American Journal of Plant Sciences
Vol.5 No.4(2014), Article ID:43254,11 pages DOI:10.4236/ajps.2014.54059
Overexpression of OsMAPK2 Enhances Low Phosphate Tolerance in Rice and Arabidopsis thaliana
![]()
College of Natural Resources and Life Science, Dong-A University, Busan, South Korea.
Email: *dhkim@dau.ac.kr
Copyright © 2014 Yeon Jae Hur, Doh Hoon Kim. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In accordance of the Creative Commons Attribution License all Copyrights © 2014 are reserved for SCIRP and the owner of the intellectual property Yeon Jae Hur, Doh Hoon Kim. All Copyright © 2014 are guarded by law and by SCIRP as a guardian.
Received October 29th, 2013; revised January 5th, 2014; accepted January 17th, 2014
KEYWORDS
Rice (Oryza sativa); Phosphorylation; MAP Kinase; Phosphate; Homeostasis
ABSTRACT
The mitogen-activated protein kinase (MAPK) cascade is the most important mechanism in environmental responses and developmental processes in plants. The OsMAPK2 gene has been found to function in plant tolerance to diverse biotic/abiotic stresses. This paper presents evidence that OsMAPK2 (Oryza sativa MAP kinase gene 2) is responsive to Pi deficiency and involved in Pi homeostasis. We found that full-length expression of OsMAPK2 was up-regulated in both rice plants and cell culture in the absence of inorganic phosphate (Pi). The transgenic rice and Arabidopsis plants overexpressing OsMAPK2 showed affected root development and increased plant Pi content compared with wild-type plants. Overexpression of OsMAPK2 controlled the expression of several Pi starvation-responsive genes. Our results indicated that OsMAPK2 enables tolerance phosphate deficiency and is involved in Pi homeostasis.
1. Introduction
Rice is a monocot model plant, as well as an important cereal crop that supplies food for more than half of the world population. Rice (Oryza sativa L.) is also a model organism to study functional genomics of monocot plants because its genome size is smaller than those of other monocot plants.
Phosphate (Pi) is one of the most important nutrients for plant growth and development, but the availability of phosphate is frequently a limiting factor for crop productivity. This is because plants preferentially take up Pi as orthophosphate, and more than 80% of soil Pi is immobilized and is not readily available to roots [1,2]. Plants respond to Pi deficiency by activating mechanisms to increase Pi bioavailability and uptake. In response to persistent Pi deficiency, plants have developed many adaptive mechanisms to enhance availability and increase the uptake of Pi, and plants exhibit morphological modifications and biochemical changes as well as regulate the expression of genes in response to cues in the phosphate signaling pathway [2,3]. Secreting acid phosphatase increases Pi bioavailability by cleaving phosphate groups esterified to organic acids [4-6]. Although some of the components of Pi-starvation signaling in plants have been identified, the overall pathways are still poorly understood. It is important to study the functions of phosphate deficient condition responsive genes to increase the productivity and distribution of crop plants.
One of the major pathways by which external stimuli are transduced into intracellular responses is the mitogenactivated protein kinase (MAPK) signaling pathway [7,8] (Figure 1). MAPK cascades are composed of three protein kinase modules: MAPKK kinases (MAPKKKs), MAPK kinases (MAPKKs) and MAPKs, which are linked in various ways to upstream receptors and downstream targets. In this phosphorylation module, a MAPKKK phosphorylates a MAPKK. MAPKKs are activated when serine and serine/threonine residues in the S/T-X3-5-S/T motif are phosphorylated by serine/threonine kinases MAPKKKs [9]. MAPK is then activated by the dual-

Figure 1. Map kinase cascade. Details are mentioned in the text.
specificity kinase MAPKK. A dual phosphorylation activation motif of MAPK is threonine and tyrosine residues in a TXY located between subdomains VII and VIII [10]. Activated MAPK is often imported into the nucleus, where it phosphorylates and activates specific downstream signaling components, such as transcription factors [11]. The activated specific MAP kinases phosphorylate various downstream targets and regulate stress and hormonal responses, innate immunity, and developmental programs [12-15].
To date, most of the reported plant MAPKs have been isolated and characterized from dicot model species such as Arabidopsis and tobacco. Arabidopsis AtMPK3 kinase expresses strongly after cold, touch and dehydration in terms of transcription level [16]. Some MAPKs have also been identified and characterized from rice (Oryza sativa; [17-21]).
The OsMAPK2 gene (also known as OsMSRMK2, OsBIMK1, or OsMAP1) has been reported, and it was demonstrated that OsMAPK2 may play a cellular role in various biotic and abiotic stresses [18-22].
In this study, we obtained transgenic rice overexpressing the full-length OsMAPK2 gene and reported the effect of OsMAPK2 in response to phosphate deficient conditions and the acquisition of Pi. These results reveal that OsMAPK2 regulates the expression of an important set of genes in response to phosphate deficient conditions.
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
Rice (Oryza sativa L. cv. Dongjin) plants were grown in a hydroponic growth facility with one-half strength Hoagland solution. Phosphorus starvation treatments were initiated three weeks after germination. The plants were transferred to an aerated hydroponics solution [5 mM Ca(NO3)4H2O, 2.5 mM K2SO4, 2 mM MgSO4∙7H2O, 500 µM KH2PO4, 0.12 mM KCl, 0.25 M MnSO4, 0.25 M H3BO3, 0.25 M ZnSO4, 20 mM CuSO4, 20 mM Na2MoO4, 9.8 mM Fe-EDTA and 0.5 mM K2SO4] with or without 500 µM Pi. Five days after transfer, leaves and roots were harvested and stored at −70˚C until further analysis. Rice cell cultures were maintained in the laboratory as described earlier [23]. Phosphorus deficiency treatments were initiated seven days after subculturing the cells. From a cell suspension, cultures growing in 25 mL Pi sufficient media were filtered through Miracloth (Calbiochem, CA) and washed with Pi deficient (0) or sufficient (500 µM) medium. The cells were transferred to the same medium used for washing. The samples were collected at the indicated times after transferring the cells to Pi-deficient medium and then centrifuged at 400 × g for 4 min; the resulting pellet was then used for RNA isolation. The deficiency of the other nutrients was imposed by removing the nutrients (N, K, and Fe) from the solution for three days. Arabidopsis plants growth conditions followed the previously described method [24]. Seven-day-old seedlings were transferred to basal medium containing 1× Murashige and Skoog (HP) micronutrients, 1/5× macronutrients, 3% sucrose, 2.5 mM MES, B5 vitamins, 1.2% agar or MS LP medium (substitute 0.6 mM K2SO4 for 1.2 M KH2PO4) for 7 days.
2.2. DNA and RNA Gel Blot Analysis
Total genomic DNA was isolated from rice leaves using the cetyltrimethyl ammonium bromide extraction method [25]. Five micrograms of genomic DNA digested with EcoRI and XhoI were separated by electrophoresis on 1.2% (w/v) agarose gels and blotted onto nylon transfer membranes (Amersham).
Total RNA was isolated from rice plants and rice cell cultures using TRIzol reagent (Sigma). Total RNA (10 µg) was separated by electrophoresis on 1.2% (w/v) denaturing formaldehyde agarose gels and blotted onto nylon transfer membranes (Amersham).
The membranes were hybridized at 65˚C for 12 h with a [α-32P]-dCTP-labeled probe in a solution containing 20% (w/v) SDS, 20 × SSPE, 100 g/L PEG (molecular weight 8000), 250 mg/L heparin, and 10 ml/L herring sperm DNA. The filters were washed twice in 2 × SSC and 0.2% (w/v) SDS at room temperature for 10 min, twice in 1 × SSC and 0.2% (w/v) SDS at 65˚C for 15 min, and twice in 0.1 × SSC and 0.2% (w/v) SDS at 65˚C for 20 min before autoradiography.
2.3. Measurement of Pi Concentration
Inorganic Pi measurement followed the previously described method [26]. A frozen sample (approximately 0.5 g) was homogenized in 1 mL of 10% (w/v) PCA, using an ice-cold mortar and pestle. The homogenate was then diluted 10 times with 5% (w/v) PCA and placed on ice for 30 min. After centrifugation at 10,000 ×g for 10 min at 4˚C, the supernatant was collected to measure the Pi, using the molybdate-blue method: 0.4% (w/v) ammonium molybdate melted in 0.5 M H2SO4 (solution A) was mixed with 10% ascorbic acid (solution B) (A:B = 6:1). Two milliliters of this solution was added to 1 mL of the sample solution and incubated in a water bath at 40˚C for 20 min. After being cooled on ice, the absorbance was measured at 820 nm.
