Advances in Microbiology
Vol.06 No.11(2016), Article ID:70642,15 pages
10.4236/aim.2016.611081
Locally Isolated Bacterial Strains with Multiple Degradation Potential Capabilities on Petroleum Hydrocarbon Pollutants
A. Bahobail1, S. M. F. Gad El-Rab2,3, G. A. Amin2,4*
1Department of Biology, Faculty of Science, Taif University, Taif, KSA
2Department of Biotechnology, Faculty of Science, Taif University, Taif, KSA
3Department of Botany and Microbiology, Faculty of Science, Assuit University, Assuit, Egypt
4Department of Microbiology, Faculty of Agriculture, Cairo University, Cairo, Egypt

Copyright © 2016 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: April 11, 2016; Accepted: September 16, 2016; Published: September 19, 2016
ABSTRACT
In the present study, 23 isolates, dominated by bacterial genera (74%) were isolated from petroleum sludge at refinery wastewater plant, Jeddah, KSA, by means of selective enrichment in nutritionally optimized refinery wastewater (NORWW) and over twelve successive transfers. Efficiency of biodegradation on complex mixture of hydrocarbons present in refinery wastewater was evidenced by changes in both total viable counts (TVC) and COD content of cultivation broth. Out of the 23 isolates three most potent isolates named BDCC-TUSA-8, BDCC-TUSA-12 and BDCC-TUSA-18 were selected for their efficient COD removal and active growth. The three isolates were tested separately in Bushnell-Haas (BH) media for their capabilities to degrade n-Hexadecane, phenol and phenanthrene, representing the major types of hydrocarbon pollutants. The results strongly indicated that all three isolates showed multiple degradation potentials with remarkably fast reaction rates. Before being recommended for future work, the three isolates were fully characterized and identified employing culture-dependent techniques such as API 20E, API 20NE and API 50CHB, and further confirmed by partial 16S rRNA gene sequencing and phylogenetic analysis as Pantoea agglomerans, Acinetobacter lwoffii and Bacillus thuringiensis respectively. The obtained potent strains provide valuable candidates if assemblages of mixed fewer strains with overall broad and complementary enzymatic capacities are to be considered in order to bring the rate and extent of petroleum biodegradation further as a cost-effective process.
Keywords:
Biodegradation, Consortium, Multiple Degradation Potentials, Acinetobacter, Bacillus, Pantoea

1. Introduction
The release of oily wastewaters into the environment is one of the major causes of environmental pollution. Petroleum refineries generate huge amounts of wastewater, 0.4 - 1.6 times the volume of processed crude oil [1] . The toxic effects of refined petroleum oils, particularly those with low boiling point alkenes, aromatic hydrocarbons including n-Hexadecane, phenol and phenanthrene are devastating. These include cancers, birth defects, endocrine disruptions, still births, nervous disorders, liver disease, depression and irregular heartbeats [2] [3] . US EPA (United States Environmental Protection Agency) designated oily wastes as hazardous wastes [4] . Physical and chemical treatments of refinery wastewater have been carried out utilizing different techniques [5] - [7] . However, these technologies are expensive and can lead to incomplete decomposition of contaminants.
Recently, attention has been paid to the use of microorganisms, owing to their diverse metabolic capabilities, to detoxify and/or remove environmental pollutants including the products of petroleum industry [8] - [10] . It is uncommon to find an organism that could effectively degrade all types of hydrocarbons present in refinery wastewater due to differences in microbial metabolic routes and pathways for degradation of all hydrocarbons [11] . As reported by Tadros et al. [12] , individual microbial strains could degrade several hydrocarbons, but preferred only one. The greater the complexity of the metabolic machinery, for a given bacterial isolate, the higher the number of specific enzyme systems present, the faster the rate of biodegradation on multiple hydrocarbons and the greater the likelihood of assembling a potential consortium composed of a relatively smaller number of effective isolates [13] with complementary set of enzyme systems. Certainly, this would contribute significantly in optimizing a cost-effective biodegradation process on complex mixture of hydrocarbons in refinery effluents, petroleum spills and other contaminated sites.
The objective of the present study was twofold: firstly, to monitor selective enrichment of indigenous microorganisms in NORWW and obtain potent species with multiple degradation potential on complex mixture of hydrocarbons; secondly, to characterize and fully identify the obtained potent isolates before they could be used in future work on control and optimization of an effective biodegradation process.
