Advances in Bioscience and Biotechnology
Vol.2 No.5(2011), Article ID:7830,4 pages DOI:10.4236/abb.2011.25049
Accumulation of amino acids deriving from pyruvate in Escherichia coli W3110 during fed-batch cultivation in a two-compartment scale-down bioreactor
![]()
1Bioprocess Engineering Laboratory, Department of Process and Environmental Engineering, University of Oulu, Oulu, Finland;
2Fachgebiet Bioverfahrenstechnik, Institut für Biotechnologie, Technische Universität Berlin, Berlin, Germany.
E-mail: jaakko.soini@oulu.fi
Received 7 September 2011; revised 20 September 2011; accepted 4 October 2011.
Keywords: E. coli; Fermentation; Scale-Down; Amino Acid
ABSTRACT
Bioprocess scale-down simulators are used to investigate the phenomena occurring in industrial scale cultivations. The aim is to simulate the large-scale conditions, characterized by long mixing times introducing local gradients into the bioreactor, in laboratory scale bioreactor. We have expanded the current physiological knowledge about the consequences of such cultivation conditions from well-known mixedacid fermentation to amino acid metabolism. Several interesting deviations were observed for amino acids derived from pyruvate when a STR-PFR scale-down simulator and a regular STR fed-batch cultivation were compared. In this article we show that the cultivation of Escherichia coli K-12 substrain W3110 in a scale-down two-compartment bioreactor has consequences on the free amino acid pools. Repeated occurrence of high glucose zones affects the biosynthesis of amino acids derived from pyruvate. This is interesting since the pyruvate based amino acids are precursors of the branched chain amino acid pathway, which is further disturbed by an unbalanced regulation caused by a frameshift mutation in the ilvG gene of E. coli K-12 strains. As a consequence the formation of non-typical amino acid norvaline was detected in the two compartment bioreactor cultivation.
1. INTRODUCTION
Industrial scale aerobic bioprocesses are characterized by lower mixing efficiency and consequently prolonged mixing times compared to laboratory scale bioprocesses. In large scale reactors more than 10,000 fold concentration difference between the feed solution and the bioreactor broth causes gradients in the reactor [1,2]. Under otherwise glucose limited conditions for E. coli the most prominent response to a glucose zone with a concentration far above the KS value is the immediate increase of the glucose uptake rate [3-7] and the accumulation of glycolytic overflow products, such as acetate. Also, if the cell density is high enough to cause oxygen limitation in such a zone, typical products of the anaerobic metabolism appear, such as lactate, formate, succinate, dihydrogen and additional carbon dioxide [4,6-8]. Despite the belief that there are major effects by the scale, only a few confirmed global differences between laboratory and large-scale cultivations have been reported.
The major difference in cultivating E. coli in large scale compared to small-scale well-mixed bioreactors is a reduced biomass yield [7,9]. Studies in chemostats [4,10,11] and fed-batch cultivations [5] have indicated that the fast response to high glucose concentration is a high glycolytic flux and this results in an immediate increase of pyruvate [4]. So far this only has been discussed in relation to consequences in the synthesis of organic acids. However, due to the central role of pyruvate one also may assume a channeling towards the amino acid synthesis pathways which are closely connected to the glycolysis, such as the synthesis of alanine and the branched chain amino acid pathway.
Earlier we have shown that many pyruvate based amino acids including alanine, valine and non-canonical amino acid norvaline accumulate in connection to rapid single shift from aerobic conditions to oxygen limitation [12]. Another earlier paper studied the accumulation of mixed-acid fermentation products in a scale-down simulator [13]. These earlier studies are brought together by this paper investigating whether the oscillatory/repeated high glucose zones have an effect on the profiles of the pyruvate-based amino acids. This is studied in a well established scaledown bioreactor system, a combined STR-PFR system containing a plug flow reactor with an integrated static mixer which allows the resolution of fast responses in the high glucose zone [6,7,14,15].