2.4. Histochemical Localization of GUS Expression
Histochemical staining for GUS activity was performed according to the protocol described by Raghothama et al. [27], with some modifications. Whole seedlings or parts of plants to be stained were incubated in GUS reaction mix (25 mg of 5-bromo-4-chloro-3-indolyl-d-glucuronide in 50 mL of 100 mM sodium Pi buffer with 0.1% [v/v] Triton X-100) for 16 h. The stained seedlings were transferred to 70% (v/v) alcohol to remove chlorophyll.
2.5. Preparation of Construct
To amplify the cDNA of OsMAPK2, a reverse transcription mix (1 μL) was used in 50 μL PCR reactions containing OsMAPK2-specific primers and high-fidelity Taq polymerase LaTaq (TaKaRa). The following PCR conditions were employed: denaturation for 30 s at 94˚C, annealing for 30 s at 55˚C, and extension for 1 min at 72˚C. After 35 cycles of amplification, the PCR product was examined by agarose gel electrophoresis with ethidium bromide staining. A PCR fragment of the expected size was purified from the gel and cloned into the plasmid vector pGemT-easy (Promega). The inserts in selected positive clones were sequenced commercially (Genotech). The OsMAPK2 gene inserted into pGemT-easy was excised by BamHI restriction digestion and cloned in the sense orientation into the BamHI sites of the pCambia1300-35S vector. The construct was introduced into Agrobacterium tumefaciens (EHA105) by electroporation.
2.6. Plant Transformation
Agrobacterium tumefaciens-mediated Arabidopsis transformation was performed using the flower vacuum infiltration procedure described by Clough and Bent [28]. Transgenic T1 plants were selected on B5 medium containing carbenicillin (100 mg/L) and hygromycin (20 mg/L) and then transferred to soil and allowed to selfpollinate. Homozygous plants were selected and subjected to growth assays under Pi-depleted conditions. Rice plants were transformed with A. tumefaciens as described by Hiei et al. [29].
2.7. qRT-PCR Analysis
The total RNA of the rice seedlings was extracted with an RNeasy Plant mini kit (Qiagen). After treatment with DNase to remove genomic DNA contamination, the first strand of cDNA was synthesized by M-MLV reverse transcriptase using 3 µg of total RNA as a template. qRT-PCR was performed with a Bio-Rad CFX system using SYBR with the following reaction conditions: 95˚C for 10 min, then 40 cycles of 95˚C for 15 s, 60˚C for 15 s and 72˚C for 15 s. Primers for qRT-PCR analysis are shown in Supplement Table S1, and actin was used as an internal control.
2.8. Statistical Analysis
The results are expressed as the mean values ± SD of at least three replicates are presented. Statistical significance between measurements for different treatments or times was analyzed using Duncan’s multiple range test.
3. Results
3.1. Genomic Organization and Expression Patterns of the OsMAPK2 Gene
To assess the exact copy number of OsMAPK2 in the rice genome, genomic Southern-blot analysis was carried out using the full-length gene as a probe. We confirmed that wild-type plants contained one copy of the OsMAPK2 gene in the O. sativa genome (Figure 2(a)). We tested whether the OsMAPK2 gene is induced in rice suspension cell culture at various time intervals. In Pi deficient conditions, the transcript level increases after 30 min and decreased after 1 hour. Northern blot analysis confirmed that in response to N, P, K, and Fe deficiency treatments, the OsMAPK2 gene was induced very rapidly and strongly (within 1 h after exposure to -N, -P, -K, -Fe nutrient medium). This gene exhibited a stronger increase in transcript abundance in response to P and Fe deficiency (Figure 2(b)), and was induced by Pi deficiency within 30 min (Figure 2(c)). Hydroponic experiments were conducted using Dongjin wild-type rice and culture solution with 500 µM Pi or 0 µM Pi for 5 days. The OsMAPK2 gene was induced strongly during Pi deficiency in roots and was expressed weakly in shoots (Figure 2(d)).
3.2. Tissue Localization of OsMAPK2
To test whether the region upstream of the alternative translation start site of the OsMAPK2 promoter regulates gene expression, we cloned approximately 1.5 kb up-
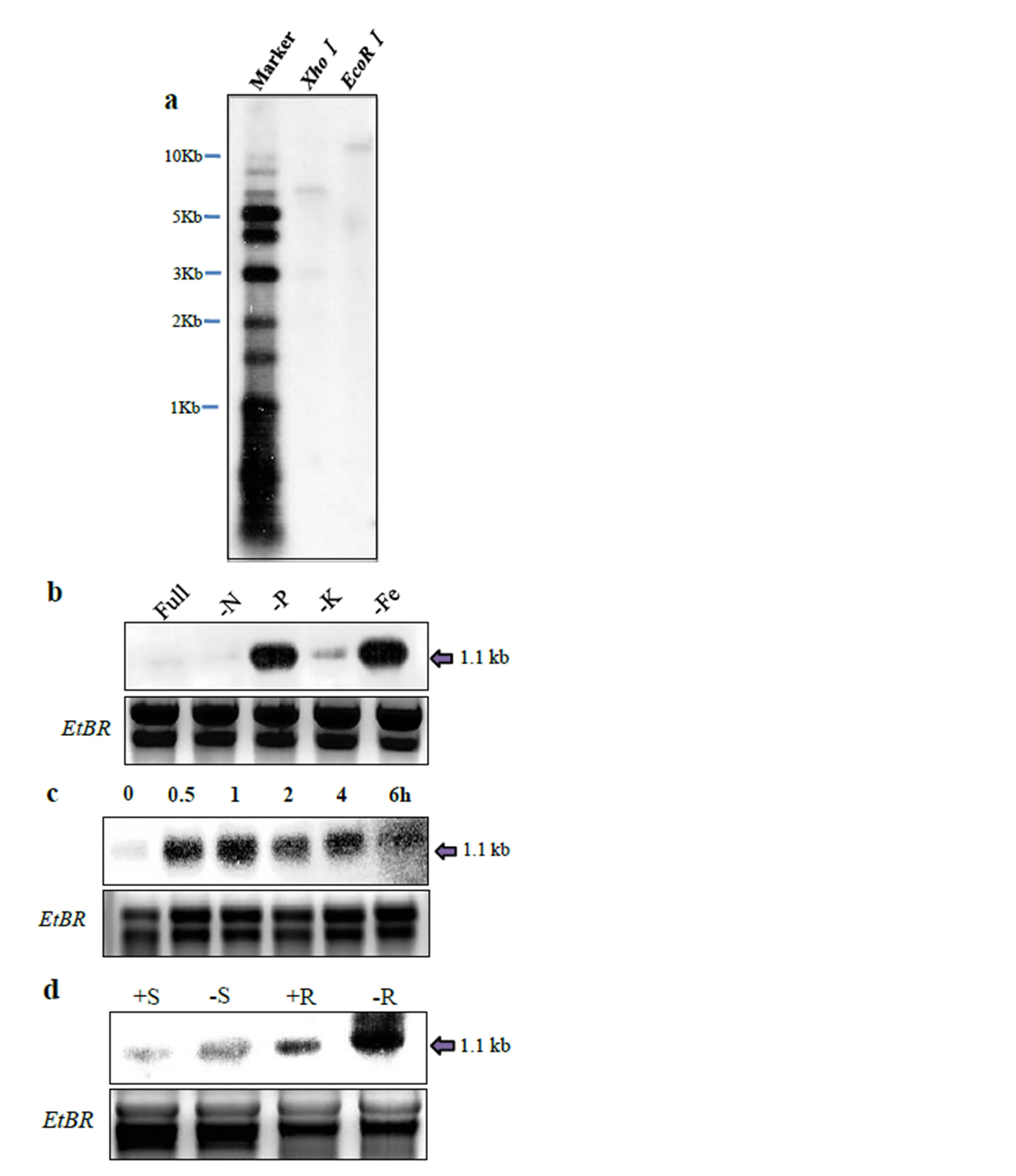
Figure 2. Hybridization analysis of OsMAPK2 genomic DNA and mRNA. (a) Southern-blot analysis for the wildtype using the OsMAPK2 gene as a probe. Five micrograms of genomic DNA were digested by XhoI and EcoRI and separated by agarose gel. Northern blot analysis of the OsMAPK2 gene; (b) Cell suspension cultures were provided with medium deficient in Pi (-P), nitrogen (-N), potassium (-K), or iron (-Fe), and a control solution with all nutrients (+) for 1 hour; (c) Expression of the OsMAPK2 gene was analyzed using RNA isolated from a rice suspension cell culture grown in the absence of Pi (−) for the indicated time; (d) Total RNA isolated from the roots and leaves of aeroponically grown plants supplied with Hoagland solution containing 500 µM Pi (+) or no Pi (−) for 5 days.
stream of the translation start sites of the OsMAPK2 gene and created transgenic lines carrying the GUS gene. Transgenic plants were confirmed for the presence of the appropriate expression cassette by PCR using specific primers (data not shown). Histochemical staining for GUS activity showed that strong GUS expression was observed in the roots of the whole plant 7 days after germination (Figure 3(a)) and in the lateral roots of transgenic plants 21 days after germination (Figure 3(d)). GUS expression was also detected in the mature spikelets in the reproductive stage (Figure 3(b)). Although weak signals appeared for the mature leaves, they were irregular and unstable (Figure 3(c)).