2. Materials & Methods
2.1. Chemicals and Materials
All reagents used were analytical grade and obtained from Sigma-Aldrich (St. Louis, MO) unless otherwise stated. Sample of petroleum sludge polluted soil was collected from a site nearby wastewater treatment plant at Jeddah refinery, Jeddah, KSA.
2.2. Wastewater and Polluted Soil
Refinery wastewater was collected from an online location, after preliminary treatment steps and before biological treatment unit, at the refinery wastewater treatment plant and used to carry out enrichment and adaptation experiment for indigenous microorganisms. It is usually stated that the ratio of BOD:N:P in the wastewater to be treated should be approximately 100:5:1 for optimum cell growth and efficient aerobic treatment [14] - [16] . The present ratio was lower; 100:1.18:0.30. This suggests that both nitrogen and phosphorous have to be added to the wastewater for effective growth of indigenous microorganisms. Based on the obtained figures, nutritionally optimized refinery wastewater (NORWW) was prepared through addition of calculated amounts of ammonium sulphate and dipotassium phosphate based on ammonium nitrogen (NH4-N) and orthophosphate 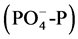 which are preferred sources for active growth and multiplication of microorganisms [17] . The pH was adjusted to 7.0.
which are preferred sources for active growth and multiplication of microorganisms [17] . The pH was adjusted to 7.0.
2.3. Enrichment of Indigenous Microorganisms in Petroleum Sludge Polluted Soil through Successive Transfers
Several Erlenmeyer flasks containing 200 ml each of NORWW were used to conduct the enrichment and adaptation process. Firstly, 2 g of petroleum sludge polluted soil, were inoculated into the first flask and incubated at 30˚C on a rotary shaker at 200 rpm. Samples were taken at regular intervals and analyzed for Chemical Oxygen Demand (COD) and population growth (OD625nm). After reaching the end of logarithmic phase, aliquot from the culture was used to inoculate a second flask containing NORWW at 5% (v/v) which was incubated as above. After twelve similar transfers, the last three transfers exhibited stabilized levels for both COD and cell growth. This suggested that complete adaptation of indigenous microorganisms and maximum degradation of hydrocarbons were reached. Up to 23 microbial isolates were selected, purified and cultured in sterile NORWW supplemented with 0.001% yeast extract as source of growth factors [17] . Finally, aliquots of the cultures were centrifuged at 1000 g for 15 min. The cell pellets obtained were suspended in glycerol (30% v/v) and frozen at −20˚C for future use.
2.4. Biodegradation Studies
2.4.1. Assessment of Individual Bacterial Isolates for Cell Growth and Biodegradation Efficiency on NORWW
Twelve bacterial isolates obtained from enrichment process were individually screened for their ability to grow and degrade NORWW. Each isolate was cultured in sterile NORWW supplemented with 0.001% of yeast extract and incubated at 30˚C for 24 h. An inoculum of 5% (v/v) from the actively growing culture was added to 500 ml Erlenmeyer flask, each containing 100 ml NORWW. The flasks were incubated on a rotary shaker at 200 rpm and 30˚C. Daily samples were taken and analyzed for both total viable counts and residual COD. Ceased uninoculated samples, were used to avoid a possible abiotic degradation.
2.4.2. Biodegradation of Various Types of Hydrocarbons by the Most Efficient Isolates
The three most efficient isolates were subsequently tested for biodegradation on various types of hydrocarbons in order to select for possible candidate(s) with multiple biodegradation capabilities. 500 ml Erlenmeyer flasks, each containing 100 ml of Bushnell-Hass media were sterilized by autoclaving at 121˚C for 20 min. Then, the flasks were charged with n-Hexadecane (0.1%), phenol (0.05%) or phenanthrene (0.5%), as sole carbon source, and inoculated with the respective isolate and incubated as above. Due to its low solubility in water, phenanthrene stock solution was prepared in dimethylsulfoxide (DMSO) [18] . Prior to use, phenanthrene was added to sterile Bushnell-Hass media to give a final concentration of 0.5%. The three hydrocarbons were selected as representatives of the major groups of hydrocarbon pollutants present in refinery wastewater [19] - [21] . To avoid a possible abiotic degradation, uninoculated samples were used. The biodegradation potential of the isolates was evaluated by monitoring growth of biomass (OD625nm) and analyzing residual hydrocarbon over the whole cultivation period.