2. MATERIALS AND METHODS
Crude cell samples for free amino acid analysis, containing both intraand extracellular fractions, were collected from the reference and scale-down cultivations from the STR compartment and frozen immediately in liquid nitrogen and stored at –20˚C until further steps. Intracellular samples from the scale-down cultivation were collected from the STR compartment and the four sampling ports of PFR compartment into precooled (20˚C) syringes containing 500 µL of 60% methanol solution to inactivate the samples. Cells were separated by centrifugation at –9˚C and 14,000 rpm for 3 min. Supernatant, containing the extracellular fraction was discarded and the pellets were stored at –20˚C until further preparation. The procedures for cell disruption by sonication, sample preparation and free amino acid analysis has been explained in detail in earlier study [12]. The residence times at four sampling ports of the PFR compartment were 15, 30, 45 and 60 s. The formation rates presented in Table 1, were calculated from the equation of a linear fit for PFR measurements. Samples were taken at three time points; 0.5, 2.0 and 5.0 h after the glucose feed start.
Glucose was the growth-limiting factor in the STR compartment during the fed-batch phase and correspondingly the culture showed a quasi linear increase of the cell density (data not shown). In the scale-down simulator the glucose concentration at the first sampling port of the PFR compartment varied between 1.0 and 1.5 g·L–1.
3. RESULTS AND DISCUSSION
As pyruvate is a key metabolite for the response to oxygen limitation and glucose excess occurring in scaledown cultivation and also an important precursor for many amino acids, we wanted to study the effect of scale-down simulator conditions for the accumulation of alanine and the branched-chain amino acids isoleucine,
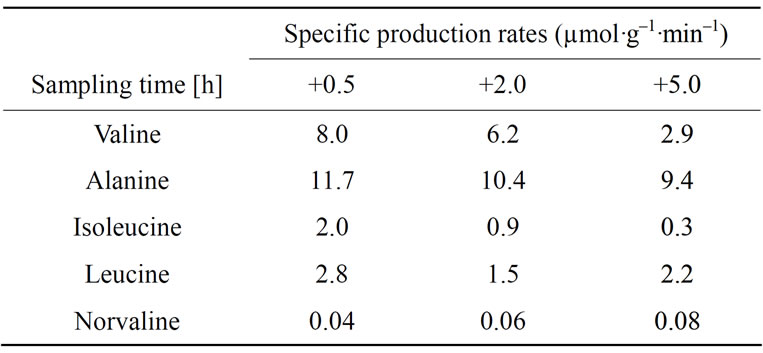
Table 1. Specific intracellular production rates for a set of amino acids.
valine and leucine. Also the non proteinogenic branched chain amino acid analogue norvaline was quantified from the crude cell extracts. The Figure 1 shows the free amino acid concentrations quantified from the reference cultivation compared with those of the scale-down bioreactor. Valine, alanine and norvaline accumulate in the scale-down simulator in comparison to the reference cultivation. The accumulation profile of valine was similar under both cultivation conditions, but the valine concentration was significantly higher in the two-compartment system. The concentration of alanine was constantly increasing in the scale-down simulator whereas a clear decline was observed for the reference cultivation. Norvaline accumulation started immediately after the PFR circulation. This is probably due to the oxygen limitation in the PFR compartment which has been earlier shown to trigger norvaline formation [12]. Leucine and isoleucine concentrations were not affected by the scale-down cultivation conditions. Isoleucine was accumulating during the batch phase and decreasing during glucose limited fed-batch phase. Leucine concentration was relatively stable in comparison to other amino acids.
To further study the dynamics of amino acid metabolism the intracellular levels (Figure 2) were determined and specific production rates (Table 1) were calculated. All studied amino acids accumulated in the PFR com-
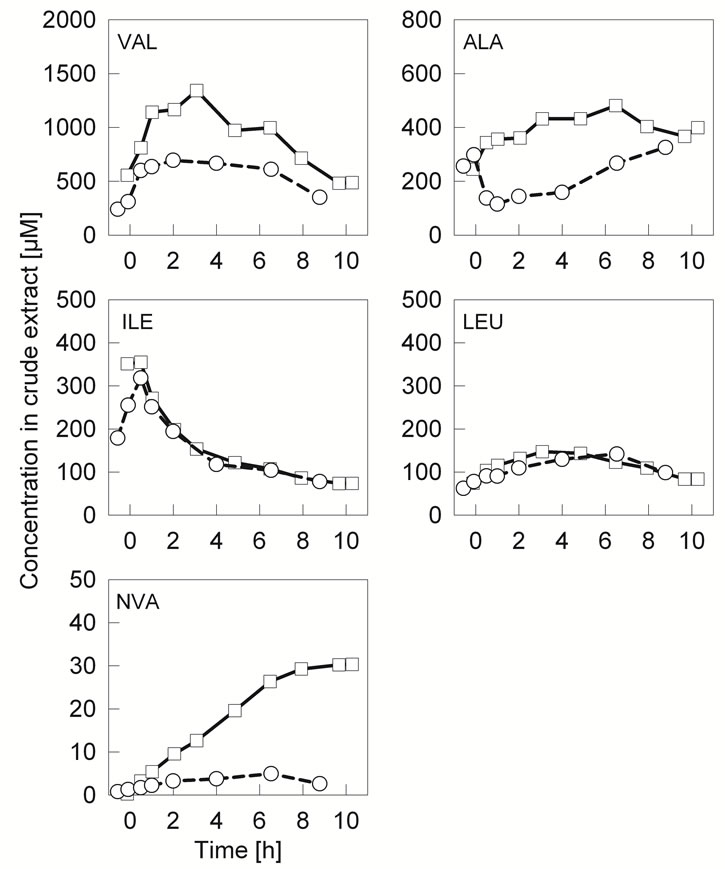
Figure 1. The accumulation profiles of a set of free amino acids in crude extracts (cells and medium) of E. coli W3110 from the cultivation in the two-compartment reactor (square) and respective reference cultivation in the STR (sphere).