3.3. Alteration of Pi Contents in OsMAPK2 Transgenic Plants
To investigate the function of this gene, we established transgenic rice plants that overexpressed OsMAPK2 (OsMAPK2-Oe) under the control of the cauliflower mosaic virus 35S constitutive promoter.
Genomic Southern-blot analysis was performed and the transgenic plants contained one copy of a hygromycin probe (Figure 4(a)). The expression levels of the transgene were elucidated by Northern-blot analysis (Figure 4(b)) and qRT-PCR (Figure 4(c)). We observed high expression in transgenic plants, but no expression level was obtained from the wild-type plant.
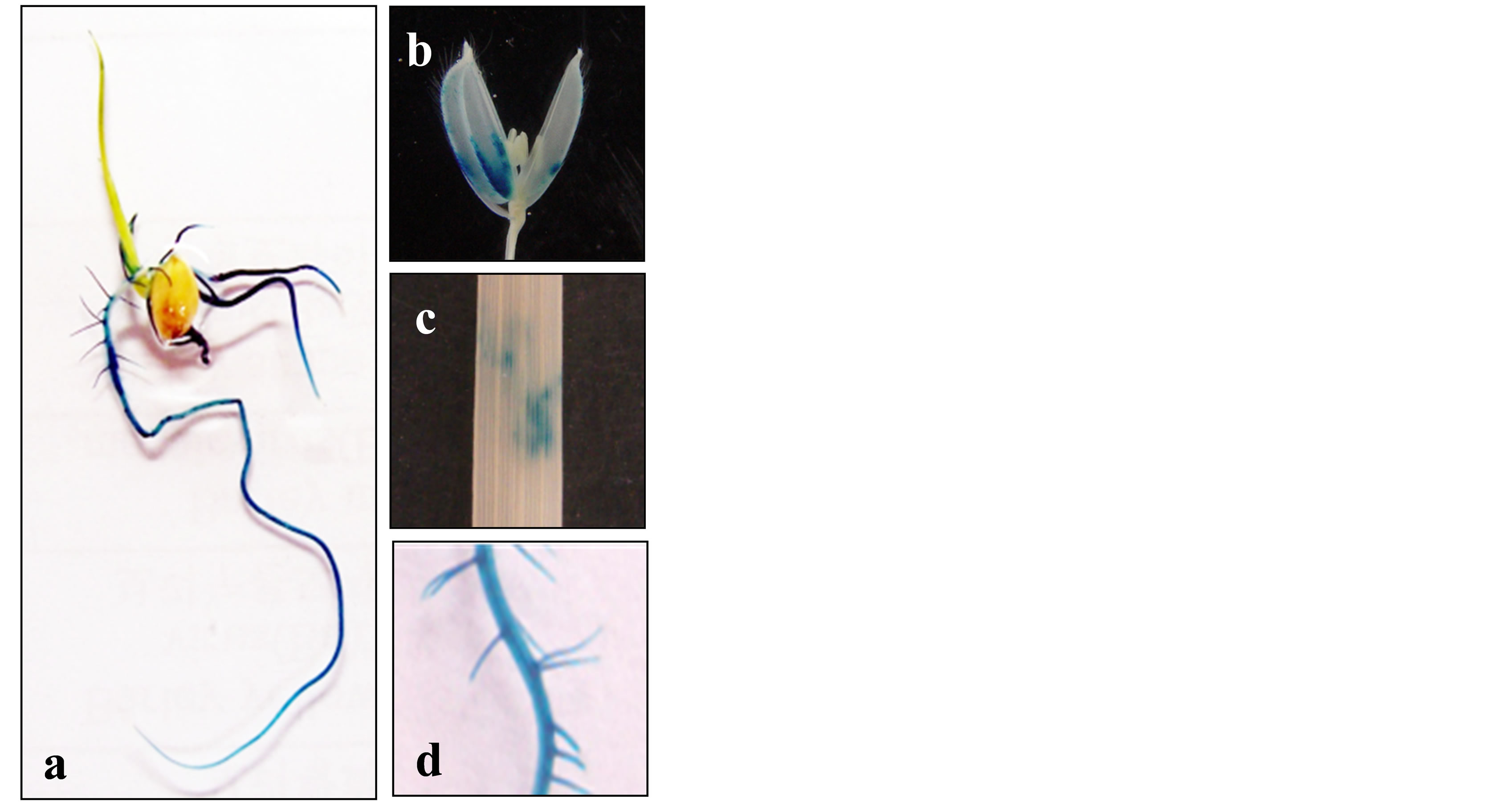
Figure 3. Histochemical analysis of OsMAPK2-GUS protein levels in transgenic rice plants. The GUS activity of OsMAPK2 of the transgenic rice plants at different developmental stages was analyzed. (a) The whole plant 7 days post-germination; (b) Mature spikelets before anthesis at flowering; (c) mature leaves; (d) a 3- to 5 cm section from the root tip at 21 days post-germination.
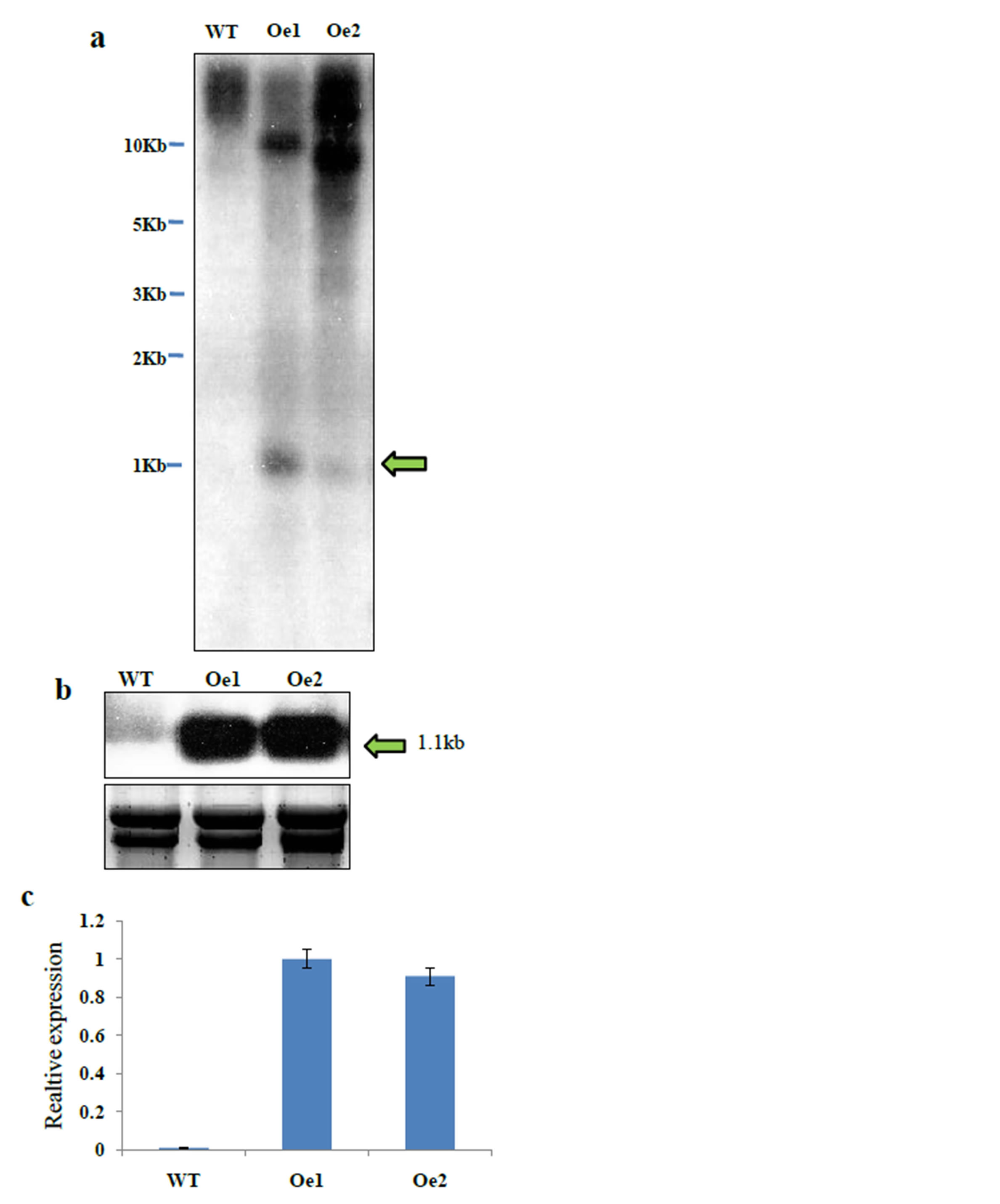
Figure 4. Molecular characterization of OsMAPK2 overexpression lines. (a) Southern blot analysis of WT and transgenic rice plants. Genomic DNA was restricted from transgenic plants by XhoI and then separated on a 1.0% agarose gel. A 1.1-kb full-length DNA fragment of the hygromycin gene was used as the probe; (b) Northern blot analysis for OsMAPK2 expression level. Total RNA extracted from the shoots of transgenic rice plants was used for analyzing the expression of OsMAPK2; (c) qRT-PCR analysis of the expression levels of two transgenic lines with overexpression of OsMAPK2. ** and * indicate significant differences relative to the control at P < 0.01 and P < 0.05, respectively.