2.5. Characterization of the Most Active Bacterial Isolates
2.5.1. Phenotypic Assays
Typical colonies of each of the 3 most active isolates were transferred to nutrient agar slants and identification was confirmed by microscopic and biochemical characterization that includes Gram stain, anaerobic utilization of glucose, Voges-Proskauer test, motility, oxidase production, catalase production, endospore formation in accordance with the Bergey’s Manual of Systematic Bacteriology [22] . Furthermore, the typical colonies were subjected to Characterization using API 20E, API 20NE and API 50CHB kits (BioMe’rieux) according to the manufacturer’s instruction. The samples were observed for 6 days. Results are given in the species descriptions.
2.5.2. Molecular Characterization
The three most active isolates were identified by 16S rDNA sequencing after extracting DNA. The sequences obtained were compared to find sequence similarity using GenBank program-Basic Local Alignment Search Tool (BLAST). The phylogenetic trees based on 16S rDNA gene sequences were constructed by the neighbour-joining method.
1) DNA isolation
In order to identify the strains, extraction of genomic DNA, amplification and analysis of 16S rRNA genes were conducted as follows: Genomic DNA was extracted from the isolated bacteria strains using the Genomic DNA Prep kit
2) PCR Amplification and Sequencing of the 16S rRNA
A universal bacterial primer set of 27F (5’-AGA GTT TGA TCC TGG CTC AG-3’) [23] and 1492R (5’-GGT TAC CTT GTT ACG ACT T-3’) [24] was used to amplify the nearly complete 16S rRNA gene. The PCR reaction was carried out in a final volume of 25 μL containing 10 - 50 ng of the template DNA, 0.4 μM of each primer, 0.75 U of EF-Taq DNA polymerase
3) Alignment and phylogenetic tree analysis
Partial 16S rRNA gene obtained sequences were compared with full sequences available in the GenBank database using a BLAST search
4) Nucleotide sequence accession number
The sequences obtained in this study were deposited in the GenBank database. The GenBank accession numbers for the nucleotide sequences are KM985370 [P. agglomerans strain gs1], KM985371 [A. lwoffii strain gs2] and KM985372 [B. thuringiensis strain gs3].
2.6. Analysis
2.6.1. Analysis of Biodegradation Variables
The samples were analyzed for residual Chemical Oxygen Demand (COD), Biological Oxygen Demand (BOD), ammonium (NH4-P), total and orthophosphate in raw wastewater and culture broth according to the standard methods for the examination of water and wastewater [29] . Orthophosphate (PO4)−3 was measured by the ascorbic acid colorimetric method and ammonia nitrogen was measured by phenate method using (Unico UV-4802 China). Cell growth was quantified by measuring the sample absorbance at 600 nm in a Beckman DU650 spectrophotometer [30] . Total viable Count (TVC) was determined by standard spread plate method with serial dilutions of the samples.
2.6.2. Analysis of Degraded Hydrocarbons
Both inoculated (biotic) and uninoculated (abiotic) experiments were performed in order to avoid any possible weathering of tested hydrocarbons and ensure the exact degradation by indigenous microorganisms. The three tested hydrocarbons were determined gas chromatographically [31] . For n-Hexadecane determinations, GC-FID; 5890 series II; Hewlett Packard with a 30-m-long wide-bore DB5 column (0.53 mm by 1 mm film thickness) was used, while for phenol and phenanthrene GC-FID using a 30-m-long DB5.625 column (0.25-mm inside diameter, 0.25-mm film thickness) was used. The injector and detector temperature were maintained at 300˚C and oven temperature was programmed to rise at 5˚C/min from 80˚C to 240˚C and to hold for 30 min. Quantitative determinations were performed against a respective authentic standard (Sigma Chemicals).
