Modern Research in Inflammation
Vol.2 No.4(2013), Article ID:39977,8 pages DOI:10.4236/mri.2013.24011
Comparison of YKL-39 and CHIT-1 expression during macrophages differentiation and polarization
![]()
1Department of Bio-Medical Sciences, University of Catania, Catania, Italy; *Corresponding Author: mdirosa@unict.it
2Department of Clinical and Molecular Biomedicine, Division of Hematology, University of Catania, Catania, Italy
3S. Immuno-Haemathology and Transfusional Medicine, Transfusional Centre “Garibaldi” Hospital, Catania, Italy
Copyright © 2013 Michelino Di Rosa et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 25 July 2013; revised 24 August 2013; accepted 1 September 2013
Keywords: YKL-39; Chitotriosidase; Human Monocyte/Macrophages; Classically Activated Macrophages; Alternatively Activated Macrophages
ABSTRACT
The chitinase-like proteins YKL-39 (chitinase 3- like-2) and Chitortriosidase (CHIT-1) are members of the chitinases family. YKL-39 expression has been associated with osteoarthritis, whereas CHIT-1 activity is regarded as a biochemical marker of macrophage activation. So far, the physiological or pathological role of YKL-39 in the inflammation is still poorly understood. We compared YKL-39 and CHIT-1 modulation during monocyte to macrophage transition and polarization. Gene expression analysis was investigated by real-time PCR from mRNA of human monocytes obtained from buffy coat of healthy volunteers, from mRNA of polarized macrophages to classically activated macrophages (or M1), obtained by interferon-γ and lipopolysaccharide exposure, and from mRNA of alternatively activated macrophages (or M2) obtained by interleukin-4 exposure. We demonstrated different variations of YKL-39 and CHIT-1 production during macrophages polarization. CHIT-1 levels gradually increase in the course of the time with a peak of expression between the fifth and the seventh day of culture. In contrast, YKL-39 expression was unaltered in the diverse stage of HMMs differentiation, but increased significantly in M1 polarized macrophages and reverted to base levels in M2 polarized macrophages. These findings indicated that the function of YKL-39 is much more restricted and selective than that exerted by CHIT-1.
1. INTRODUCTION
YKL-39 (or chitinase 3-like 2, YKL-39) is a human chitinase-like protein which contains Glyco-18 domain with no catalytic activity but with putative lectin properties [1]. Although YKL-39 is homologous to the family 18 chitinases in the human genome, it has been reported to lack any chitinase activity. Recently, it has been found that human YKL-39 possesses a chitinase-like fold. Strikingly, the chitinase activity of YKL-39 was recovered by reverting two non-conservative substitutions in the active site to those found in the active enzymes, suggesting that YKL-39 is a pseudo-chitinase with retention of chitinaselike ligand-binding properties [2]. YKL-39 displayed anabolic properties in cartilage [3,4]. It was originally identified as an abundantly secreted protein in primary culture of human articular chondrocytes [3]. YKL-39 mRNA is significantly up-regulated in cartilage of patients with osteoarthritis [4]. Since the expression of YKL-39 was upregulated both in early degenerative and late stage of osteoarthritis, it has been recognized as a biochemical marker for the activation of chondrocytes and the progress of the osteoarthritis in human [5]. Proteomic analysis confirmed that YKL-39 is secreted by human osteoarthritic cartilage in culture [6]. Two physiological activities of YKL-39 might contribute to the disease progression: the former is the induction of autoimmune response [7,8]; the second is its involvement in tissue remodeling. Several mammalian Glyco_18 domain containing proteins are expressed by macrophages in various physiological situations [9]. Enzymatically active chitotriosidase is expressed by mature monocyte derived macrophages, lung macrophages and Gaucher cells [10- 12]. Gaucher cells are abnormal lipid-laden macrophages formed in tissues of Gaucher disease patients. Gaucher cells can be classified as a variation of alternatively activated macrophages [13]. Consequently CHIT-1 is used as a highly specific biomarker for lysosomal storage disorder [14]. Later the levels of this enzyme were found elevated in Niemann-Pick diseases [15], but also in other pathological conditions including β-thalassaemia [16], sarcoidosis [17], multiple sclerosis [18], atherosclerosis [10] and in parasitic infections such as plasmodium falciparum malaria [19]. Interestingly, other investigations have suggested that CHIT-1 may be involved in the progression of nonalcoholic steatohepatitis [20,21] and could have a crucial role even in pathological conditions, such as coronary artery disease [22], acute ischemic stroke [23] cerebrovascular dementia (CVD) and Alzheimer’s disease (AD) [24]. Overall, these data underline the notion that CHIT-1 acts as an important factor in inflammation [11]. To our knowledge, so far, there is only one report suggesting that YKL-39 can be expressed on a very low level in macrophages differentiated in the presence of IFNγ and IL-4 [25]. Numerous degenerative diseases are associated with a deficiency or alteration in macrophage function [26]. Macrophages play a crucial role in regulating the initiation, amplification, and resolution of innate immune responses. Macrophages can be classically activated (M1), in the presence of interferon (IFN)-γ and lipopolysaccharide (LPS), while in the presence of interleukin (IL)-4 and (IL)-13, macrophages undergo alternative activation or are skewed toward an M2 phenotype of alternatively activated macrophages (M2). Macrophage polarization deeply alters the immune properties of these cells as shown by the potent anti-microbial properties of M1 macrophages versus the marked tissue repair properties of M2 macrophages [27]. It has been reported that not only elevated levels of YKL-39 observed during chronic inflammation can be assigned to the activation of synovial cells, but also macrophages can contribute to the YKL-39 production [28]. The aim of our study was to compare the modulation YKL-39 expression with CHIT-1 expression during macrophage activation and their polarization in M1 and M2.
2. MATERIALS AND METHODS
2.1. Cells
Human monocyte-derived macrophages (HMMs) were isolated from fresh buffy coat of healthy volunteers as previously described [29]. The buffy coat were diluted with phosphate-buffered saline (PBS) supplemented with 2.5 mM EDTA and layered onto Ficoll-Hypaque gradients (Gibco, Invitrogen, Milan, Italy). After 30 min of centrifugation at 400 × g at room temperature, the mononuclear cells were collected, washed twice with PBS and placed in plastic Petri dishes at a concentration of 1 × 106 - 2 × 106 cells/cm surface areas in Iscove’s medium supplemented with 2 mM glutamine, and 50 mg/ml of penicillin/streptomycin. The cells were further purified using CD14 magnetic beads (Miltenyi Biotec) according to the manufacturer’s instructions. The HMMs were cultured in on Iscove’s medium supplemented with 10% of fetal calf serum (FCS) 2 mM glutamine and 1% of penicillin/streptomycin (Invitrogen, Milan, Italy) [29].
