/j.watres.2009.02.011
- Morbt, N., Tomm, J., Feltens, R., Mogel, I., Kalkhof, S., Murugesan, K., Wirth, H., Vogt, C., Binder, H., Lehmann, I. and von Bergen, M. (2010) Chlorinated benzenes cause concomitantly oxidative stress and induction of apoptotic markers in lung epithelial cells (A549) at nonacute toxic concentrations. Journal of Proteome Research, 10, 363- 378. doi:10.1021/pr1005718
- Ogata, M., Taguchi, T., Hirota, N., Shimada, Y. and Nakae, S. (1991) Quantitation of urinary chlorobenzene metabolites by HPLC: Concentrations of 4-chlorocatechol and chlorophenols in urine and of chlorobenzene in biological specimens of subjects exposed to chlorobenzene. International Archives of Occupational and Environmental Health, 63, 121-128. doi:10.1007/BF00379075
- Nair, R.S., Barter, J.A., Schroeder, R.E., Knezevich, A. and Stack, C.R. (1987) A two-generation reproduction study with monochlorobenzene vapor in rats. Fundamental and Applied Toxicology, 9, 678-686. doi:10.1016/0272-0590(87)90174-6
- Aiso, S., Takeuchi, T., Arito, H., Nagano, K., Yamamoto, S. and Matsushima, T. (2005) Carcinogenicity and chronic toxicity in mice and rats exposed by inhalation to paradichlorobenzene for two years. Journal of Veterinary Medical Science, 67, 1019-1029. doi:10.1292/jvms.67.1019
- Yamazaki, K., Aiso, S., Matsumoto, M., Kano, H., Arito, H., Nagano, K., Yamamoto, S. and Matsushima, T. (2006) Carcinogenicity and chronic toxicity of 1,4-dichloro-2- nitrobenzene in rats and mice by two years feeding. Industrial Health, 44, 230-243. doi:10.2486/indhealth.44.230
- Sharma, S., Mukhopadhyay, M. and Murthy, Z.V.P. (2010) Degradation of 4-chlorophenol in wastewater by organic oxidants. Industrial & Engineering Chemistry Research, 49, 3094-3098. doi:10.1021/ie9018066
- Zhao, Y., Qin, F., Boyd, J.M., Anichina, J. and Li, X.F. (2010) Characterization and determination of chloroand bromo-benzoquinones as new chlorination disinfection byproducts in drinking water. Analytical Chemistry, 82, 4599-4605. doi:10.1021/ac100708u
- Qin, F., Zhao, Y.Y., Zhao, Y., Boyd, J.M., Zhou, W. and Li, X.F. (2010) A toxic disinfection by-product, 2,6-dichloro-1,4-benzoquinone, identified in drinking water. Angewandte Chemie International Edition, 49, 790-792. doi:10.1002/anie.200904934
- Yin, L., Niu, J., Shen, Z. and Chen, J. (2010) Mechanism of reductive decomposition of pentachlorophenol by Tidoped beta-Bi2O3 under visible light irradiation. Environmental Science & Technology, 44, 5581-5586. doi:10.1021/es101006s
- Yin, L., Shen, Z., Niu, J., Chen, J. and Duan, Y. (2010) Degradation of pentachlorophenol and 2,4-dichlorophenol by sequential visible-light driven photocatalysis and laccase catalysis. Environmental Science & Technology, 44, 9117-9122. doi:10.1021/es1025432
- Snyder, R. and Hedli, C.C. (1996) An overview of benzene metabolism. Environmental Health Perspectives, 104, 1165-1172.
- Bolton, J.L., Trush, M.A., Penning, T.M., Dryhurst, G. and Monks, T.J. (2000) Role of quinones in toxicology. Chemical Research in Toxicology, 13, 135-160. doi:10.1021/tx9902082
- Kondrova, E., Stopka, P. and Soucek, P. (2007) Cytochrome P450 destruction by benzene metabolites 1,4- benzoquinone and 1,4-hydroquinone and the formation of hydroxyl radicals in minipig liver microsomes. Toxicology in Vitro, 21, 566-575. doi:10.1016/j.tiv.2006.11.002
- Bodell, W.J., Pathak, D.N., Levay, G., Ye, Q. and Pongracz, K. (1996) Investigation of the DNA adducts formed in B6C3F1 mice treated with benzene: Implications for molecular dosimetry. Environmental Health Perspectives, 104, 1189-1193.