3. Results and Discussions
3.1. Isolation of COD Removing Microorganisms from Refinery Wastewater
By enrichment of indigenous microorganisms, twelve successive transfers in NORWW were performed. Growth of indigenous bacteria was enhanced many folds, and a total of twenty three microbial isolates with the ability to grow on complex mixture of hydrocarbons present in NORWW as carbon and energy sources and representing different colony morphologies were obtained. These include seventeen (73.9%) isolates as bacteria, 4 (17.4%) yeasts and 2 (8.7%) as fungi. The purified and isolated seventeen bacterial isolates were named after the Biotechnology Department Culture Collection, Taif University, Saudi Arabia as BDCC-TUSA-4 to BDCC-TUSA-20 and were screened individually for their capabilities to remove COD from NORWW (Table 1).
The main finding was that COD removal correlated very well with cell growth of microbial population present over 7 days of cultivation. The higher the cell growth (OD625nm values) the more efficient was the COD removal. Isolate BDCC-TUSA-12 exhibited the highest growth and COD removal efficiency (0.403 and 92% resp.), followed by BDCC-TUSA-8 (0.365 and 91.1% resp.) and BDCC-TUSA-18 recorded the 3rd highest performances (0.304 and 89% resp.). Much lower performances were found with
Table 1. Growth and COD removal by indigenous bacterial isolates on NORWW.
remaining isolates. Similar biodegradation potentials on crude oil [32] and other hydrocarbon contaminated sites [33] [34] were reported for indigenous microorganisms but with much lower rates. Addition of nitrogen and phosphorus during enrichment process might have played an important role in acceleration of biodegradation in the present study.
3.2. Screening for Potent Isolate(s) with Multiple Biodegradation Potentials
The 3 most efficient bacterial isolates, named BDCC-TUSA-8, BDCC-TUSA-12 and BDCC-TUSA-18, exhibiting active growth and effective COD removal of more than OD625nm 0.3 and 80% respectively (Table 1) were selected for biodegradation experiments. Three different hydrocarbons representing the major constituents of hydrocarbon pollutants [17] were used as target substrates, namely n-Hexadecane, phenol and phenanthrene (Figure 1).
Figure 1. Cell growth and utilization of phenanthrene (a), phenol (b) and D-Hexadecane (c) by isolate BDCC-TUSA-8, isolate BDCC-TUSA-12 and isolate BDCC-TUSA-18 in batch cultures. Open symbols for cell growth and closed symbols for hydrocarbon utilization.
Generally, the three isolates degraded all hydrocarbons and grew well indicating multiple biodegradation potentials but with different efficiencies. Isolate BDCC-TUSA-8 showed preference to aromatic hydrocarbons and isolate BDCC-TUSA-18 to alkenes while isolate BDCC-TUSA-12 showed active degradation on all tested hydrocarbons. The fastest degradation rate and the most efficient growth were recorded for isolate BDCC-TUSA-18 on n-Hexadecane with almost complete consumption and maximum cell growth (OD625nm) of 0.475 on day 4. It is well-known that the linear alkenes are considered to be especially easily biodegradable by microbes [34] [35] . Similar performances on consumption of phenol and phenanthrene were achieved by the other two isolates but with isolate BDCC-TUSA-8 exhibiting faster (day 3) and complete degradation on both hydrocarbons with relatively higher cell growth. The multiple biodegradation capabilities shown by the three indigenous isolates could be attributed to the enrichment approach employed which allow long exposure to NORWW with its diverse ingredients and/or to a possible unique genetic makeup for each individual isolates during that long exposure.
Consortia of hydrocarbon degraders displayed metabolic versatility and superiority compared very favorably to single pure isolates for efficient biodegradation on aromatic hydrocarbons [35] , crude oil and diesel [36] and petroleum contaminated soil [37] . However, costs for mass preparation of each individual member of the consortium contribute significantly to the success of biodegradation process. The obtained three indigenous bacterial isolates with multiple biodegradation potentials can contribute positively to establish an effective consortium with much smaller number of isolates and consequently considerable reduction in preparation costs.