2.2. Macrophage Differentiation
Macrophages were obtained by culturing monocytes for 7 days in Iscove’s medium supplemented with 10% FBS, 2 mM glutamine, 1% of penicillin/streptomycin (Invitrogen, Milan, Italy) and 5 ng/ml M-CSF in Petri dishes at a density 1 × 106 - 2 × 106 cells/cm2. Macrophage polarization was obtained as previously described [30]. In brief, the culture medium was removed and cells were cultured for an additional 18 h in RPMI 1640 supplemented with 5% FBS and LPS (50 ng/ml) plus IFN-γ (100 U/ml) (for M1 polarization) or IL-4 (20 ng/ml) (for M2 polarization) (Peproteck, Milan, Italy). Five different cell types were generated: freshly isolated monocytes (Mono T0), cells at intermediate differentiation (3 days of culture: Mono T3), resting fully differentiated macrophages (7 days of culture: macrophages), classical activated macrophages (M1), alternative activated macrophages (M2). Macrophage polarization was confirmed by real-time PCR detecting the expression for CXCL11 and CCL18, markers of M1 and M2 respectively (data not shown). Additionally we detected the transcriptional levels of TNF-α and superoxide dismutase (SOD2) in order to ascertain that monocytes macrophages were not activated during differentiation. The polarization of macrophages in M1 induced by LPS (50 ng/ml) and IFN-γ (100 U/ml) resulted in an increase of both TNF-α and SOD2 expression (data not shown). In contrast, both TNF-α and SOD2 expression was reduced in M2 polarized macrophages (data not shown).
2.3. Gene Expression Analysis by Real-Time PCR
Total RNA was extracted from cells using TRIzol reagent (Invitrogen, Milan, Italy). RNA was further purified using RNeasy Mini columns and DNAse I digestion (QIAGEN, Milan, Italy) to remove any contaminating DNA, and subjected to real-time quantitative PCR in triplicate. For reverse transcription-polymerase chain reaction (RT-PCR), 2 µg of total RNA was reverse-transcribed with High-Capacity cDNA Reverse Trascription Kit (Applied Biosystems, Monza, Italy) in a 20 ml reaction solution. Real-time fluorescence PCR, based on SYBR Green, was carried out in a 30 ml final volume containing 1 SYBR Green PCR Master Mix (Applied Biosystems, Monza, Italy), 200 nM forward and 200 nM reverse primers (Table 1) and 20 ng of cDNA. For each gene, generation of the correct PCR product was confirmed by melting curve analysis, and fold difference in the abundance of mRNA (compared to the housekeeping gene GAPDH) was then calculated using the ΔCt method with the assumption of ideal primer efficiencies. Thermal cycling was performed in MicroAmp Optical 96-well reaction plate (Applied Biosystems, Monza, Italy) with MicroAmp optical caps (Applied Biosystems, Monza, Italy) using the ABI PRISM 7700 sequence detection system (Applied Biosystems, Monza, Italy). Amplification was carried out with the following conditions: 50˚C for 2 min, 95˚C for 10 min, and 30 cycles each of 95˚C for 15 sec and 60˚C for 1 min. Data are presented as mean % ± S.D. of at least three independent experiments. Differences were analyzed by Student t test, with p < 0.05 being considered statistically significant.
2.4. DNA Sequence Analysis by Bioinformatic Tools
Potential control elements and consensus sites were analyzed by using MatInspector Professional software [31], which locates matches by comparing DNA sequences with weighted matrix descriptions of functional binding sites, based on the TRANSFAC database (http://www.biobase.de) [32]. MatInspector assigns a core similarity (CS) of 1 when the (usually four) highest conserved bases of a matrix (“core sequence”) match exactly in the sequence. The matrix similarity (MS) takes into account all bases over the matrix length. A “good” match usually has an MS of >0.8 [31]. Site mutations in the promoter sequence were analyzed routinely to verify that mutagenesis did not result in the generation of new potential binding sites. The FastM computer program [33] was applied to build models based on information about the individual elements involved, their strand orientation, their sequential order, and the distances between the elements. To analyze the human, vertebrate (rodent/other mammalian/other vertebrate), and viral databases for potential co-regulated promoters, the Model Inspector Professional program was applied [34] (Genomatix, Munich, Germany; http://genomatix.gsf.de).
2.5. Statistical Analysis
Data are expressed as mean ± standard error (SE). Significance was assessed by one-way analysis of variance (ANOVA). p < 0.05 was considered to be statistically significant.
3. RESULTS
3.1. Effect of LPS and IFN-γ on the Expression of YKL-39 and CHIT-1 in Undifferentiated Monocytes
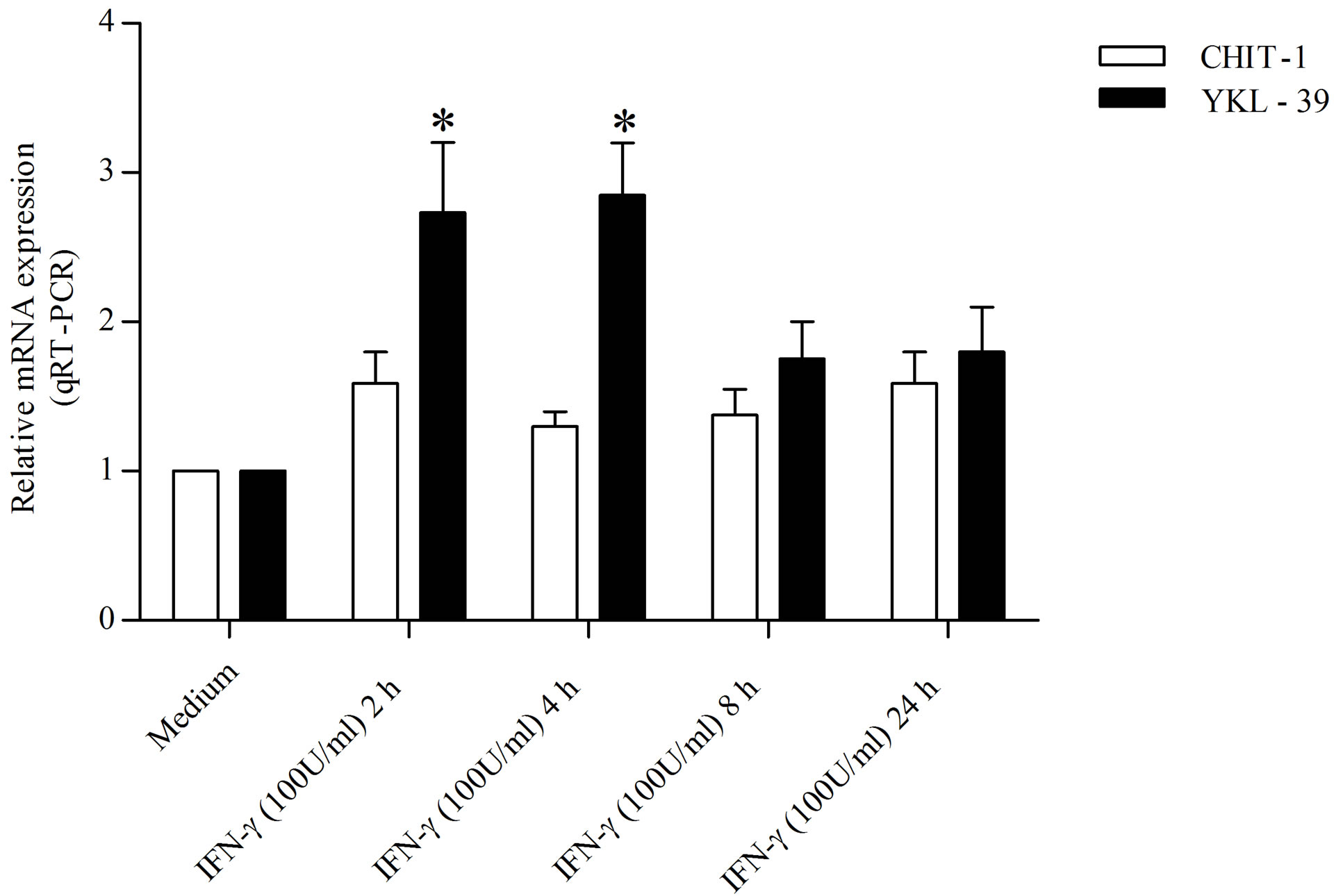
In order to detected the action carried out by LPS and IFN-γ on the expression of YKL-39 and CHIT-1 in undifferentiated monocytes, we treated the cells at different time points (2, 4, 8, and 24 h) maintaining the concentration used for the polarization in M1. We found that the treatment with IFN-γ (100 U/ml) increased YKL-39 expression by 2.69 and 2.83, over that of the control within 4 and 8 h, respectively. In contrast, IFN-γ (100 U/ml) barely induced CHIT-1expression by 1.67 fold at 2 h, by 1.21 and 1.34 at 4 and 8 h respectively and by 1.59 at 24 h compared to the control (Figure 1(a)). Similarly, the treatment with LPS (50 ng/ml) increased YKL-39 expression by 1.87, 2.12, 2.39 and 2.41 fold over that of the control within 2, 4, 8 and 24 h respectively. The expression of CHIT-1 under the effect of with LPS (50 ng/ml) was reduced by 0.31 fold within 2 h compared to the control and hold steady within 24 h (Figure 1(b)).