To evaluate the potential function of OsMAPK2, the growth performance and Pi contents in leaves and roots of the OsMAPK2 and WT rice plants were investigated in a Hoagland solution culture with Pi sufficient and Pi deficient conditions for 4 weeks (Figure 5(a)). The shoot and root fresh weight of the OsMAPK2-Oe plants in HP conditions were 6.1% and 9.6% higher than in wild-type
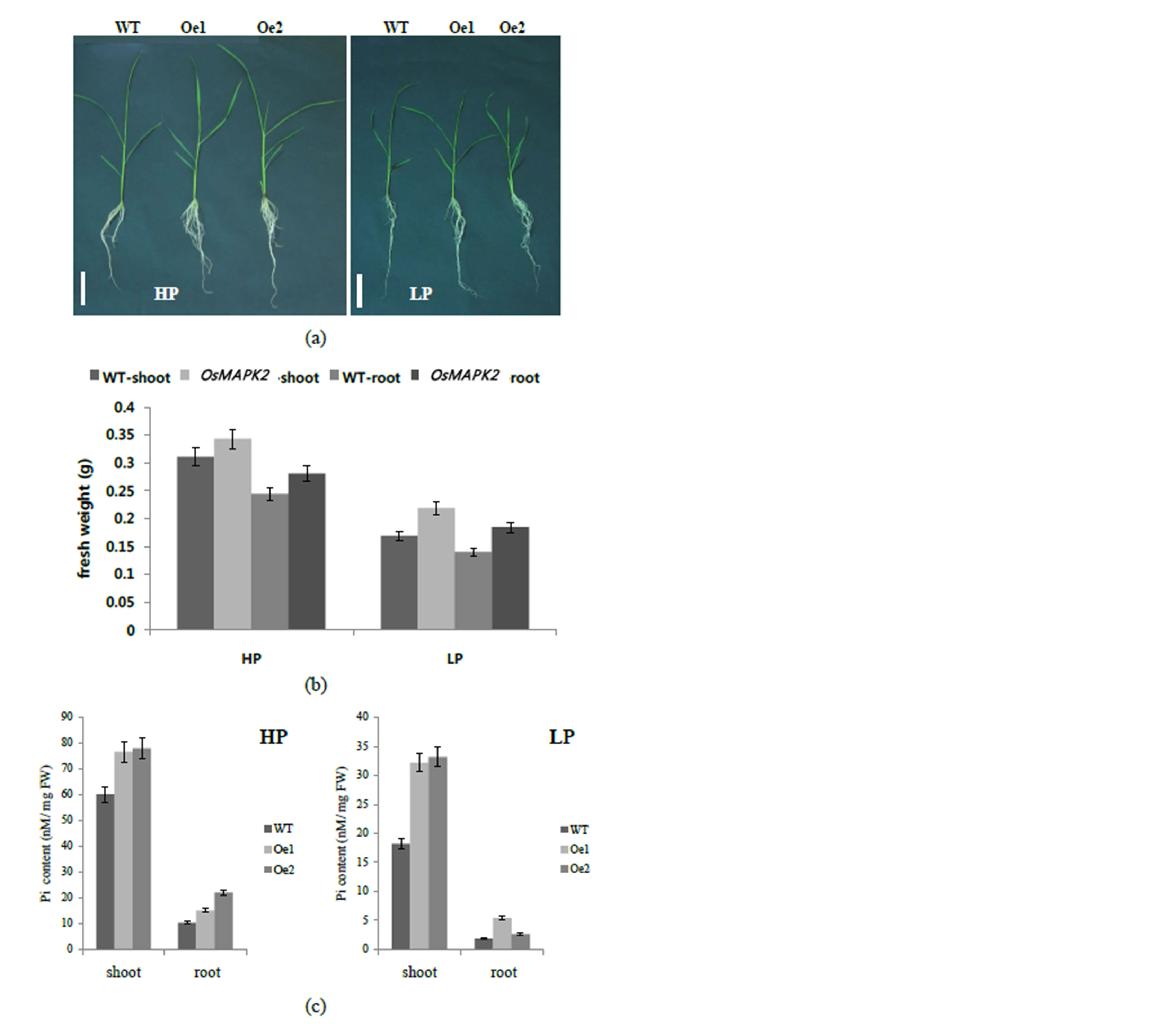
Figure 5. Phenotypic analysis of the OsMAPK2 transgenic lines and wild-type (WT) rice plants a Growth of 30-day-old seedlings of wild-type and OsMAPK2 in hydroponic solutions with Pi sufficient (HP) and Pi deficient (LP) conditions for 3 weeks. Scale bars represent 5 cm; (b) Fresh shoot and root weight of WT and OsMAPK2 transgenic rice plants under HP and LP conditions. Five plants per line were measured. Error bars indicate the SD (n = 5); (c) Pi contents in the shoots and roots of wild-type (WT) and OsMAPK2 (Oe1, Oe2) plants in hydroponic solutions with Pi sufficient (HP) and Pi deficient (LP) conditions for 3 weeks. Error bars indicate the SD (n = 4). ** and * indicate significant differences relative to the control at P < 0.01 and P < 0.05, respectively.
plants, respectively (Figure 5(b)). In LP conditions, the shoot and root fresh weight of the OsMAPK2-Oe plants was approximately 28% higher than that of wild-type plants. The Pi contents from the transgenic lines and wild-type were remarkably different. The Pi contents in shoots of the OsMAPK2-Oe plants grown at HP or LP condition were 26% and 66% higher than those in wildtype plants, respectively (Figure 5(c)). These results suggest that there may be a different regulatory mechanism downstream of OsMAPK2 that controls Pi homeostasis.
3.4. Expression of Pi Starvation-Induced Genes in WT and OsMAPK2 Transgenic Plants
The expression patterns of well-characterized Pi-starvation-inducible genes (PSI) in rice were examined by qRT-PCR analysis in the roots of 14-day-old OsMAPK2- Oe and WT plants grown under both Pi-sufficient and deficient conditions for 7 days (Figure 6). We examined the expression of OsIPS1 {a member of the Mt4/TPSI1 family in rice that is induced strongly by phosphate starvation [30]}, OsSPX1 {a negative factor for the accumulation of Pi in leaves [31]}, OsPHO2 {the potential ortholog of AtPHO2; its mutant is identified as a Pi overaccumulator [32]}, and the phosphate transporters (OsPT2, OsPT4 and OsPT8). The expression level of these PSI genes was controlled by overexpression of OsMAPK2. While OsMAPK2-Oe decreased the expression levels of OsIPS, OsSPX1, OsPT2, OsPT4 and OsPT8, the expression level of OsPHO2 was increased in Pi deficient condition. It could be that due to the higher internal Pi contents, the response of the PSI genes is weaker in the OsMAPK2-Oe plants compared to WT plants.