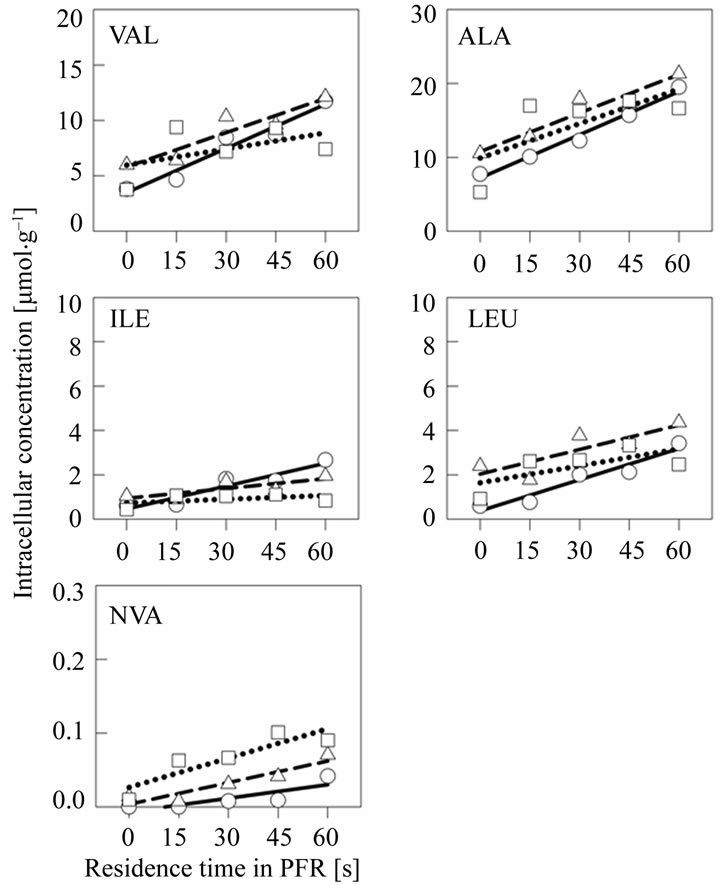
Figure 2. A set of intracellular free amino acid concentrations in scale-down cultivation samples from the cultivation of E. coli W3110 in the two-compartment reactor. The samples were taken 0.5 h (sphere and solid line), 2 h (triangle and dashed line) and 5 h (square and dotted line) after the PFR recirculation was initiated at cell density of 19 g·L–1 (CDW). The peristaltic pump was set to flow rate 1 L·min–1, and this rate was used to approximate the residence times in the PFR compartment shown on X-axis.
partment, i.e. their synthesis was stimulated by the glucose pulse. Highest production rates were measured for alanine and valine 0.5 h after the glucose feed start. Leucine and isoleucine production was observed on intracellular level even if the accumulation on crude cell extract level was not detected. In later sampling points with higher cell density, the specific production rates for alanine, valine, isoleucine and leucine decreased. Norvaline was produced but in relatively low quantities. However, the production rate of norvaline increased with increasing cell density and the highest production rate 0.08 µmol·g–1·min–1 was measured at the latest time point. In Table 2 the ratios of amino acid concentrations intracellular and crude cell fraction are presented.