3.2. Effect of IL-4 on the Expression of YKL-39 and CHIT-1 in Undifferentiated Monocytes
To examine the effect of IL-4 on YKL-39 and CHIT-1 expression in undifferentiated monocytes, we treated the cells at different time points (2, 4, 8, 24 h) by using the concentration used for the polarization of macrophages in M2. Treatment with IL-4 (20 ng/ml) inhibited YKL-39 expression. We found that the expression of YKL-39 underwent by 6.02, 8.12, 9.16 and 9.03 fold decreaseswithin 2, 4, 8 and 24 h respectively (Figure 2). In contrast IL-4 treatment was able to modulate CHIT-1 expression in-

Table 1. Primer pairs used in real-time PCR analysis.
 (a)
(a)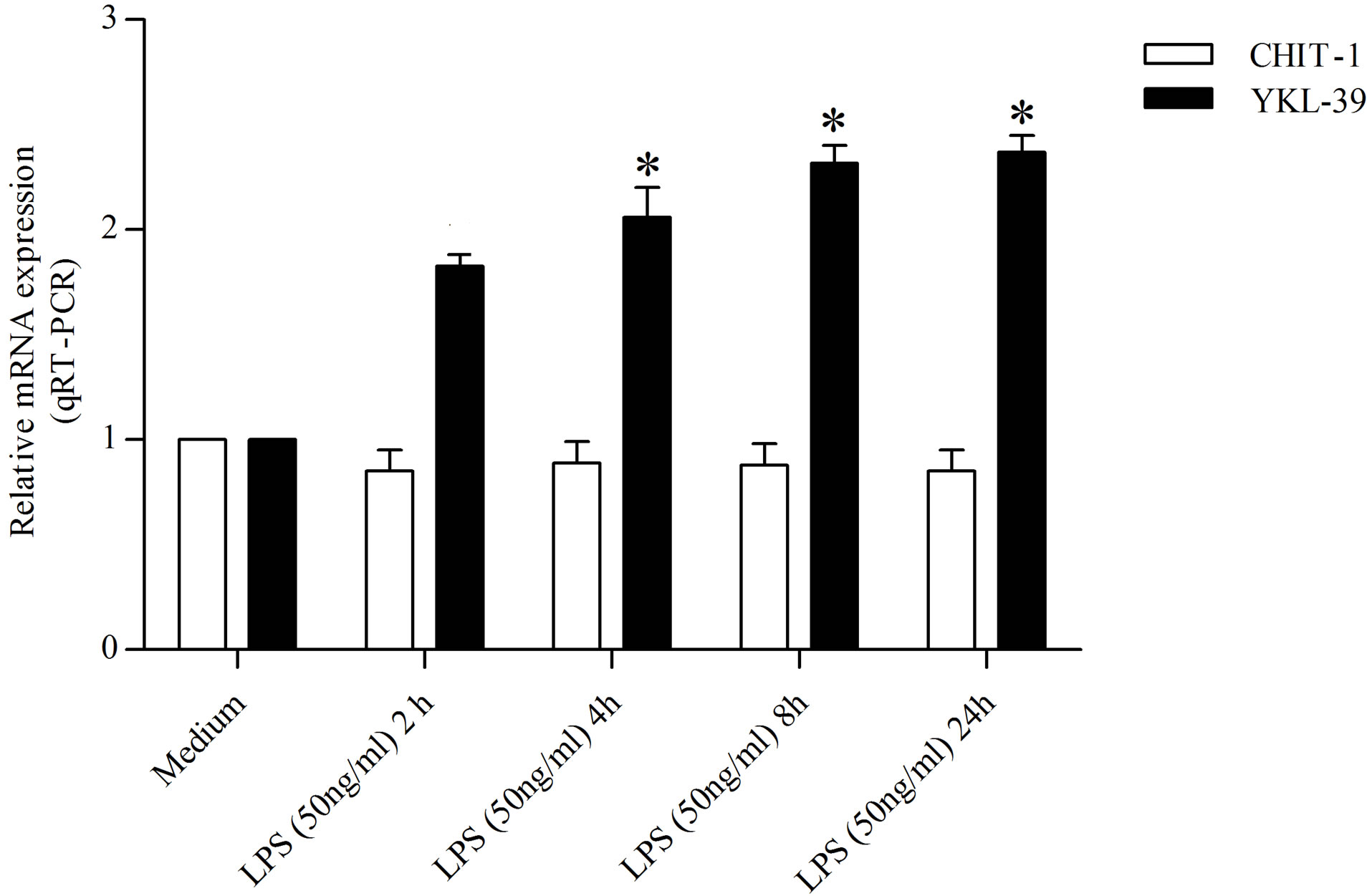 (b)
(b)
Figure 1. Detection of YKL-39 and CHIT-1 mRNA level by real time RT-PCR in undifferentiated monocytes, untreated and treated for 2, 4, 8, and 24 h with (a) IFN-γ (100 U/ml) (b) LPS (50 ng/ml). All RT-PCR values are given as ΔCt values, which correspond to the difference of the maximum number of RT-PCR cycles and the value obtained for each individual sample. Statistical analysis was performed by student’s t-test. *p < 0.05 of treated cells versus control.
ducing an increase in the early hours of treatment (5.30 fold and 5.5 fold increase within 2 and 4 h respectively) compared to the control cells (Figure 2). Whereas a significant reduction of CHIT-1 expression was detected within 8 and 24 h (1.04 fold decrease and 5.66 fold decrease, respectively) compared to the untreated cells (Figure 2).