3.5. Generation of Transgenic Arabidopsis Plants Overexpressing OsMAPK2
To determine whether OsMAPK2 confers similar effects
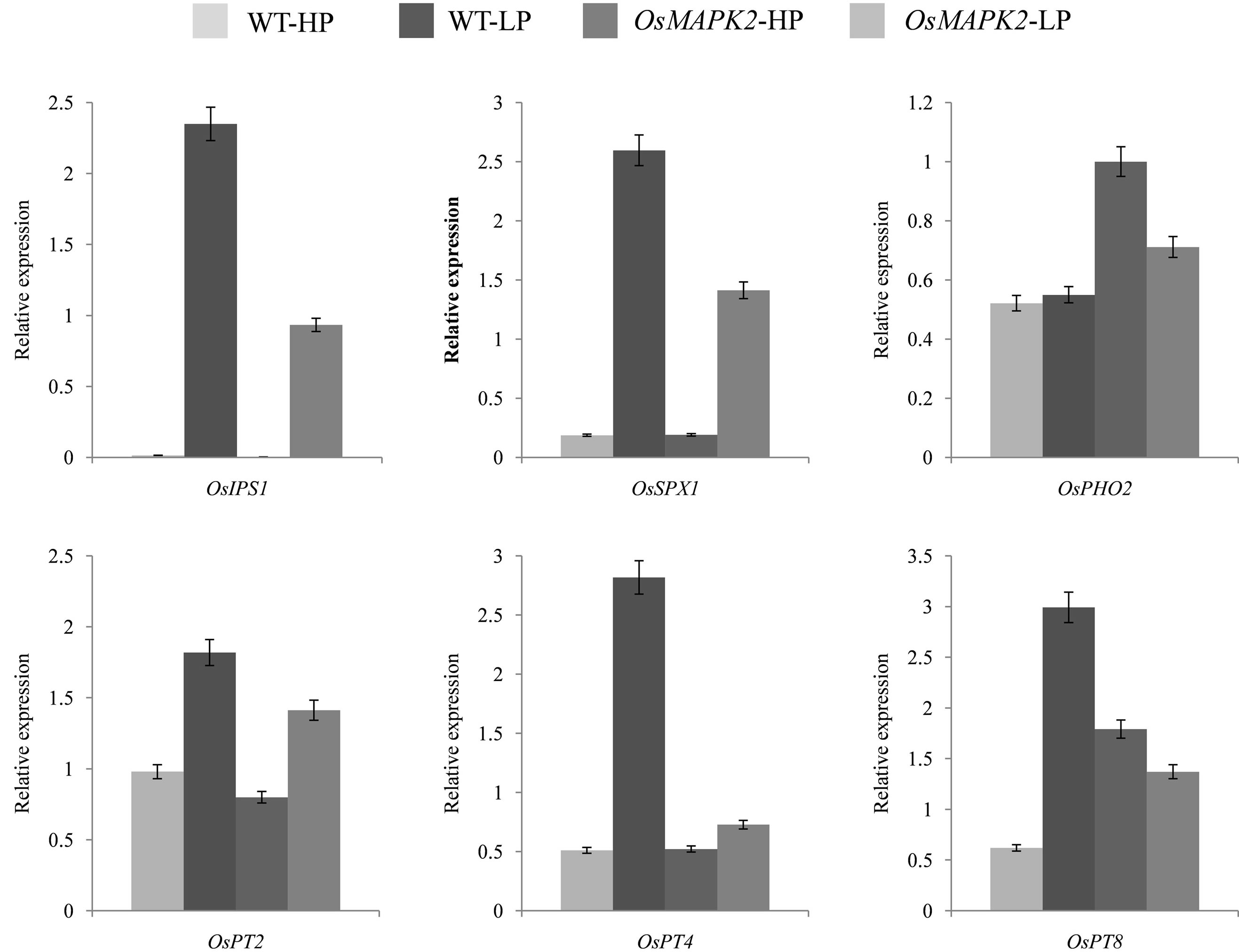
Figure 6. Real-time PCR analysis of Pi starvation-induced genes in the seedlings of wild-type (WT) and OsMAPK2 rice plants under (A) Pi sufficient (HP) and (B) Pi deficient (LP) conditions for five days. The rice Actin1 gene was amplified as the internal control. ** and * indicate significant differences relative to the control at P < 0.01 and P < 0.05, respectively.
in Arabidopsis, we transformed Arabidopsis with the OsMAPK2 gene driven by the 35S constitutive promoter. Overexpression of OsMAPK2 was confirmed by PCR and Northern blot analysis, and both transgenic lines showed significantly enhanced levels of OsMAPK2 mRNA transcripts compared with control wild-type plants (data not shown).
Seven day-old seedlings of OsMAPK2-Oe Arabidopsis and WT plants grown in one-half-strength Murashige and Skoog medium were transferred to Murashige and Skoog medium supplemented with high (HP) and low (LP) Pi (1.25 and 0.0125 mM) for 7 days (Figure 7(a)).
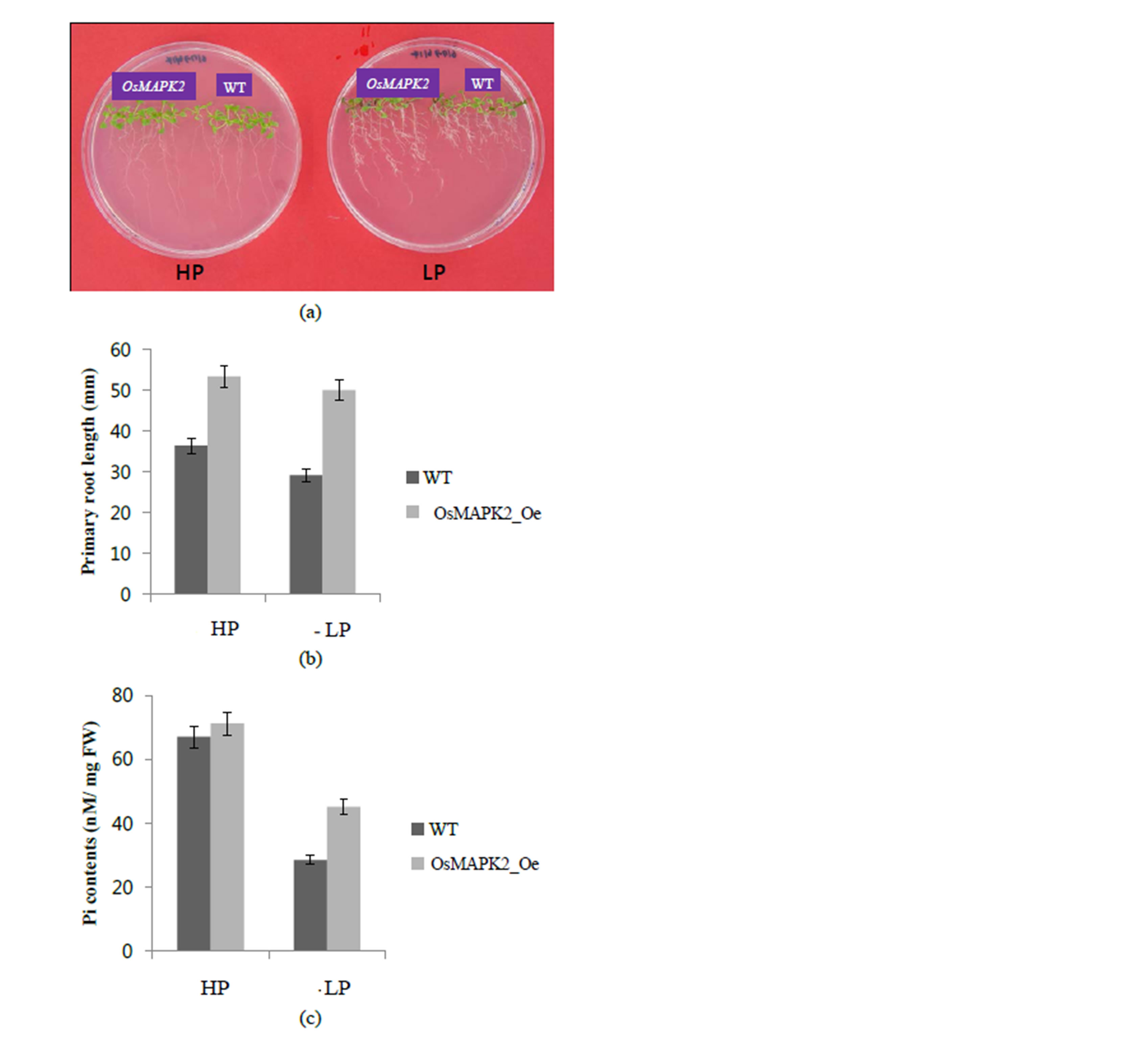
Figure 7. Growth performance of wild-type (WT) and OsMAPK2 transgenic Arabidopsis plants. (a) Phenotype comparison of the WT and transgenic plants. The 7-day-old seedlings germinated on MS medium were transferred to HP or LP medium for another 7 d; (b) Primary root length and (c) Pi contents in the wild type and OsMAPK2 transgenic plants in HP or LP conditions. ** and * indicate significant differences relative to the control at P < 0.01 and P < 0.05, respectively.