3.3. Identification of the Most Active Bacterial Isolates
3.3.1. Phenotype Characterization
Based on morphological, biochemical features obtained from tests performed according to Bergey’s Manual of Determinative Bacteriology and API 20E and API 50CHB kits, isolate BDCC-TUSA-8 was tentatively identified as Pantoea spp., isolate BDCC-TUSA-12 as Acinetobacter spp. and isolate BDCC-TUSA-18 as Bacillus spp. Colonies of isolate BDCC-TUSA-8 were yellow round. Cells were short rods, Gram negative, motile, catalase positive, oxidase negative and nitrate positive. API 20E profile is included in Table 2. For isolate isolate BDCC-TUSA-12, colonies were translucent round. Cells were short rods, Gram negative, motile, catalase positive, oxidase negative and nitrate negative. API 20E profile is presented in Table 3. Colonies of isolate BDCC-TUSA-18 were white round. Cells were long rods, Gram positive, motile, catalase positive, oxidase negative, nitrate positive. Profiles of API 20E and API 50CHB are tabulated in Table 4.
3.3.2. Genotype Characterization
The use of 16S rRNA in identification of hydrocarbon degrading bacteria has been reported to be more sensitive and reliable compared with culture-dependant characterization techniques [38] - [40] . Therefore, the three isolates were further identified by partial sequencing of the PCR amplified 16S rDNA gene fragment (about 1500 bp) and used in a BLAST search in order to find a homology with other 16S RNA sequences. Comparing sequences of 16S rRNA gene of the three isolates with sequences in GenBank revealed that Bacillus sp. isolate was similar to B. thuringiensis strain Bt407and Acinetobacter sp. isolate was similar to A. lwoffii strain JCM 6840 with similarity of 97% and 99% respectively, while Pantoea sp. demonstrated 99% identity to P. agglomerans DSM 3493.
Table 2. Morphological, physiological and biochemical characteristics of isolate BDCC-TUSA-8.
+, positive; −, negative.
Table 3. Morphological, physiological and biochemical characteristics of isolate BDCC-TUSA-12.
+, positive; −, negative.
A phylogenetic tree was constructed for each individual isolate based on 16S RNA gene sequences. It was found that, each isolate shared one clad cluster with the respective identified bacterial strain as mentioned above (Figures 2-4). Therefore, phylogenetic analysis further confirmed the three isolates as P. agglomerans, A. lwoffii and B. thuringiensis respectively.
To test the relationship among the three isolated bacterial strains, a sample of sequences that showed high identity to each isolate were used to construct a general phylogenetic tree (Figure 5). It showed that each isolated strain lied on a separate clade with their relatives which confirm the obtained characterization.
Similar hydrocarbon degrading bacterial species have demonstrated high production
Table 4. Morphological, physiological and biochemical characteristics of isolate BDCC-TUSA-18.
+, positive; −, negative.
Figure 2. The phylogenetic relationships between the strain gs1 and other 16S rRNA sequences of the most closely related bacteria species.
Figure 3. The phylogenetic relationships between the strain gs2 and other 16S rRNA sequences of the most closely related bacteria species.
Figure 4. The phylogenetic relationships between the strain gs3 and other 16S rRNA sequences of the most closely related bacteria species.
of surfactants and emulsifiers which greatly enhance degradation efficiency through rapid promotion of substrate availability and uptake mechanisms [41] , and as such obtained strains of B. thuringiensis, P. agglomerans and A. lwoffii may have such potential [32] [42] [43] . Together with their demonstrated multiple degradation potentials, the three obtained bacterial strains could significantly contribute in the development of a cost-effective bioremediation process on petroleum oil contaminated environments.
Figure 5. A phylogenetic tree of isolated bacterial based on the nucleotide sequences of 16S rRNA genes was constructed by neighbor-joining method. The scale bar shows the genetic distance. The number presented next to each node shows the percentage bootstrap value of 1000 replicates. The Klebsiella quasipneumoniae was treated as the out-group. The GenBank accession numbers of the bacteria are presented in parentheses.
4. Conclusion
It can be concluded that, screening for indigenous bacterial isolates from petroleum sludge enriched in NORWW, resulted in the isolation of three potent isolates with multiple degradation potentials and remarkably fast reaction rates on various types of hydrocarbon, namely, n-Hexadecane, phenol and phenanthrene. All the three isolates could degrade and grow on tested hydrocarbons and achieve almost complete degradation. The potent strains were fully characterized and identified as P. agglomerans, A. Iwoffii and B. thuringiensis. Work is in progress in order to assess for a possible surfactant secretion by the obtained strains that could enhance biodegradation efficiency, and also to optimize nutritional and environmental parameters that ensure vigorous mixed growth of the three isolates and the constitutive expression of catalyzing enzymes that can work in a complementary action. Certainly, this would be of great importance when such approach is to be considered for establishing a cost-effective bioremediation process not only for the treatment of refinery wastewater but also for any other hydrocarbon impacted environments as well.
