Advances in Microbiology
Vol.4 No.5(2014), Article ID:45245,15 pages
DOI:10.4236/aim.2014.45032
Suppression of Methane Gas Emissions and Analysis of the Electrode Microbial Community in a Sediment-Based Bio-Electrochemical System
Yoshiyuki Ueno1*, Yoji Kitajima2
1Kajima Technical Research Institute, Tokyo, Japan
2Kajima Corporation, Tokyo, Japan
Email: *uenoyo@kajima.com
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/


Received 5 March 2014; revised 4 April 2014; accepted 13 April 2014
The effects of bioelectrochemical systems (BESs) for the suppression of methane gas emissions from sediment were examined using a laboratory-scale reactor system. Methane gas emissions from acetate were suppressed by approximately 36% from control based on the installation of a BES in which carbon-graphite electrodes were buried in sediment and arbitrarily set at certain oxidative potentials (+300 mV vs Ag/AgCl) using a potentiostat. Meanwhile, methane gas emissions increased in the BES reactor where the electrode potential was set at −200 mV. Results obtained from pyrotag sequencing analysis of the microbial community on the surface of the buried electrodes targeting 16S rRNA genes demonstrated that the genus Geobacter had drastically propagated in a sample from the reactor where the electrodes were buried. Quantitative analysis of 16S rRNA genes of archaea also revealed that the archaeal population had decreased to approximately 1/6 of its original level on the electrode of the BES set at +300 mV. This implied that the oxidation-reduction potential (ORP) in the sediment was raised to the inhibition level for methanogenesis in the vicinity of the buried electrode. Analysis of electron flux in the experiment revealed that electrons intrinsically used for methanogenesis were recovered via current generation in the sediment where a potential of +300 mV was set for the electrode, although most electrons donated from acetate were captured by oxygen respiration and other electron-accepting reactions. These results imply that BES technology is suitable for use as a tool for controlling redox-dependent reactions in natural environments, and that it also brought about changes in the microbial population structure and methanogenic activity in sediment.
Keywords:Microbial Fuel Cell, Bio-Electrochemical System, Methanogenesis, Electricigens, Green House Gas

Bioelectrochemical system (BES) application has attracted attention as a new approach to biological energy conversion [1] . Microorganisms interact with electrodes through electrons, which are either removed or supplied via an electrical circuit. The most popular type of BES is the microbial fuel cells (MFCs), which typically consist of an anode, a cathode and a membrane separating their two compartments. A microorganism-catalyzing oxidation process occurs at the anode and a reduction process occurs at the cathode. When a BES is operated in MFC mode, electricity can be recovered.
Sediment microbial fuel cells (SMFCs) are an application of MFCs. The SMFC system utilizes the natural potential gradient between sediment and the upper water phase. Electrons released from the microbial oxidation of organic compounds flow from the anode in the sediment to the cathode in water through an external circuit [2] .
The production of energy in the form of readily usable electrical power is the most direct potential application of SMFCs, and these systems were originally expected to allow in situ electricity generation for small electrical appliances. However, the average amount of sustainable power that has been obtained from SMFCs is in the order of 10 - 100 mW per square meter of anodic electrode footprint area (EFA) [2] , meaning that they do not supply sufficient electricity to power electrical equipment and cannot be used as an electricity generation system with present technology levels. However, although such cells may not be suitable for producing electrical energy, SMFC and/or BES technology can be employed for various other purposes. In SMFCs, the anode has a significant effect on dissolved organic matter oxidation, in which oxygen availability is a limiting factor. It is assumed that using electrodes as a way of supplying oxidation power to sediment helps to enhance the oxidation rates of organic compounds and/or pollutants in sediment.
Methane is a greenhouse gas (GHG) whose global warming potential (GWP) is 20 times higher than that of carbon dioxide [3] . Between 0.3 and 300 mg/m2/day of methane gas is emitted depending on the type of soil and its potential cultivation type. By way of example, a maximum of 2.9 mg/m2/day has been noted for rice paddy soil [4] . Agriculture, landfills and wastewater treatment plants are also sources of methane gas emissions. In these natural environments, methane gas usually evolves via microbial metabolisms that depend on thermodynamics and stoichiometry. Although hydrolysis and fermentation in the degradation steps of complex organic matter are not thought to be directly mediated by an active anode, the anodic process stimulates the final steps in the breakdown of such matter, with acetate presumed to be the most important electron donor because of its pivotal role in organic matter degradation by anaerobic microbial consortia [5] .
Methane gas emissions are reduced in situations with higher redox potential [6] [7] as seen in the presence of an anode or an electrode with arbitrarily controlled potential and a redox potential sufficiently higher than that of the corresponding redox couples. The standard redox potential of the CO2/CH4 couple is −0.244 V versus SHE [8] , and the corresponding methanogenesis process is generally found not to occur at soil redox potentials above −0.150 V versus SHE [7] .
It has been successfully demonstrated that an electron-capturing anode can suppress methanogenesis in an MFC reactor fed with cellulose and inoculated with rice paddy field soil as a microbial seed [9] . A previous study by the authors also highlighted the potential of BESs to reduce methane gas emissions from sediment [10] . Relatively low concentrations of such emissions were observed when electrodes were buried in sediment and set at certain oxidative potentials. Electrode installation and potential control were seen to trigger changes in microbial activity in the sediment. The relative abundance of archaea largely decreased in the vicinity of the anode, and significant mitigation of methane gas evolution activity in the sediment was found.