There was an increase in the primary root length of the OsMAPK2 plants compared with the wild-type plants, 36% and 72% under HP and LP conditions, respectively (Figure 7(b)). Regarding the Pi contents, the transgenic plants showed slightly increased Pi compared with wildtype plants under HP conditions, and a 42% increased under LP conditions (Figure 7(c)). Our results suggest that OsMAPK2 also plays a role in regulating the accumulation of Pi in Arabidopsis plants.
4. Discussion
MAPK cascades are evolutionarily conserved signaling modules that play an important role in plant responses to multiple stresses. Protein phosphorylation/dephosphorylation modifications are essential in intracellular signaling pathways. Recent studies showed that plant MAPKs are activated by many kinds of abiotic stresses and pathogens [20,33,34].
Earlier studies suggested that OsMAPK2 could be induced during multiple stresses [19]. In this study, through transgenic analysis, we investigated the role of the OsMAPK2 gene, which is involved in regulating root development and Pi homeostasis. To the best of our knowledge, OsMAPK2 is the first member of the MAP kinase genes to be identified as having Pi stress tolerance in plants.
Protein kinase activity has been examined in many studies of kinase [35-37]. Previous study demonstrated that the OsMAPK2 can phosphorylate itself [38]. OsMAPK2 was induced within the first 30 min to 1 hour in rice suspension cell culture during Pi deficient conditions and then decreased. In addition, OsMAPK2 is responsive to both Pi and Fe deficiency. This result showed OsMAPK2 could also play an important role in the regulation of Fe nutrient deficiency. We have shown that OsMAPK2 is expressed in both seedling leaves and roots under normal conditions, and higher relative amounts of OsMAPK2 transcripts were detected in the roots during Pi deficient conditions compared to those in leaves.
First, we generated transgenic rice plants that overexpress OsMAPK2 under the control of the CaMV 35S promoter. After characterization of the transgenic rice plants, we generated transgenic Arabidopsis plants to confirm the function of OsMAPK2 in dicotyledonous plants. We observed an increase of the Pi content in shoots and roots of both the OsMAPK2-Oe rice and Arabidopsis plants. OsMAPK2-Oe rice and Arabidopsis plants had increased the fresh weight of transgenic plants compared with wild-type plants. Changes in root architecture increase the ability of a plant to overcome nutrient stresses [39]. As a result of the changes of root architecture, the Pi content of OsMAPK2-Oe plants was higher than that of WT plants in both HP and LP conditions. The increasing rate of fresh weight under LP conditions between the transgenic rice plants and WT plants was higher than under HP conditions. And the increasing rates in Pi content in transgenic plants under LP conditions were similar to those under HP conditions. These results indicate that the increased Pi content of transgenic plants is strongly related with the root development.
The expression level of several PSI genes was reduced in the OsMAPK2-Oe plants. Previous study of plant phosphate responsive gene (OsPHR2) showed that OsPHR2 overexpressing rice plants exhibited enhanced root elongation and proliferated root hair growth, and some Pi transporters were up-regulated under HP conditions [40]. These results implicate the increase in Pi content in OsPHR2-Oe plants. The expression level of PSI genes regulated by OsMAPK2 could be reduced by the hypothetical intracellular Pi sensing mechanism [41]. A similar effect has also been observed in case of ZAT6, which was identified as an Arabidopsis transcription factor functioning in the Pi starvation response [42]. Arabidopsis plants overexpressing ZAT6 showed altered root architecture and the expression of several Pi starvationresponsive genes also decreased in ZAT6 overexpressing plants.
We tested the expression level of 6 phosphate starvation-induced genes. OsIPS1 has a rapid and specific response to Pi starvation [30]. AtIPS negatively affects plant Pi uptake in Arabidopsis [43], but the function of OsIPS1 remains unclear. OsSPX1 is a negative regulator in both shoots and roots. Suppression of OsSPX1 increased leaf Pi content and also increased the expression of Pi transporters in roots [44]. Because OsIPS1 and OsSPX1 are up-regulated in the roots of the pho2 mutant [45], it is possible that the expression of OsIPS1 and OsSPX1 decreased and the expression of OsPHO2 increased with the over-expression of OsMAPK2 compared with wild-type. We also tested the expression of phosphate transporters upor down-regulation upon overexpression of OsMAPK2. Most of the phosphate transporters are induced by LP conditions; the expression levels of OsPT2, 4 and 8 were decreased compared with WT under LP conditions. This result indicated that the transgenic plants contain high Pi contents even under LP conditions.
Although many plant MAPK genes localize in both the cytoplasm and the nucleus, OsMAPK2 has been shown to localize in the chloroplast. The chloroplast is an important organelle that involved in cellular signaling and the phosphorylation network in plants. STN7 and STN8 are well-characterized kinases that are involved in light dependent thylakoid-protein phosphorylation. The loss-offunction mutants showed changes in chloroplast and nuclear gene expression [46,47]. This result suggests that protein phosphorylation in the chloroplast may be associated with the regulation of plastid and nuclear gene expression [48]. Tissue-specific expression analysis using the promoter-GUS fusion construct showed that the expression of OsMAPK2 was predominantly localized to roots and reproductive organs, such as the mature spikelet. Plant root parts are likely to be involved in Pi uptake from the soil solution, and root-specific expression of OsMAPK2 could enhance Pi stress tolerance. The study of rice OsMAPK2 transgenic plants may provide an idea of why the overexpression of OsMAPK2 genes in Arabidopsis could lead to higher Pi contents and increasing primary root length.
In conclusion, our results suggest that OsMAPK2 regulates root development and Pi homeostasis. Overexpression of OsMAPK2 also affected the expression of downstream genes that play an important role in the phosphate signaling pathway in rice. When plants expressing OsMAPK2 were grown under low phosphate conditions, the root parts could improve absorption of phosphates and enhance low phosphate tolerance.
Acknowledgements
This work was supported by a grant from the Next-Generation BioGreen 21 Program (Plant Molecular Breeding Center No. PJ008035), Rural Development Administration, Republic of Korea.