Acknowledgements
The authors are grateful to Department of Scientific Research Affairs, Taif University, K.S.A. for the financial support for this work.
Cite this paper
Gómez, R.J.V., Restrepo, G.E.C., Herrera, C.X.M., Ospina, V., Uribe, S.I. and Robledo, S.M. (2016) Antagonistic Effect of Bacteria Isolated from the Digestive Tract of Lutzomyia evansi against Pro- mastigotes of Leishmania infantum, Antimicrobial Activities and Susceptibility to Antibiotics. Advances in Microbiology, 6, 760-775. http://dx.doi.org/10.4236/aim.2016.610075
References
- 1. Ishak, S., Malakahmad, A. and Isa, M.H. (2012) Refinery Wastewater Biological Treatment: A Short Review. Journal of Scientific and Industrial Research, 71, 251-256.
- 2. Chen, C.S., Hseu, Y.C., Liang, S.H., Jar-Yi, K. and Ssu, C.C. (2008) Assessment of Genotoxicity of Methyl-tert-Butyl Ether, Benzene, Toluene, Ethylbenzene, and Xylene to Human Lymphocytes Using Comet Assay. Journal of Hazardous Materials, 153, 351-356.
http://dx.doi.org/10.1016/j.jhazmat.2007.08.053 - 3. Lewis, C., Chris, P. and Tamara, G. (2008) Reproductive Toxicity of the Water Accommodated Fraction (WAF) of Crude Oil in the Polychaetes Arenicola marina (L.) and Nereis virens (Sars). Aquatic Toxicology, 90, 73-81.
http://dx.doi.org/10.1016/j.aquatox.2008.08.001 - 4. Zhu, X., Albert, D., Venosa, M., Suidan, T. and Lee, K. (2001) US Environmental Protection Agency, USA, Guidelines for the Bioremediation of Marine Shorelines and Freshwater Wetlands.
- 5. El-Naas, M.H., Al-Zuhair, S., Al-Lobanery, A. and Makhlouf, S. (2009) Assessment of Electrocoagulation for the Treatment of Petroleum Refinery Wastewater. Journal of Environmental Management, 21, 180-185.
http://dx.doi.org/10.1016/j.jenvman.2009.08.003 - 6. Hami, M.L., Al-Hshimi, M.A. and Al-Doori, M.M. (2007) Effect of Activated Carbon on BOD and COD Removal in a Dissolved Air Flotation Unit Treating Refinery Wastewater. Desalination, 216, 116-122.
http://dx.doi.org/10.1016/j.desal.2007.01.003 - 7. Yan, L., Bo-Ma, H., Wang, Y. and Chen, Y. (2011) Electrochemical Treatment of Petroleum Refinery Wastewater with Three-Dimensional Multi-Phase Electrode. Desalination, 276, 397-402.
http://dx.doi.org/10.1016/j.desal.2011.03.083 - 8. Mara, K., Decorosi, F., Viti, C., Giovannetti, L., Papaleo, M., Maida, I., Perrin, E., Fondi, M., Vaneechoutte, M., Nemec, A., van den Barselaar, M., Dijkshoorn, L. and Fani, R. (2012) Molecular and Phenotypic Characterization of Acinetobacter Strains Able to Degrade Diesel Fuel. Research in Microbiology, 163, 161-172.
http://dx.doi.org/10.1016/j.resmic.2011.12.002 - 9. Rojo, F. (2009) Degradation of Alkanes by Bacteria. Environmental Microbiology, 11, 2477-2490.
http://dx.doi.org/10.1111/j.1462-2920.2009.01948.x - 10. Zhang, Z.Z., Gai, L.X., Hou, Z.W., Yang, C.Y. Ma, C.Q., Wang, Z.G., Sun, B.P., He, X.F., Tang, H.Z. and Xu, P. (2010) Characterization and Biotechnological Potential of Petroleum-Degrading Bacteria Isolated from Oil-Contaminated Soils. Bioresource Technology, 101, 8452-8456.
http://dx.doi.org/10.1016/j.biortech.2010.05.060 - 11. Fritsche, W. and Hofrichter, M. (2008) Investigation of the Bioremediation Potential of Aerobic Zymogenous Microorganisms in Soil for Crude Oil Biodegradation. Biotechnology, 11, 144-150.