3.3. Expression of YKL-39 and CHIT-1 during in Vitro Differentiation and Polarization of Peripheral Blood Monocytes
In order to compare the modulation of YKL-39 expression and CHIT-1 on primary human monocyte-tomacrophage maturation and subsequent polarization into M1 or M2 cells we used quantitative real time RT-PCR. The levels of CHIT-1 during the differentiation of mono-
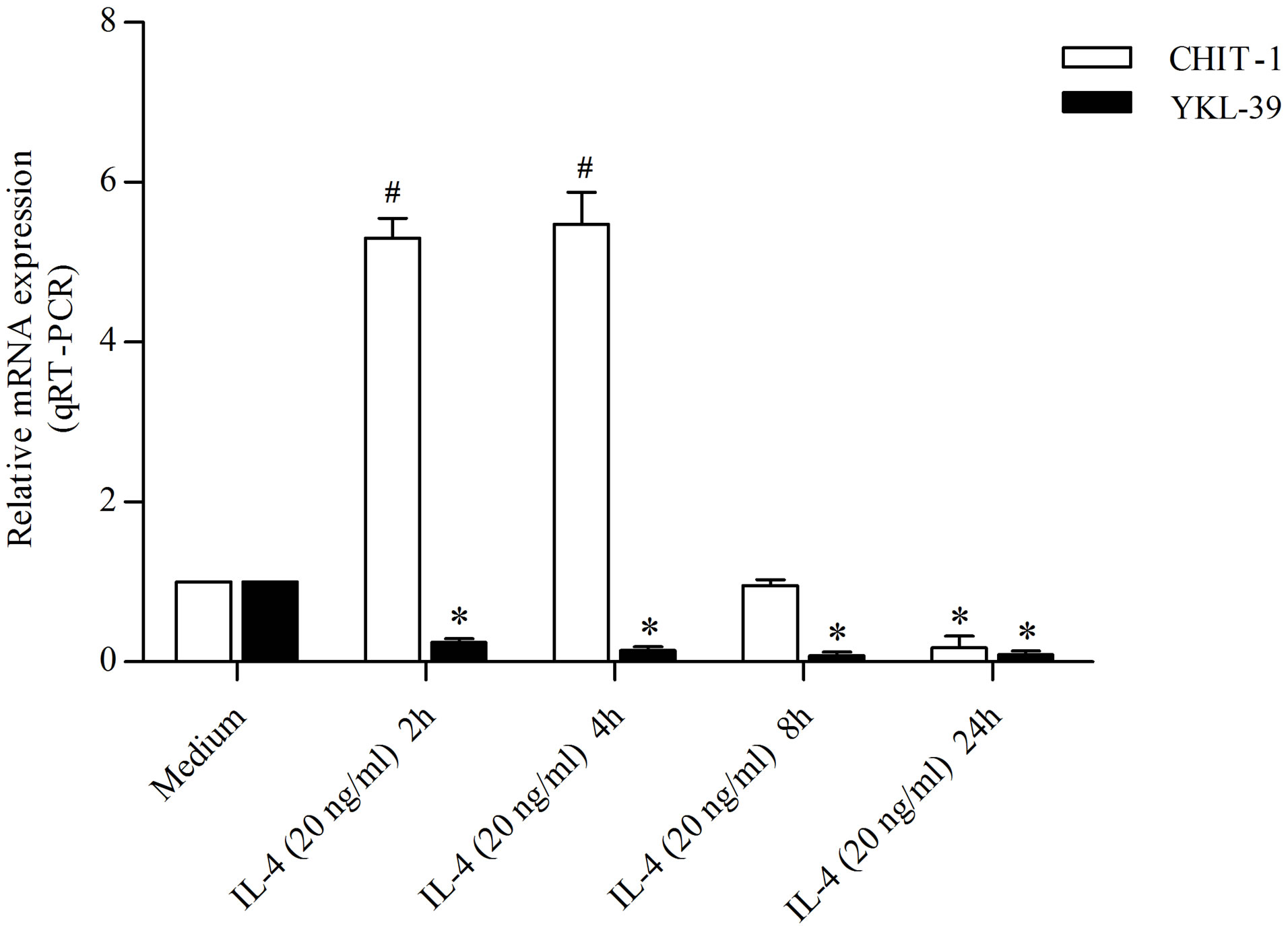
Figure 2. Detection of YKL-39 and CHIT-1 mRNA level by real time RT-PCR in M2-alternative activated macrophages, untreated and treated for 2, 4, 8, and 24 h with IL-4 (20 ng/ml). All RT-PCR values are given as ΔCt values, which correspond to the difference of the maximum number of RT-PCR cycles and the value obtained for each individual sample. Statistical analysis was performed by student’s t-test; *p < 0.05 and #p < 0.001 of treated cells versus control.
cytes into macrophages and subsequent macrophages polarization into M1 and M2 showed significant changes. After 3 days of culture a fold increase of 2.87 was observed compared to the day 0 (Figure 3). On the fifth day levels of CHIT-1 expression were increased by 15.12, and on the seventh day, the expression of CHIT-1 reached an increase by 15.75 fold. In a previous study quantitative evaluation of CHIT-1 protein confirmed that CHIT-1 enhancement is a mechanism operant with macrophages differentiation and polarization [35]. In contrast, YKL-39 expression was unchanged in the diverse stage of HMMs differentiation. Following polarization in M1 macrophages YKL-39 levels showed a significant increase (14.72 fold) compared to day 0. In contrast, CHIT-1 displayed an opposite trend compared to YKL-39 (Figure 3). In fact, CHIT-1 expression showed a fold increase of 6.62 compared to day 0. In M2 macrophages CHIT-1 was more expressed than in M1 showing a fold increase by 7.52 compared to day 0 (Figure 3) whereas YKL-39 expression reverted to the basal levels.