REFERENCES
- G. Welp, U. Herms and G. Brümmer, “Einfluß von Bodenreaktion, Redoxbedingungen und Organischer Substanz auf die Phosphatgehalte der Bodenlösung,” Zeitschrift für Pflanzenernährung und Bodenkunde, Vol. 146, No. 1, 1983, pp. 38-52. http://dx.doi.org/10.1002/jpln.19831460106
- I. C. R. Holford, “Soil Phosphorus: Its Measurement, and Its Uptake by Plants,” Australian Journal of Soil Research, Vol. 35, No. 2, 1997, pp. 277-239. http://dx.doi.org/10.1071/S96047
- K. G. Raghothama, “Phosphate Acquisition,” Annual Review of Plant Physiology and Plant Molecular Biology, Vol. 50, 1999, pp. 665-693. http://dx.doi.org/10.1146/annurev.arplant.50.1.665
- W. K. Gardner, D. G. Parbery and D. A. Barber, “Proteoid Root Morphology and Function in Linus Albus,” Plant Soil, Vol. 60, 1983, pp. 143-147. http://dx.doi.org/10.1007/BF02377120
- D. S. Lipton, R. W. Blanchar and D. G. Blenvins, “Citrate, Malate and Succinate Concentration in Exudates from P-Sufficient and P-Stressed Medicago sativa L. Seedlings,” Plant Physiology, Vol. 85, No. 2, 1987, pp. 315- 317. http://dx.doi.org/10.1104/pp.85.2.315
- E. Hoffland, R. V. D. Boogaard, J. A. Nelemans and G. R. Findenegg, “Biosynthesis and Root Exudation of Citric and Malic Acids in Phosphate-Starved Rape Plants,” New Phytologist, Vol. 122, No. 4, 1992, pp. 675-680. http://dx.doi.org/10.1111/j.1469-8137.1992.tb00096.x
- S. Mishra, S. Srivastava, R. D. Tripathi, R. Kumar, C. S. Seth and D. K. Gupta, “Lead Detoxification by Coontail (Ceratophyllum demersum L.) Involves Induction of Phytochelatins and Antioxidant System in Response to Its Accumulation,” Chemosphere, Vol. 65, No. 6, 2006, pp. 1027-1039. http://dx.doi.org/10.1016/j.chemosphere.2006.03.033
- S. Zhang and D. F. Klessig, “Salicylic Acid Activates a 48-kD MAP Kinase in Tobacco,” The Plant Cell, Vol. 9, No. 5, 1997, pp. 809-824.
- C. Jonak, L. Okresz, L. Bogre and H. Hirt, “Complexity, Cross Talk and Integration of Plant MAP Kinase Signaling,” Current Opinion in Plant Biology, Vol. 5, No. 5, 2002, pp. 415-424. http://dx.doi.org/10.1016/S1369-5266(02)00285-6
- R. Seger and E. G. Krebs, “The MAPK Signaling Cascade,” The FASEB Journal, Vol. 9, No. 9, 1995, pp. 726- 735.
- A. V. Khokhlatchev, “Phosphorylation of the MAP Kinase ERK2 Promotes Its Homodimerization and Nuclear Translocation,” Cell, Vol. 93, No. 4, 1998, pp. 605-615. http://dx.doi.org/10.1016/S0092-8674(00)81189-7
- R. W. Innes, “Mapping out the Roles of MAP Kinases in Plant Defense,” Trends in Plant Science, Vol. 6, No. 9, 2001, pp. 392-394. http://dx.doi.org/10.1016/S1360-1385(01)02058-1
- G. Tena, T. Asai, W. L. Chiu and L. Sheen, “Plant Mitogen-Activated Protein Kinase Signaling Cascades,” Current Opinion in Plant Biology, Vol. 4, No. 5, 2001, pp. 392-400. http://dx.doi.org/10.1016/S1369-5266(00)00191-6
- P. J. Krysan, P. J. Jester, J. R. Gottwald and M. R. Sussman, “An Arabidopsis MAPKK Kinase Gene Family Encodes Essential Positive Regulators of Cytokinesis,” The Plant Cell, Vol. 14, No. 5, 2002, pp. 1109-1120. http://dx.doi.org/10.1105/tpc.001164
- D. Matsuoka, T. Nanmori, K. Sato, Y. Fukami, U. Kikkawa and T. Yasuda, “Activation of AtMek1, an Arabidopsis Mitogen-Activated Protein Kinase Kinase, in Vitro and in Vivo: Analysis of Active Mutants Expressed in E. coli and Generation of the Active form in Stress Response in Seedlings,” The Plant Journal, Vol. 29, No. 5, 2002, pp. 637-647. http://dx.doi.org/10.1046/j.0960-7412.2001.01246.x
- T. Mizoguchi, K. Irie, T. Hirayama, N. Hayashida, K. Yamaguchi-Shinozaki, K. Matsumoto and K. Shinozaki, “A Gene Encoding a Mitogen-Activated Protein Kinase Is Induced Simultaneously with Genes for a Mitogen-Activated Protein Kinase and S6 Ribosomal Protein Kinase by Touch, Cold, and Water Stress in Arabidopsis Thaliana,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 93, No. 2, 1996, pp. 765-769. http://dx.doi.org/10.1073/pnas.93.2.765
- C. He, S. H. T. Fong, D. Yang and G. L. Wang, “BWMK1, a Novel MAP Kinase Induced by Fungal Infection and Mechanical Wounding in Rice,” Molecular Plant-Microbe Interactions, Vol. 12, No. 12, 1999, pp. 1064-1073. http://dx.doi.org/10.1094/MPMI.1999.12.12.1064
- L. Xiong, M. W. Lee, M. Qi and Y. Yang, “Identification of Defense-Related Rice Genes by Suppression Subtractive Hybridization and Differential Screening,” Molecular Plant-Microbe Interactions, Vol. 14, No. 5, 2001, pp. 685-692. http://dx.doi.org/10.1094/MPMI.2001.14.5.685
- G. K. Agrawal, R. Rakwal and H. Iwahashi, “Isolation of Novel Rice (Oryza sativa L.) Multiple Stress Responsive MAP Kinase Gene, OsMSRMK2, Whose mRNA Accumulates Rapidly in Response to Environmental Cues,” Biochemical and Biophysical Research Communications, Vol. 294, No. 5, 2002, pp. 1009-1016. http://dx.doi.org/10.1016/S0006-291X(02)00571-5
- H. J. Huang, S. F. Fu, Y. H. Tai, W. C. Chou and D. D. Huang, “Expression of Oryza sativa MAP Kinase Gene Is Developmentally Regulated and Stress-Responsive,” Physiologia Plantarum, Vol. 114, No. 4, 2002, pp. 958-963. http://dx.doi.org/10.1034/j.1399-3054.2002.1140410.x
- J. Q. Wen, K. Oono and R. Imai, “Two Novel MitogenActivated Protein Signaling Components, OsMEK1 and OsMAP1 Are Involved in Moderate Low-Temperature Signaling Pathway in Rice,” Plant Physiology, Vol. 129, No. 4, 2002, pp. 1880-1891. http://dx.doi.org/10.1104/pp.006072
- F. Song and R. M. Goodman, “OsBIMK1, a Rice MAP Kinase Gene Involved in Disease Resistance Responses,” Planta, Vol. 215, No. 6, 2002, pp. 997-1005. http://dx.doi.org/10.1007/s00425-002-0794-5
- S. M. Yu, Y. H. Kuo, G. Sheu, Y. J. Sheu and L. F. Liu, “Metabolic Derepression of Alpha-Amylase Gene Expression in Suspension-Cultured Cells of Rice,” The Journal of Biological Chemistry, Vol. 266, No. 31, 1991, pp. 21131-21137.
- K. Miura, A. Rus, A. Sharkhuu, S. Yokoi, A. S. Karthikeyan, K. G. Raghothama, D. W. Baek, Y. D. Koo, J. B. Jin, R. A. Bressan, D. J. Yun and P. M. Hasegawa, “The Arabidopsis SUMO E3 Ligase SIZ1 Controls Phosphate Deficiency Responses,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 102, No. 21, 2005, pp. 7760-7765. http://dx.doi.org/10.1073/pnas.0500778102
- W. Zhang, D. McElroy and R. Wu, “Analysis of Rice Act1 5’ Region Activity in Transgenic Rice Plants,” The Plant Cell, Vol. 3, No. 11, 1991, pp. 1155-1165.