- 12. Tadros, M.G. and Hughes, J.B. (1997) Degradation of Polycyclic Aromatic Hydrocarbons (PAHs) by Indigenous Mixed and Pure Cultures Isolated from Coastal Sediments. Biotechnology and Applied Biochemistry, 63-65, 865-871.
http://dx.doi.org/10.1007/BF02920482 - 13. Atlas, R.M. (1995) Bioremediation of Petroleum Pollutants. International Biodeterioration and Biodegradation, 35, 317-327.
http://dx.doi.org/10.1016/0964-8305(95)00030-9 - 14. Ammary, B.Y. (2004) Nutrients Requirements in Biological Industrial Wastewater Treatment. African Journal of Biotechnology, 3, 236-241.
http://dx.doi.org/10.5897/AJB2004.000-2042 - 15. Chan, Y.J., Chong, M.F. and Law, C.L. (2010) Biological Treatment of Anaerobically Digested Palm Oil Mill Effluent (POME) Using a Lab-Scale Sequencing Batch Reactor (SBR). Journal of Environmental Management, 91, 1738-1746.
http://dx.doi.org/10.1016/j.jenvman.2010.03.021 - 16. Maier, R.M. (1999) Chapter 14: Biochemical Cycling, In: Maier, R.M., Pepper, I.L. and Gerba, C.P., Eds., Environmental Microbiology, Vol. 9, Academic Press, New York, 319-346.
- 17. Pirt, J.S. (1975) Principles of Microbe and Cell Cultivation. Blackwell Scientific Publications, Cornwall, 4-44.
- 18. Liu, X., Mao, X., Yang, J., Barry, D. and Li, L. (2006) Experiments and Modeling of Phenanthrene Biodegradation in the Aqueous Phase by a Mixed Culture. Journal of Environmental Sciences, 8, 147-153.
- 19. Al-Godah, A.N. (2012) Bioremediation of Crude Oil-Contaminated Effluents from an Oil Refinery in Jeddah. MSc Thesis, King Abdulaziz University, Jeddah.
- 20. Al-Lighaibi, S.S. (2000) Petroleum Hydrocarbons in Sediments from Jeddah Coast of Saudi Red Sea following the 1996 Fuel Oil Spill. Journal of Marine Science, 11, 27-41.
- 21. Ollivier, B. and Magot, B. (2005) Petroleum Microbiology. American Society for Microbiology. ASM Press, Washington DC.
- 22. Holt, J.G., Kreieg, N.R., Sneath, P.H., Staley J.T. and Williams, S.T. (1994) Bergey’s Manual of Determinative Bacteriology. William and Wilkins Co., Baltimore, 559-564.
- 23. Lane, D.J. (1991) 16S/23S rRNA Sequencing. In: Stackebrandt, E. and Goodfellow, M., Eds., Nucleic Acid Techniques in Bacterial Systematic, John Wiley and Sons, New York, 115-175.
- 24. Turner, S., Pryer, K.M., Miao, V.P. and Palmer, J.D. (1999) Investigating Deep Phylogenetic Relationships among Cyanobacteria and Plastids by Small Subunit rRNA Sequence Analysis. Journal of Eukaryotic Microbiology, 46, 327-338.
http://dx.doi.org/10.1111/j.1550-7408.1999.tb04612.x - 25. Tamura, K., Stecher, G., Peterson, D., Filipski, A. and Kumar, S. (2013) MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution, 30, 2725-2729.
http://dx.doi.org/10.1093/molbev/mst197 - 26. Jukes, T.H. and Cantor, C.R. (1969) Evolution of Protein Molecules. In: Munro, H.N., Ed., Mammalian Protein Metabolism, Academic Press, New York, 21-132.
http://dx.doi.org/10.1016/B978-1-4832-3211-9.50009-7 - 27. Saitou, N. and Nei, M. (1987) The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Molecular Biology and Evolution, 4, 406-425.
- 28. Felsenstein, J. (1985) Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution, 39, 783-791.
http://dx.doi.org/10.2307/2408678 - 29. APHA (2005) Standard Methods for the Examination of Water and Wastewater. 21st Edition, American Public Health Association/American Water Works Association/Water Environment Federation, Washington DC.