3.4. Analysis of YKL-39 Promoter Module Using MatInspector Modeling
Functionally related promoters often exhibit a defined core organization of binding sites conserved in orientation and in distance. This condition is true even when the promoter sequences do not show significant overall sequence similarity [36]. The advances in silico analysis of promoter structure have allowed characterization of this higher-order organization and the subsequent search for potential co-regulated genes. To elucidate the mechanism
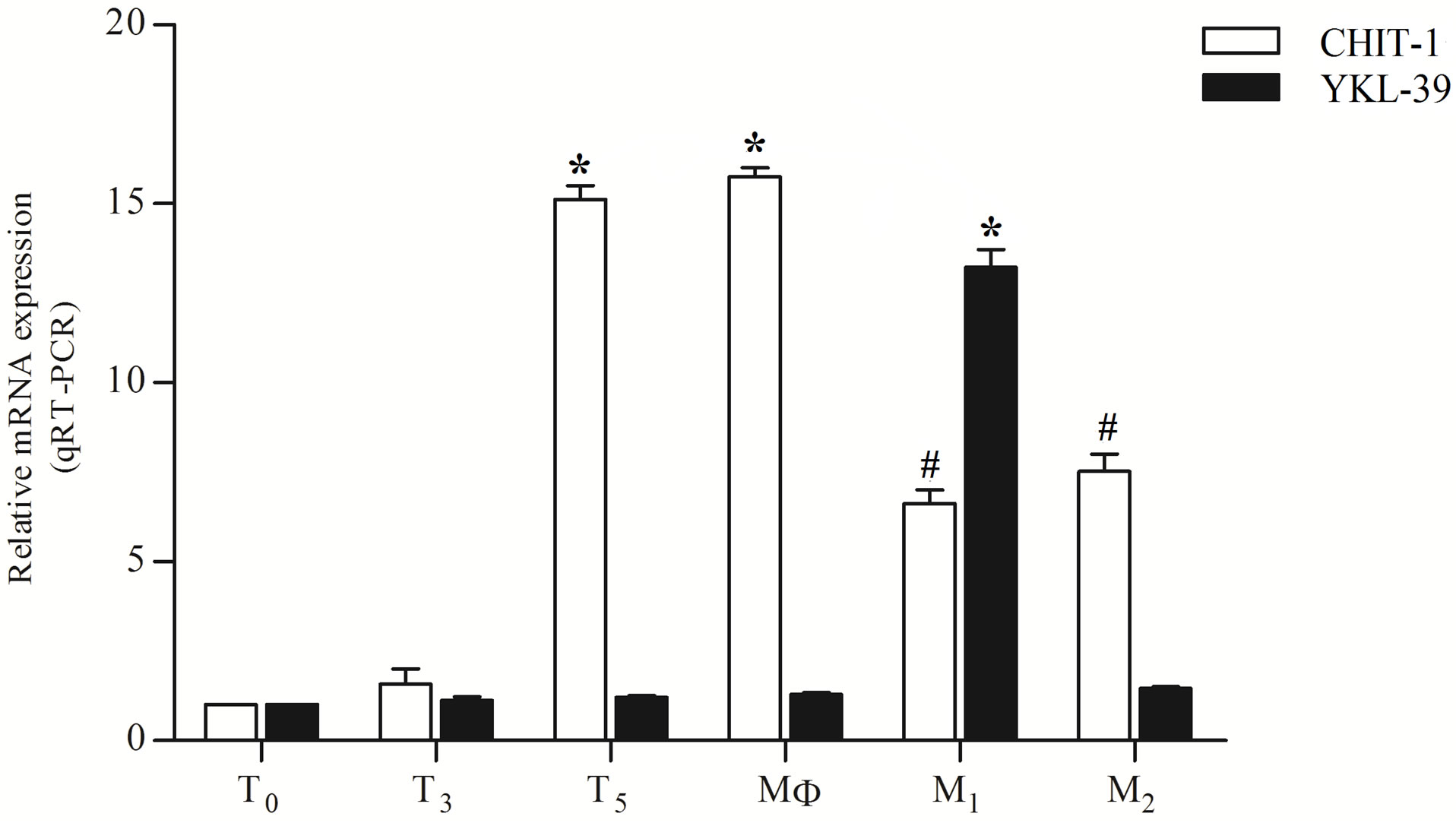
Figure 3. Real-time PCR of YKL-39 and CHIT-1 expression during human monocyte/macrophages differentiation and polarization. T0, T3, T5 (monocyte/macrophages at day 0, 3, 5); MΦ 7 (macrophages at day 7), M1: classical activated macrophages, M2: alternative activated macrophages. All RT-PCR values are given as ΔCt values, which correspond to the difference of the maximum number of RT-PCR cycles and the value obtained for each individual sample. Statistical analysis was performed by student’s t test. *p < 0.001 and #p < 0.05 of differentiated or polarized cells versus day 0 cells.
of different regulation of CHI3L-2 by IFN-γ and IL-4 we analyzed the promoter region of YKL-39 to search the presence of STATs consensus sequences. As shown in Figure 4(a), the YKL-39 promoter contains consensus sequences with defined core and matrix parameters for STAT1, STAT3, STAT5, STAT6 and NF-κB [31]. A total of two sequence for NF-κB (one in positive strand and one in negative strand), for STAT5 (in positive strand) and STAT1 (two in negative strand), one for STAT 3 (positive strand) and for STAT6 (positive strand) were found (Figure 4(b)). The core and matrix similarity scores for the various transcription factors weight matrices in the YKL-39 promoter are shown Figure 4(c). For generating the models, moderate similarity thresholds were used (CS > 0.75, MS optimized for the individual weight matrices) for each element. Matches on both strands of the DNA were recognized. Some flexibility was introduced into the models when determining space between the elements. ModelInspector was previously used to search for CHIT-1 promoter in which we found consen-
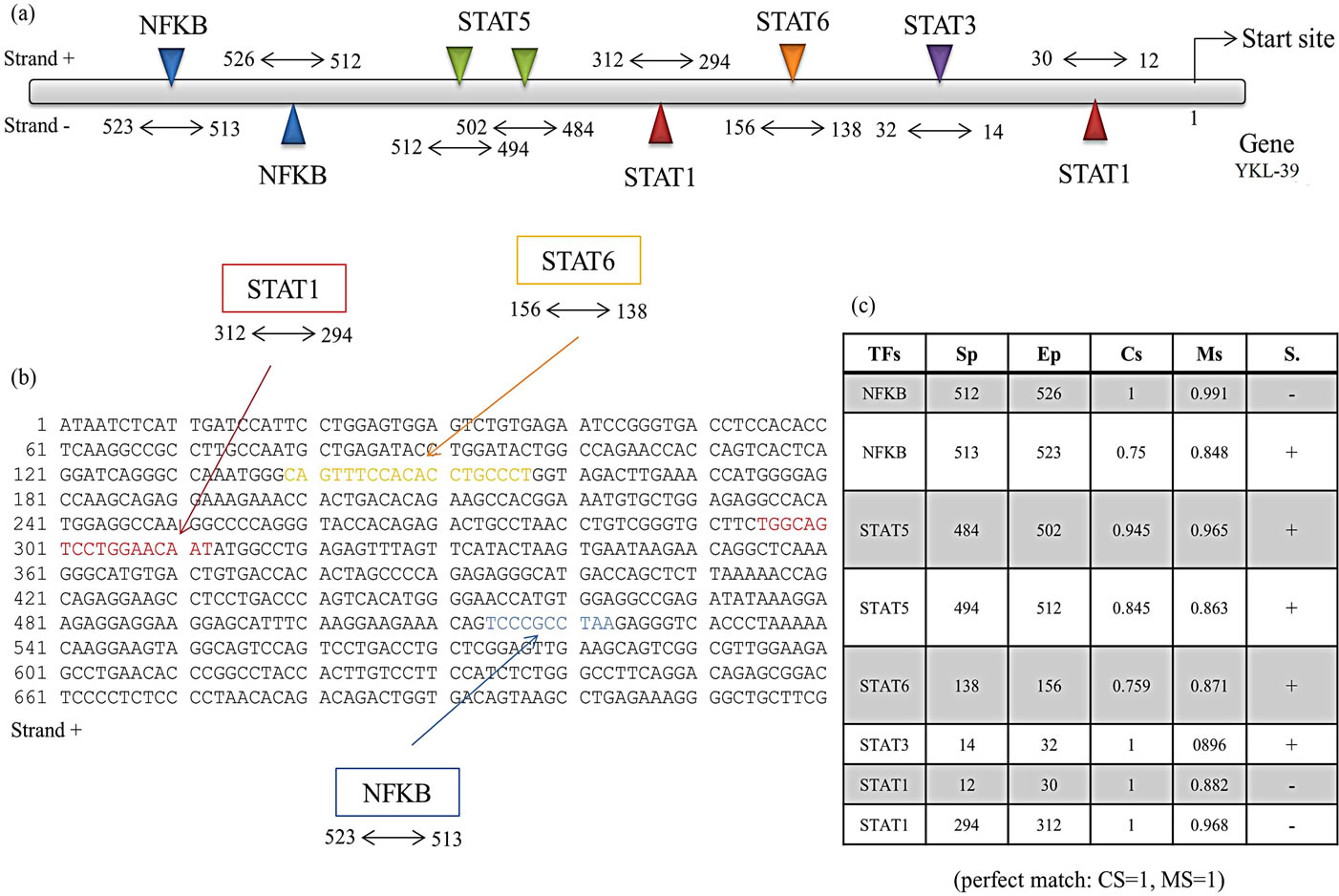
Figure 4. Design of FastM computer models. (a) Schematic representation of YKL-39 promoter, with the related transcription factors weight matrix parameters obtained from a MatInspector analysis. The matrix similarity regions are boxed, and the core and matrix similarity scores (CS, MS) state how close the YKL-39 promoter sequence matches the weight matrix (perfect match: CS = 1, MS = 1); (b) Sequence of the positive strand of the promoter of chit1 in box with the recognition sequences for transcription factors; (c) Table for transcription factors with the following parameters: Tfs, transcription factor, Sp, start position, Ep, end position, Cs, core similarity, Ms, matrix similarity, S, strand. Some flexibility was introduced for the distances between the elements, and strand orientation was not restricted.
sus sequences with defined core and matrix parameters for STAT1, STAT3 and STAT6 [37].
4. Discussion
In this study we compared the modulation of expression of both YKL-39 and CHIT-1 during monocyte/ macrophages differentiation and polarization. The results reported here show that the effective maturation of monocytes into macrophages is shown by the increase of CHIT-1 levels in the course of the time and whit a peak of expression between the fifth and the seventh day of culture. In contrast, YKL-39 expression doesn’t show significant changes in the diverse stages of HMMs differentiation, but increases significantly in M1 polarized macrophages, whereas in M2 reverted to base levels. In contrast to CHIT-1, YKL-39 has only be found to be expressed by synovial fibroblasts during osteoarthritis (OA) [4,5]. Nevertheless, more recently was demonstrated that YKL-39 mRNA was expressed on a very low level in macrophages differentiated in the presence of IFN-γ or IL-4 [25]. Demonstration of very low levels of YKL-39 transcripts during macrophages differentiation is consistent, in part, with our results. Here we present the evidence, that only classically activated or M1 macrophages are able to produce YKL-39 and that