- M. Nanamori, T. Shinano, J. Wasaki, T. Yamamura, I. M. Rao and M. Osaki, “Low Phosphorus Tolerance Mechanisms: Phosphorus Recycling and Photosynthate Partitioning in the Tropical Forage Grass, Brachiaria Hybrid Cultivar Mulato Compared with Rice,” Plant and Cell Physiology, Vol. 45, No. 4, 2004, pp. 460-469. http://dx.doi.org/10.1093/pcp/pch056
- K. G. Raghothama, A. Maggio, M. L. Narasimhan, A. K. Kononowicz, G. L. Wang, M. P. D’Urzo, P. M. Hasegawa and R. A. Bressan, “Tissue-Specific Activation of the Osmotin Gene by ABA, C2H4 and NaCl Involves the Same Promoter Region,” Plant Molecular Biology, Vol. 34, No. 3,1997, pp. 393-402. http://dx.doi.org/10.1023/A:1005812217945
- S. J. Clough and A. F. Bent, “Floral Dip: A Simplified Method for Agrobacterium Mediated Transformation of Arabidopsis thaliana,” The Plant Journal, Vol. 16, No. 6, 1998, pp. 735-743. http://dx.doi.org/10.1046/j.1365-313x.1998.00343.x
- Y. Hiei, S. Ohta, T. Komari and T. Kumashiro, “Efficient Transformation of Rice (Oriza sativa) Mediated by Agrobacterium and Sequence Analysis of the Boundaries of the T-DNA,” The Plant Journal, Vol. 6, No. 2, 1994, pp. 271-282. http://dx.doi.org/10.1046/j.1365-313X.1994.6020271.x
- X. L. Hou, P. Wu, F. C. Jiao, Q. J. Jia, H. M. Chen, J. Yu, X. W. Song and K. K. Yi, “Regulation of the Expression of OsIPS1 and OsIPS2 in Rice via Systemic and Local Pi Signaling and Hormones,” Plant Cell & Environment, Vol. 28, No. 3, 2005, pp. 353-364. http://dx.doi.org/10.1111/j.1365-3040.2005.01272.x
- C. Wang, S. Ying, H. Huang, K. Li, P. Wu and H. Shou, “Involvement of OsSPX1 in Phosphate Homeostasis in Rice,” The Plant Journal, Vol. 57, No. 5, 2009, pp. 895- 904. http://dx.doi.org/10.1111/j.1365-313X.2008.03734.x
- R. Bari, B. D. Pant, M. Stitt and W.-R. Scheible, “PHO2, microRNA399, and PHR1 Define a Phosphate-Signaling Pathway in Plants,” Plant Physiology, Vol. 141, No. 3, 2006, pp. 988-999. http://dx.doi.org/10.1104/pp.106.079707
- L. Zhang, D. Xi, S. Li, Z. Gao, S. Zhao, J. Shi, C. Wu and X. Guo, “A Cotton Group C MAP Kinase Gene, GhMPK2, Positively Regulates Salt and Drought Tolerance in Tobacco,” Plant Molecular Biology, Vol. 77, No. 1-2, 2011, pp. 17-31. http://dx.doi.org/10.1007/s11103-011-9788-7
- X. Kong, J. Pan, M. Zhang, X. Xing, Y. Zhou, Y. Liu, D. Li and D. Li, “ZmMKK4, a Novel Group C Mitogen-Activated Protein Kinase Kinase in Maize (Zea mays), Confers Salt and Cold Tolerance in Transgenic Arabidopsis,” Plant Cell & Environment, Vol. 34, No. 8, 2001, pp. 1291-1303. http://dx.doi.org/10.1111/j.1365-3040.2011.02329.x
- Y. H. Cheong, B. C. Moon, J. K. Kim, C. Y. Kim, M. C. Kim, I. H. Kim, C. Y. Park, J. C. Kim, B. O. Park, S. C. Koo, H. W. Yoon, W. S. Chung, C. O. Lim, S. Y. Lee and M. J. Cho, “BWMK1, a Rice Mitogen-Activated Protein Kinase, Locates in the Nucleus and Mediates Pathogenesis-Related Gene Expression by Activation of a Transcription Factor,” Plant Physiology, Vol. 132, No. 4, 2003, pp. 1961-1972. http://dx.doi.org/10.1104/pp.103.023176
- X. S. Huang, T. Luo, X.-Z. Fu, Q.-J. Fan and J.-H. Liu, “Cloning and Molecular Characterization of a MitogenActivated Protein Kinase Gene from Poncirus Trifoliata Whose Ectopic Expression Confers Dehydration/Drought Tolerance in Transgenic Tobacco,” Journal of Experimental Botany, Vol. 62, No, 14, 2011, pp. 5191-5206. http://dx.doi.org/10.1093/jxb/err229
- L. Xiong and Y. Yang, “Disease Resistance and Abiotic Stress Tolerance in Rice Are Inversely Modulated by an Abscisic Acid-Inducible Mitogen-Activated Protein Kinase,” The Plant Cell, Vol. 15, No. 3, 2003, pp. 745-759. http://dx.doi.org/10.1105/tpc.008714
- J. B. Heo, Y. B. Yi and J. D. Bahk, “Rice GDP Dissociation Inhibitor 3 Inhibits OsMAPK2 Activity through Physical Interaction,” Biochemical and Biophysical Research Communications, Vol. 414. No. 4, 2011, pp. 814- 819. http://dx.doi.org/10.1016/j.bbrc.2011.10.018
- J. López-Bucio, A. Cruz-Ramírez and L. Herrera-Estrella, “The Role of Nutrient Availability in Regulating Root Architecture,” Current Opinion in Plant Biology, Vol. 6, No. 3, 2003, pp. 289-287. http://dx.doi.org/10.1016/S1369-5266(03)00035-9
- J. Zhou, F. Jiao, Z. Wu, X. Wang, X. He, W. Zhong and P. Wu, “OsPHR2 Is Involved in Phosphate-Starvation Signaling and Excessive Phosphate Accumulation in Shoots of Plants,” Plant Physiology, Vol. 146, No. 4, 2008, pp. 1673-1686. http://dx.doi.org/10.1104/pp.107.111443
- S. Abel, A. C. Ticconi and C. A. Delatorre, “Phosphate Sensing in Higher Plants,” Physiologia Plantarum, Vol. 115, No. 1, 2002, pp. 1-8. http://dx.doi.org/10.1034/j.1399-3054.2002.1150101.x
- B. N. Devaiah, V. K. Nagarajan and K. G. Raghothama, “Phosphate Homeostasis and Root Development in Arabidopsis Are Synchronized by the Zinc Finger Transcription Factor ZAT6,” Plant Physiology, Vol. 145, No. 1, 2007, pp. 147-159. http://dx.doi.org/10.1104/pp.107.101691
- J. M. Franco-Zorrilla, E. Gonzalez, R. Bustos, F. Linhares, A. Leyva and J. Paz-Ares, “The Transcriptional Control of Plant Responses to Phosphate Limitation,” Journal of Experimental Botany, Vol. 55, No. 396, 2004, pp. 285- 293. http://dx.doi.org/10.1093/jxb/erh009
- C. Wang, S. Ying, H. Huang, K. Li, P. Wu and H. Shou, “Involvement of OsSPX1 in Phosphate Homeostasis in Rice,” The plant Journal, Vol. 57, No. 5, 2009, pp. 895- 904. http://dx.doi.org/10.1111/j.1365-313X.2008.03734.x
- F. Liu, Z. Wang, H. Ren, C. Shen, Y. Li, H. Q. Ling, C. Wu, X. Lian and P. Wu, “OsSPX1 Suppresses the Function of OsPHR2 in the Regulation of Expression of OsPT2 and Phosphate Homeostasis in Shoots of Rice,” The Plant Journal, Vol. 62, No. 3, 2010, pp. 508-517. http://dx.doi.org/10.1111/j.1365-313X.2010.04170.x
- V. Bonardi, P. Pesaresi, T. Becker, E. Schleiff, R. Wagner, T. Pfannschmidt, P. Jahns and D. Leister, “Photosystem II Core Phosphorylation and Photosynthetic Acclimation Require Two Different Protein Kinases,” Nature, Vol. 437, No. 7062, 2005, pp. 1179-1182. http://dx.doi.org/10.1038/nature04016
- J. D. Rochaix, “Role of Thylakoid Protein Kinases in Photosynthetic Acclimation,” FEBS Letters, Vol. 581, No. 152, 2007, pp. 2768-2775. http://dx.doi.org/10.1016/j.febslet.2007.04.038
- S. Baginsky and W. Gruissem, “The Chloroplast Kinase Network: New Insights from Large-Scale Phosphoproteome Profiling,” Molecular Plant, Vol. 2, No. 6, 2009, pp. 1141-1153. http://dx.doi.org/10.1093/mp/ssp058
Abbreviations
cDNA: complementary DNA
GUS: b-glucuronidase HP: high phosphate LP: low phosphate M-MLV reverse transcriptase: moloney murine leukemia virus reverse transcriptase mRNA: messenger RNA Oe: over-expression PCA: perchloric acid PCR: polymerase chain reaction qRT-PCR: quantitative real time polymerase chain reaction SD: standard deviation v/v: volume per volume WT: wild type w/v: weight per volume
Supplemental Information
Table S1. Primers used in real time PCR analysis.
NOTES
*Corresponding author.