- Buben, A., Narasimhan, N. and Hanzlik, R.P. (1988) Effects of chemical and enzymic probes on microsomal covalent binding of bromobenzene and derivatives. Evidence for quinones as reactive metabolites. Xenobiotica, 18, 501-510. doi:10.3109/00498258809041687
- Anichina, J., Zhao, Y., Hrudey, S.E., Le, X.C. and Li, X.F. (2010) Electrospray ionization mass spectrometry characterization of interactions of newly identified water disinfection byproducts halobenzoquinones with oligodeoxynucleotides. Environmental Science & Technology, 44, 9557-9563. doi:10.1021/es1024492
- Nguyen, T.N., Bertagnolli, A.D., Villalta, P.W., Buhlmann, P. and Sturla, S.J. (2005) Characterization of a deoxyguanosine adduct of tetrachlorobenzoquinone: Dichlorobenzoquinone-1,N2-etheno-2’-deoxyguanosine. Chemical Research in Toxicology, 18, 1770-1776. doi:10.1021/tx050204z
- Slaughter, D.E. and Hanzlik, R.P. (1991) Identification of epoxideand quinone-derived bromobenzene adducts to protein sulfur nucleophiles. Chemical Research in Toxicology, 4, 349-359. doi:10.1021/tx00021a015
- Hanzlik, R.P. (1986) Chemistry of covalent binding: Studies with bromobenzene and thiobenzamide. Advances in Experimental Medicine and Biology, 197, 31-40. doi:10.1007/978-1-4684-5134-4_3
- Zaborska, W., Krajewska, B., Kot, M. and Karcz, W. (2007) Quinone-induced inhibition of urease: Elucidation of its mechanisms by probing thiol groups of the enzyme. Bioorganic Chemistry, 35, 233-242. doi:10.1016/j.bioorg.2006.11.001
- Meade, S.J., Miller, A.G. and Gerrard, J.A. (2003) The role of dicarbonyl compounds in non-enzymatic crosslinking: A structure-activity study. Bioorganic & Medicinal Chemistry, 11, 853-862. doi:10.1016/S0968-0896(02)00564-3
- Kim, J., Vaughn, A.R., Cho, C., Albu, T.V. and Carver, E.A. (2012) Modifications of ribonuclease A induced by p-benzoquinone. Bioorganic Chemistry, 40, 92-98. doi:10.1016/j.bioorg.2011.11.002
- Reznick, A.Z. and Packer, L. (1994) Oxidative damage to proteins: Spectrophotometric method for carbonyl assay. Methods in Enzymology, 233, 357-363. doi:10.1016/S0076-6879(94)33041-7
- Laemmli, U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680-685. doi:10.1038/227680a0
- Weber, K. and Osborn, M. (1969) The reliability of molecular weight determinations by dodecyl sulfate polyacrylamide gel electrophoresis. The Journal of Biological Chemistry, 244, 4406-4412.
- Yuasa, J., Yamada, S. and Fukuzumi, S. (2008) One-step versus stepwise mechanism in protonated amino acidpromoted electron-transfer reduction of a quinone by electron donors and two-electron reduction by a dihydronicotinamide adenine dinucleotide analogue. Interplay between electron transfer and hydrogen bonding. Journal of the American Chemical Society, 130, 5808-5820. doi:10.1021/ja8001452
- Fukuzumi, S., Fujii, Y. and Suenobu, T. (2001) Metal ion-catalyzed cycloaddition vs hydride transfer reactions of NADH analogues with p-benzoquinones. Journal of the American Chemical Society, 123, 10191-10199. doi:10.1021/ja016370k
- Gorner, H. (2005) Photoreactions of 1,4-naphthoquinones: Effects of substituents and water on the intermediates and reactivity. Photochemical & Photobiological Sciences, 81, 376-383. doi:10.1562/2004-08-11-RA-270.1
- Verrax, J., Delvaux, M., Beghein, N., Taper, H., Gallez, B. and Buc Calderon, P. (2005) Enhancement of quinone redox cycling by ascorbate induces a caspase-3 independent cell death in human leukaemia cells. An in vitro comparative study. Free Radical Research, 39, 649-657. doi:10.1080/10715760500097906
- Roginsky, V.A., Barsukova, T.K and Stegmann, H.B. (1999) Kinetics of redox interaction between substituted quinones and ascorbate under aerobic conditions. Chemico-Biological Interactions, 121, 177-197. doi:10.1016/S0009-2797(99)00099-X
- Person, M.D., Mason, D.E., Liebler, D.C., Monks, T.J. and Lau, S.S. (2005) Alkylation of cytochrome c by (glutathion-S-yl)-1,4-benzoquinone and iodoacetamide demonstrates compound-dependent site specificity. Chemical Research in Toxicology, 18, 41-50. doi:10.1021/tx049873n
- Hanzlik, R.P., Harriman, S.P. and Frauenhoff, M.M. (1994) Covalent binding of benzoquinone to reduced ribonuclease. Adduct structures and stoichiometry. Chemical Research in Toxicology, 7, 177-184. doi:10.1021/tx00038a010