in response to IFN-γ and LPS stimulation undifferentiated monocytes were able to produce YKL-39. It has been shown that Th1 cells predominate in the synovium of patients with OA [38]. Consequently, IFN-γ which is one of the prevailing cytokine in OA tissues is able to induce the production of YKL-39 by monocyte-derived macrophages. In patients with OA, the prevalence of autoantibodies to YKL-39 and other auto-antigens on early stages of disease implied that the autoimmune response occurs during the initial phase of cartilage degeneration [7]. This finding suggests that synovial fibroblasts do not represent the sole source of YKL- 39 in OA. A study of Gratchev et al. showed that combination of IL-4 and TGF-β had strong stimulatory effect on the expression of YKL-39 in macrophage cultures [28]. Nevertheless they reported that IFN-γ has inhibitory effect on the production of YKL-39 by monocyte-derived macrophages, which is in contrast with our results. The “classically” polarized macrophages or “M1” resemble to Th1 cells. Like Th1 cells they are considered pro-inflammatory since they secrete proinflammatory cytokines and an array of cytotoxic molecules that help the clearance of invading pathogens and stimulate the acquired immune response. In our study, verifying the effect that LPS or IFN-γ individually exerts on YKL-39 and CHIT-1 expression in undifferentiated monocytes, we found that while treatment with LPS was unable to enhance CHIT- 1 expression, it started to induce YKL-39 expression after 2 h of treatment and went on over the time. Instead IFN-γ treatment enhanced YKL-39 expression at 2 h and reached the higher expression at 4 h. In contrast, IFN-γ was ineffective on CHIT-1 expression. These results suggest that monocytes recruited to the sites of chronic inflammation can be a source of YKL-39 themselves. Numerous evidences report the central role of CHIT-1 in the expanding spectrum of disorders suggesting that overproduction of CHIT-1 could exert deleterious effect in many degenerative disorders [24]. This concept is also sustained by our previous findings in which we observed that genetic variation within the CHIT-1 gene was strongly associated with human longevity and with several phenotypes of healthy aging [39], and that a functional polymorphism in the CHIT-1 gene protects from NAFLD progression [40]. Alternative or “M2” macrophages, reproduce the Th2 response of T cells and are considered anti-inflammatory; they express an array of phagocytic receptors, anti-inflammatory cytokines and release products that promote tissue regeneration and healing. Interestingly, in undifferentiated monocytes IL-4 treatment induced a significant increase on CHIT-1 expression, whereas was ineffective on YKL-39. As recently reported the presence of several recognition sites for STAT-6 on CHIT-1 promoter confirmed the up-regulation observed after treatment of macrophages with IL-4 [37]. In contrast, in the present study the in silico analysis of YKL-39 promoter sequences shows two STAT1 and two NF-κB binding site confirming and clarifying the YKL-39 induction during differentiation of macrophages into M1. Moreover, the finding of several recognition sites for NF-κB and STAT1 in YKL-39 promoter confirms the up-regulation observed after treatment of macrophages with LPS and IFN-γ. Moreover, analyzing the CS and MS we found sequences for STAT1 (induced by IFN-γ) and sequences for NF-κB (induced by LPS), with a higher CS and MS for the perfect match. In contrast there are recognition sites for IL4 in positive strand showing CS and MS very low. The lack of YKL-39 expression in undifferentiated monocytes IL-4-treated and in M2 polarized macrophages indicated that the function of these chitinase is much more restricted and selective than that exerted by CHIT-1. In conclusion the findings presented here confirm that certainly YKL-39 has a crucial role as an inducer of immune responses related to arthritis. On the contrary, the hypothesis that YKL-39 might serve as a useful biomarker to detect macrophage-specific response in pathologies like tumor, atherosclerosis and Alzheimer disease must be accepted with prudence. Further investigations are indispensable to pinpoint the biological activity of YKL-39 in inflammation and degenerative diseases.