The conditions in the two-compartment scale-down reactor caused interesting deviations in the concentration profiles of amino acids deriving from pyruvate. The drastically accumulating valine is interesting because of the valine toxicity in K-12 strains of E. coli [16]. Valine accumulates rapidly up to a concentration clearly over 1 mM. The accumulation of valine could lead to growth inhibition and it also partly explains the rapid decrease
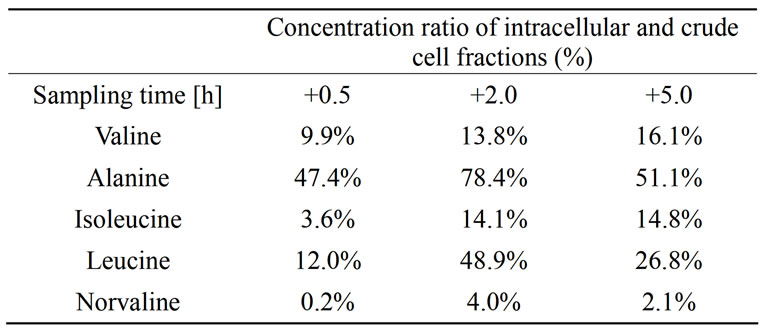
Table 2. Ratio of the free amino acid concentrations intracellular fraction related to the crude cell fraction (combined intraand extracellular fraction).
of isoleucine since the valine toxicity prevents the isoleucine biosynthesis.
Furthermore, in the scale-down cultivation the nonproteinogenic amino acid analogue norvaline was produced. Norvaline formation occurs simultaneously with valine accumulation and declined isoleucine biosynthesis and specific production rate was highest with the highest cell density. As the norvaline formation is connected to the scale-down cultivations, one could hypothesize that it may be synthesized in large-scale industrial cultivations if zones with high glucose concentration occur, which are simulated in our scale-down bioreactor. This would be interesting to study, as norvaline is known to be incorporated into certain recombinant proteins if it accumulates in the cell [17]. Another interesting finding was the rapid increase of intracellular leucine and isoleucine concentrations in the PFR compartment in contradiction to crude cell results is STR samples, which showed no increase. This could be explained by further re-assimilation as is the case for common anerobic metabolites such as acetate and lactate.
Amino acids deriving from pyruvate showed interesting accumulation profiles when E. coli W3110 cells were cultivated in a scale-down simulator consisting of a wellstirred STR compartment and a PFR compartment, characterized by a high glucose concentration and oxygen limitation. Valine, alanine and the non-proteinogenic amino acid norvaline were produced in PFR compartment and accumulated in the STR compartment. Especially the accumulation of norvaline should be considered in large-scale recombinant protein processes. Leucine and isoleucine were produced in the PFR compartment based on intracellular measurements but did not accumulate in the STR compartment. This might indicate that leucine and isoleucine are further re-assimilated as is the case with the common anaerobic markers acetate and lactate, which are produced under oxygen limitation but rapidly consumed in the aerobic environment of the STR.
REFERENCES
- Enfors, S.O., Jahic, M., Rozkov, A., Xu, B., Hecker, M., Jürgen, B., Krüger, E., Schweder, T., Hamer, G., O’Beirne, D., Noisommit-Rizzi, N., Reuss, M., Boone, L., Hewitt, C., McFarlane, C., Nienow, A., Kovacs, T., Trägårdh, C., Fuchs, L., Revstedt, J., Friberg, P.C., Hjertager, B., Blomsten, G., Skogman, H., Hjort, S., Hoeks, F., Lin, H.Y., Neubauer, P., Van der, L.R., Luyben, K., Vrabel, P. and Manelius, Å. (2001) Physiological responses to mixing in large scale bioreactors. Journal of Biotechnology, 85, 175-185. doi:10.1016/S0168-1656(00)00365-5
- Lapin, A., Schmid, J. and Reuss, M. (2006) Modeling the dynamics of E-coli populations in the three-dimensional turbulent field of a stirred-tank bioreactor—A structuredsegregated approach. Chemical Engineering Science, 61, 4783-4797. doi:10.1016/j.ces.2006.03.003
- Chassagnole, C., Noisommit-Rizzi, N., Schmid, J.W., Mauch, K. and Reuss, M. (2002) Dynamic modeling of the central carbon metabolism of Escherichia coli. Biotechnology and Bioengineering, 79, 53-73. doi:10.1002/bit.10288