- 30. Koch, A.L. (1981) Growth Measurement. In: Gerhardt, P., Murray, R., Costilow, R., et al., Eds., Manual of Methods for General Bacteriology, American Society for Microbiology, New York, 179-207.
- 31. Mishra, S., Yot, J. and Lal, B. (2001) In Situ Bioremediation Potential of an Oily Sludge Degrading Bacterial Consortium. Current Microbiology, 43, 328-337.
http://dx.doi.org/10.1007/s002840010311 - 32. Sarma, A. and Sarma, H. (2010) Enhanced Biodegradation of Oil Products by Some Microbial Isolate Supplemented with Heavy Metals. International Journal of Botany, 6, 441-448.
http://dx.doi.org/10.3923/ijb.2010.441.448 - 33. Ojo, O.A. (2006) Petroleum-Hydrocarbon Utilization by Native Bacterial Population from a Wastewater Canal Southwest Nigeria. African Journal of Biotechnology, 5, 333-337.
- 34. Ozaki, S., Kishimoto, N. and Fujita, T. (2007) Change in the Predominant Bacteria in a Microbial Consortium Cultured on Media Containing Aromatic and Saturated Hydrocarbons as the Sole Carbon Source. Microbes and Environments, 22, 128-135.
http://dx.doi.org/10.1264/jsme2.22.128 - 35. Abdel-Megeed, A., Al-Harbi, N. and Al-Deyab, S. (2010) Hexadecane Degradation by Bacterial Strains Isolated from Contaminated soils. African Journal of Biotechnology, 44, 7487-7494.
- 36. Leahy, J.G. and Colwell, R.R. (1990) Microbial Degradation of Hydrocarbons in the Environment. Applied and Environmental Microbiology, 54, 305-315.
- 37. Parakash, A., Bisht, S., Singh, J., Teotia, P., Kela, R. and Kumar, V. (2014) Biodegradation Potential of Petroleum Hydrocarbons by Bacteria and Mixed Bacterial Consortium Isolated from Contaminated sites. Turkish Journal of Engineering and Environmental Sciences, 38, 41-50.
http://dx.doi.org/10.3906/muh-1306-4 - 38. Amin, G.A. (2010) A Potent Biosurfactant Producing Bacterial Strain for Application in Enhanced Oil Recovery Applications. Journal of Petroleum & Environmental Biotechnology, 1, 104-109.
http://dx.doi.org/10.4172/2157-7463.1000104 - 39. Babu, K.S., Jyothi, K., Nancy, C.K. and Kashyap, A. (2012) Identification and Isolation of Hydrocarbon Degrading Bacteria by Molecular Characterization. Helix, 2, 105-111.
- 40. Al-Awadhi, H., Dashti, N. and Khanafer, S. (2013) Bias Problems in Culture Independent Analysis of Environmental Bacterial Communities: A Representative Study on Hydrocarbon Oclastic Bacteria. Springer Plus, 2, 369-373.
http://dx.doi.org/10.1186/2193-1801-2-369 - 41. Cho, H.S., Moon, H.S., Kim, M., Nam, K. and Kim, J.Y. (2011) Biodegradability and Biodegradation Rate of Poly(Caprolactone) Starch Blend and Poly(Butane Succinate) Biodegradable Polymer under Aerobic and Anaerobic Environment. Waste Manage, 31, 475-480.
http://dx.doi.org/10.1016/j.wasman.2010.10.029 - 42. Jacobucci, D.F., Oriani, M.R. and Durrant, L.R. (2009) Reducing COD Level on Oily Effluent by Utilizing Biosurfactant-Producing Bacteria. Brazilian Archives of Biology and Technology, 52, 1037-1042.
http://dx.doi.org/10.1590/S1516-89132009000400029 - 43. Kim, P.I., Bai, H., Bai, D., Chae, H., Chungm, S., Kim, Y., Park, R. and Chi, Y.-T. (2004) Purification and Characterization of a Lipopeptide Produced by Bacillus thuringiensis CMB26. Journal of Applied Microbiology, 97, 942-949.
http://dx.doi.org/10.1111/j.1365-2672.2004.02356.x