5. ACKNOWLEDGEMENTS
We thank Dr. Filomena Biazzo of the Transfusional Centre “Garibaldi” Hospital, Catania, S. Immuno-Haemathology and Transfusional Medicine, for providing buffy coats.
REFERENCES
- Funkhouser, J.D. and Aronson Jr., N.N. (2007) Chitinase family GH18: Evolutionary insights from the genomic history of a diverse protein family. BMC Evolutionary Biology, 7, 96. http://dx.doi.org/10.1186/1471-2148-7-96
- Schimpl, M., Rush, C.L., Betou, M., Eggleston, I.M., Recklies, A.D. and van Aalten, DM. (2012) Human YKL- 39 is a pseudo-chitinase with retained chito-oligosaccharide-binding properties. Biochemical Journal, 446, 149- 157. http://dx.doi.org/10.1042/BJ20120377
- Hu, B., Trinh, K., Figueiira, W.F. and Price, P. (1996) Isolation and sequence of a novel human chondrocyte protein related to mammalian members of the chitinase protein family. The Journal of Biological Chemistry, 271, 19415-19420. http://dx.doi.org/10.1074/jbc.271.32.19415
- Steck, E., Breit, S., Breusch, S.J. Axt, M. and Richter, W. (2002) Enhanced expression of the human chitinase 3- like 2 gene (YKL-39) but not chitinase 3-like 1 gene (YKL-40) in osteoarthritic cartilage. Biochemical and Biophysical Research Communications, 299, 109-115. http://dx.doi.org/10.1016/S0006-291X(02)02585-8
- Knorr, T., Obermayr, F., Bartnik, E., Zien, A. and Aigner, T. (2003) YKL-39 (chitinase 3-like protein 2), but not YKL-40 (chitinase 3-like protein 1), is up regulated in osteoarthritic chondrocytes. Annals of the Rheumatic Diseases, 62, 995-998. http://dx.doi.org/10.1136/ard.62.10.995
- De Ceuninck, F., Marcheteau, E., Berger, S., Caliez, A., Dumont, V., Raes, M., Anract, P., Leclerc, G, Boutin, J.A. and Ferry, G. (2005) Assessment of some tools for the characterization of the human osteoarthritic cartilage proteome. Journal of Biomolecular Techniques, 16, 256-265.
- Du, H., Masuko-Hongo, K., Nakamura, H., Xiang, Y., Bao, C.D., Wang, X.D., Chen, S.L., Nishioka, K. and Kato, T. (2005) The prevalence of autoantibodies against cartilage intermediate layer protein, YKL-39, osteopontin, and cyclic citrullinated peptide in patients with earlystage knee osteoarthritis: Evidence of a variety of autoimmune processes. Rheumatology International, 26, 35- 41. http://dx.doi.org/10.1007/s00296-004-0497-2
- Tsuruha, J., Masuko-Hongo, K., Kato, T., Sakata, M., Nakamura, H., Sekine, T., Takigawa, M. and Nishioka, K. (2002) Autoimmunity against YKL-39, a human cartilage derived protein, in patients with osteoarthritis. Journal of Rheumatology, 29, 1459-1466.
- Kzhyshkowska, J., Gratchev, A. and Goerdt, S. (2007) Human chitinases and chitinase-like proteins as indicators for inflammation and cancer. Journal of Biomarker Insights, 2, 128-146.
- Boot, R.G., van Achterberg, T.A., van Aken, B.E., Renkema, G.H., Jacobs, M.J., Aerts, J.M. and de Vries, C.J. (1999) Strong induction of members of the chitinase family of proteins in atherosclerosis: Chitotriosidase and human cartilage gp-39 expressed in lesion macrophages. Arteriosclerosis, Thrombosis, and Vascular Biology, 19, 687-694. http://dx.doi.org/10.1161/01.ATV.19.3.687
- Malaguarnera, L. (2006) Chitotriosidase: The yin and yang. Cellular and Molecular Life Sciences, 63, 3018- 3029. http://dx.doi.org/10.1007/s00018-006-6269-2
- Hall, A.J., Morroll, S., Tighe, P., Götz, F. and Falcone, F.H. (2008) Human chitotriosidase is expressed in the eye and lacrimal gland and has an antimicrobial spectrum different from lysozyme. Microbes and Infection, 10, 69- 78. http://dx.doi.org/10.1016/j.micinf.2007.10.007
- Boven, L.A., van Meurs, M., Boot, R.G., Mehta, A., Boon, L., Aerts, J.M. and Laman, J.D. (2004) Gaucher cells demonstrate a distinct macrophage phenotype and resemble alternatively activated macrophages. American Journal of Clinical Pathology, 122, 359-369. http://dx.doi.org/10.1309/BG5VA8JRDQH1M7HN
- Aerts, J.M., Hollak, C.E., van Breemen, M., Maas, M., Groener, J.E. and Boot, R.G. (2005) Identification and use of biomarkers in Gaucher disease and other lysosomal storage diseases. Acta Paediatrica Supplement, 94, 43-46. http://dx.doi.org/10.1080/08035320510028094
- Brinkman, J., Wijburg, F.A., Hollak, C.E., Groener, J.E., Verhoek, M., Scheij, S., Scheij, S., Aten, J., Boot, R.G. and Aerts, J.M. (2005) Plasma chitotriosidase and CCL18: Early biochemical surrogate markers in type B NiemannPick disease. Journal of Inherited Metabolic Disease, 28, 13-20. http://dx.doi.org/10.1007/s10545-005-4416-9
- Barone, R., Malaguarnera, L., Angius, A. and Musumeci, S. (2003) Plasma chitotriosidase activity in patients with beta-thalassemia. American Journal of Hematology, 72, 285-286. http://dx.doi.org/10.1002/ajh.10294
- Bargagli, E., Bennett, D., Maggiorelli, C., Di Sipio, P., Margollicci, M., Bianchi, N. and Rottoli, P. (2013) Human chitotriosidase: A sensitive biomarker of sarcoidosis. Journal of Clinical Immunology, 33, 264-270. http://dx.doi.org/10.1007/s10875-012-9754-4
- Comabella, M., Domínguez, C., Rio, J., Martín-Gallán, P., Vilches, A., Vilarrasa, N., Espejo, C. and Montalban, X. (2009) Plasma chitotriosidase activity in multiple sclerosis. Clinical Immunology, 131, 216-222. http://dx.doi.org/10.1016/j.clim.2008.12.004
- Malaguarnera, L., Simporè, J., Prodi, D.A., Angius, A., Sassu, A., Persico, I., Barone, R. and Musumeci, S. (2003) 24-bp duplication in exon 10 of human chitotriosidase gene from the sub-Saharan to the Mediterranean area: Role of parasitic diseases and environmental conditions. Genes and Immunity, 4, 570-574. http://dx.doi.org/10.1038/sj.gene.6364025
- Malaguarnera, L., Di Rosa, M., Zambito, A.M., Dell’Ombra, N., Di Marco, R. and Malaguarnera, M. (2006) Potential role of chitotriosidase gene in nonalcoholic fatty liver disease evolution. American Journal of Gastroenterology, 101, 2060-2069. http://dx.doi.org/10.1111/j.1572-0241.2006.00680.x