- De Mey, M., Taymaz-Nikerel, H., Baart, G., Waegeman, H., Maertens, J., Heijnen, J.J. and van Gulik, W.M. (2010) Catching prompt metabolite dynamics in Escherichia coli with the BioScope at oxygen rich conditions. Metabolic Engineering, 12, 477-487. doi:10.1016/j.ymben.2010.04.003
- Lin, H.Y., Mathiszik, B., Xu, B., Enfors, S.O. and Neubauer, P. (2001) Determination of the maximum specific uptake capacities for glucose and oxygen in glu-coselimited fed-batch cultivations of Escherichia coli. Biotechnology and Bioengineering, 73, 347-357. doi:10.1002/bit.1068
- Neubauer, P., Häggström, L. and Enfors, S.O. (1995) Influence of substrate oscillations on acetate formation and growth yield in Escherichia coli glucose-limited fed-batch cultivations. Biotechnology and Bioengineering, 47, 139-146. doi:10.1002/bit.260470204
- Xu, B., Jahic, M., Blomsten, G. and Enfors, S.O. (1999) Glucose overflow metabolism and mixed-acid fermentation in aerobic large-scale fed-batch processes with Escherichia coli. Applied Microbiology and Biotechnology, 51, 564-571. doi:10.1007/s002530051433
- Xu, B., Jahic, M. and Enfors, S.O. (1999) Modeling of overflow metabolism in batch and fed-batch cultures of Escherichia coli. Biotechnology Progress, 15, 81-90. doi:10.1021/bp9801087
- Hewitt, C.J., Onyeaka, H., Lewis, G., Taylor, I.W. and Nienow, A.W. (2007) A comparison of high cell density fed-batch fermentations involving both induced and non-induced recombinant Escherichia coli under wellmixed small-scale and simulated poorly mixed largescale conditions. Biotechnology and Bioengineering, 96, 495-505. doi:10.1002/bit.21127
- Schaub, J., Mauch, K. and Reuss, M. (2008) Metabolic flux analysis in Escherichia coli by integrating isotopic dynamic and isotopic stationary 13C labeling data. Biotechnology and Bioengineering, 99, 1170-1185. doi:10.1002/bit.21675
- Lara, A.R., Taymaz-Nikerel, H., Mashego, M.R., Van Gulik, W.M., Heijnen, J.J., Ramirez, O.T. and Van Winden, W.A. (2009) Fast dynamic response of the fermentative metabolism of Escherichia coli to aerobic and anaerobic glucose pulses. Biotechnology and Bioengineering, 104, 1153-1161. doi:10.1002/bit.22503
- Soini, J., Falschlehner, C., Liedert, C., Bernhardt, J., Vuoristo, J. and Neubauer, P. (2008) Norvaline is accumulated after a down-shift of oxygen in Escherichia coli W3110. Microbial Cell Factories, 7, 30. doi:10.1186/1475-2859-7-30
- Soini, J., Ukkonen, K. and Neubauer, P. (2008) High cell density media for Escherichia coli are generally designed for aerobic cultivations—Consequences for large-scale bioprocesses and shake flask cultures. Microbial Cell Factories, 7, 26. doi:10.1186/1475-2859-7-26
- George, S, Larsson, G. and Enfors, S.O. (1993) A scale-down 2-compartment reactor with controlled substrate oscillations—Metabolic response of Saccharomyces Cervisiae. Bioprocess Engineering, 9, 249-257. doi:10.1007/BF01061530
- Neubauer, P., Ahman, M., Tornkvist, M., Larsson, G. and Enfors, S.O. (1995) Response of guanosine tetraphosphate to glucose fluctuations in fed-batch cultivations of Escherichia coli. Journal of Biotechnology, 43, 195-204. doi:10.1016/0168-1656(95)00130-1
- Andersen, D.C., Swartz, J., Ryll, T., Lin, N. and Snedecor, B. (2001) Metabolic oscillations in an E. coli fermentation. Biotechnology and Bioengineering, 75, 212-218. doi:10.1002/bit.10018
- Apostol, I., Levine, J., Lippincott, J., Leach, J., Hess, E., Glascock, C.B., Weickert, M.J. and Blackmore, R. (1997) Incorporation of norvaline at leucine positions in recombinant human hemoglobin expressed in Escherichia coli. The Journal of Biological Chemistry, 272, 28980-28988. doi:10.1074/jbc.272.46.28980