- Malaguarnera, L., Di Rosa, M., Zambito, A.M., Dell’Ombra, N., Nicoletti, F. and Malaguarnera, M. (2006) Chitotriosidase gene expression in Kupffer cells from patients with non-alcoholic fatty liver disease. Gut, 55, 1313- 1320. http://dx.doi.org/10.1136/gut.2005.075697
- Artieda, M., Cenarro, A., Gañán, A., Lukic, A., Moreno, E., Puzo, J., Pocoví, M. and Civeira, F. (2007) Serum chitotriosidase activity, a marker of activated macrophages, predicts new cardiovascular events independently of Creactive protein. Cardiology, 108, 297-306. http://dx.doi.org/10.1159/000099099
- Palasik, W., Fiszer, U., Lechowicz, W., Czartoryska, B., Krzesiewicz, M. and Lugowska, A. (2005) Assessment of relations between clinical outcome of ischemic stroke and activity of inflammatory processes in the acute phase based on examination of selected parameters. European Neurology, 53, 188-193. http://dx.doi.org/10.1159/000086355
- Di Rosa, M., Dell’Ombra, N., Zambito, A.M., Malaguarnera, M., Nicoletti, F. and Malaguarnera, L. (2006) Chitotriosidase and inflammatory mediator levels in Alzheimer’s disease and cerebrovascular dementia. European Journal of Neuroscience, 23, 2648-2656. http://dx.doi.org/10.1111/j.1460-9568.2006.04780.x
- Kzhyshkowska, J., Mamidi, S., Gratchev, A., Kremmer, E., Schmuttermaier, C., Krusell, L., Haus, G., Utikal, J., Schledzewski, K., Scholtze, J. and Goerdt, S. (2006) Novel stabilin-1 interacting chitinase-like protein (SICLP) is up-regulated in alternatively activated macrophages and secreted via lysosomal pathway. Blood, 107, 3221-3228. http://dx.doi.org/10.1182/blood-2005-07-2843
- Ross, R., Ross, X.L., Ghadially, H., Lahr, T., Schwing, J., Knop, J. and Reske-Kunz, AB. (1999) Mouse langerhans cells differentially express an activated T cell attracting CC chemokine. Journal of Investigative Dermatology, 113, 991-998. http://dx.doi.org/10.1046/j.1523-1747.1999.00803.x
- Martinez, F.O., Sica, A. and Mantovani, A.M. (2008) Macrophage activation and polarization. Frontiers in Bioscience, 13, 453-461. http://dx.doi.org/10.2741/2692
- Gratchev, A., Schmuttermaier, C., Mamidi, S., Gooi, L., Goerdt, S. and Kzhyshkowska, J. (2008) Expression of osteoarthritis marker YKL-39 is stimulated by transforming growth factor beta (TGF-beta) and IL-4 in differentiating macrophages. Journal of Biomarker Insights, 3, 39- 44.
- Malaguarnera, L., Imbesi, R., Di Rosa, M., Scuto, A., Castrogiovanni, P., Messina, A. and Sanfilippo, S. (2005) Action of prolactin, IFN-gamma, TNF-alpha and LPS on heme oxygenase-1 expression and VEGF release in human monocytes/macrophages. International Immunopharmacology, 5, 1458-1469. http://dx.doi.org/10.1016/j.intimp.2005.04.002
- Fagone, P., Di Rosa, M., Palumbo, M., De Gregorio, C., Nicoletti, F. and Malaguarnera, L. (2012) Modulation of heat shock proteins during macrophage differentiation. Inflammation Research, 61, 1131-1139. http://dx.doi.org/10.1007/s00011-012-0506-y
- Quandt, K., Frech, K., Karas, H., Wingender, E. and Werner, T. (1995) MatInd and MatInspector: New fast and versatile tools for detection of consensus. Matches in nucleotide sequence data. Nucleic Acids Research, 23, 4878-4884. http://dx.doi.org/10.1093/nar/23.23.4878
- Heinemeyer, T., Chen, X., Karas, H., Kel, A.E., Kel, O.V., Liebich, I., Meinhardt, T., Reuter, I., Schacherer, F. and Wingender, E. (1999) Expanding the TRANSFAC database towards an expert system of regulatory molecular mechanisms. Nucleic Acids Research, 27, 318-322. http://dx.doi.org/10.1093/nar/27.1.318
- Klingenhoff, A., Frech, K., Quandt, K. and Werner, T. (1999) Functional promoter modules can be detected by formal models independent of overall nucleotide sequence similarity. Bioinformatics, 15, 180-186. http://dx.doi.org/10.1093/bioinformatics/15.3.180
- Frech, K., Danescu-Mayer, J. and Werner, T. (1997) A novel method to develop highly specific models for regulatory units detects a new LTR in GenBank which contains a functional promoter. Journal of Molecular Biology, 270, 674-687. http://dx.doi.org/10.1006/jmbi.1997.1140
- Di Rosa, M., Malaguarnera, G., De Gregorio, C., D’Amico, F., Mazzarino, M.C. and Malaguarnera, L. (2013) Modulation of chitotriosidase during macrophage differentiation. Cell Biochemistry and Biophysics, 66, 239-247. http://dx.doi.org/10.1007/s12013-012-9471-x
- Lavorgna, G., Boncinelli, E., Wagner, A. and Werner, T. (1998) Detection of potential target genes in silico? Trends in Genetics, 14, 375-376. http://dx.doi.org/10.1016/S0168-9525(98)01551-0
- Di Rosa, M., Malaguarnera, G., De Gregorio, C., Drago, F. and Malaguarnera, L. (2013) Evaluation of CHIT3L-1 and CHIT-1 expression in differentiated and polarized macrophages. Inflammation, 36, 482-492. http://dx.doi.org/10.1007/s10753-012-9569-8
- Ishii, H., Tanaka, H., Katoh, K., Nakamura, H., Nagashima, M. and Yoshino, S. (2002) Characterization of infiltrating T cells and Th1/Th2-type cytokines in the synovium of patients with osteoarthritis. Osteoarthritis Cartilage, 10, 277-281. http://dx.doi.org/10.1053/joca.2001.0509
- Malaguarnera, L., Ohazuruike, L.N., Tsianaka, C., Antic, T., Di Rosa, M. and Malaguarnera, M. (2010) Human chitotriosidase polymorphism is associated with human longevity in Mediterranean nonagenarians and centenarians. Journal of Human Genetics, 55, 8-12. http://dx.doi.org/10.1038/jhg.2009.111
- Di Rosa, M., Mangano, K., De Gregorio, C., Nicoletti, F. and Malaguarnera, L. (2013) Association of chitotriosidase genotype with the development of nonalcoholic fatty liver disease. Hepatology Research, 43, 267-275. http://dx.doi.org/10.1111/j.1872-034X.2012.01063.x