These results imply that BES technology can be used as a tool for controlling redox-dependent reactions in natural environments, and that it also brings about changes in the microbial population structure as well as methanogenic activity in sediment. In this study, the suppression effect of methane gas emissions was quantitatively analyzed by considering electron flux to methane and current generation. The community structure of microorganisms on the electrode surface, which was enriched by arbitrarily setting potential, was also investigated and characterized via pyrotag sequencing.
2.1. Reactor Apparatus
BES reactors were made using 2-liter PET (polyethylene terephthalate) bottles as reservoirs for sediment created by mixing freshwater sediment from a fish breeding facility with andosol, zeolite and sand. The bottles were filled with approximately 0.89 L of sediment (equivalent to a depth of approx. 10 cm). The electrodes were made of plain carbon graphite (4 × 5 × 0.7 cm; electrode footprint area: 86 cm2) and insulating wire was affixed to them with electrically conductive epoxy (H20E silver epoxy, Epotek, USA). A modified silicone sealant (SM- 447, Cemedine Co. Ltd., Japan) was applied after the epoxy had dried to protect it and any exposed wire from contact with water or sediment. An electrode was buried at the bottom of the sediment, and tap water with a 1/100 concentration of the basal medium was gently added to a depth of 8.5 cm over the sediment and allowed to permeate the particles overnight. Another electrode was set in the overlying water. The basal medium contained the following material amounts per liter: KH2PO4: 0.91 g; Na2HPO4∙12H2O: 2.39 g; NH4Cl: 0.5 g; MgCl2∙6H2O: 0.18 g; mineral solution: 5.0 ml; vitamin solution: 1.0 ml [11] .
The electrodes were connected to a potentiostat (HA-151B, Hokuto Denko Co. Ltd., Japan), and the potential of the buried one was set arbitrarily. The electrode wires were connected with a resistor (1210 W) between them when the BES reactor was used in SMFC mode, and the voltage across the resistor was monitored every 10 min using a data logger. Electrodes that were connected neither via an external circuit nor via a potentiostat were used as a control system. The BES reactors were contained in a hood chamber, and methane gas concentration in the air phase of the reactor was continuously measured using a photoacoustic multi-gas monitor (Model 1312, Innova, Denmark). Total amount of the methane gas emission during experimental period was determined from the methane gas concentration and sampling volume for determination. The BES reactors were installed in a water bath at 30˚C (Figure 1).
Electron recovery in the experiment was calculated on the basis of the amount of acetate decomposed and the amounts of current measured and methane evolved [12] .
2.2. DNA Extraction
Sediment or biofilm that was spread onto the surface of electrodes with a spatula was taken from the reactors and suspended in phosphate buffered saline (PBS). Any loose soil on the electrodes was removed before the biofilm sample was taken. DNA was extracted using the MagNA Pure System (Roche, USA). A MagNA Pure LC DNA Isolation Kit III (Roche, USA) was used according to the manufacturer’s instructions expect for one
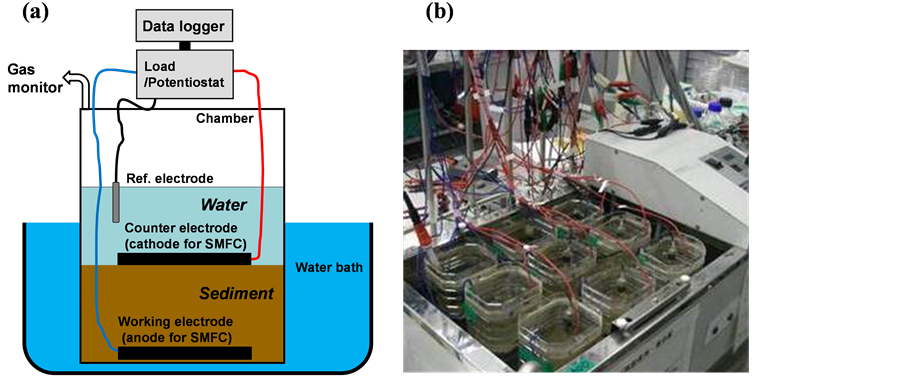
Figure 1. Schematic drawing of the BES reactor (a) and the appearance of the reactors in a water bath when hood chambers for gas collection were detached from the reactors (b).
modification in which the sample was agitated with a bead crusher (3200 r/min) for 30 sec using 0.1 mm of glass beads after solubilization using a lysis buffer and proteinase K treatment.
2.3. Pyrotag Sequencing of 16S rRNA Gene Amplicons
PCR amplification of 16S rRNA gene fragments (V4 region) was performed using the primers ad-tag-515F (5’-CGTATCGCCTCCCTCGCGCCATCAGXXXXXX GTGCCAGCMGCCGCGGTAA-3’) and ad-806R (5’- CTATGCGCCTTGCCAGCCCGCTCAG GGACTACHVGGGTWTCTAAT) [13] , where the underlined sequences are adaptors for pyrosequencing and XXXXXX is an arbitrary tag sequence for sample identification [14] .
An AmpliTaq Gold® DNA Polymerasekit (Life Technologies, California, USA) was used for the PCR reactions. The PCR mixture contained 36.5 μL of sterilized water, 5 μL of PCR buffer, 5 μL of dNTP mix, 1 μL of each of the forward and reverse primers (0.2 μM) and 1.0 μL of genomic DNA. Reactions were held at 95˚C for 10 min to denature the DNA, with amplification proceeding for 28 cycles at 95˚C for 45 s, 55˚C for 60 sand 72˚C for 90 s. A final extension of 10 min at 72˚C was added to ensurecomplete amplification.
A composite sample for sequencing was created by combining equimolar ratios of amplicons from the individual samples followed by gel purification (illustra GFX PCR DNA and Gel Band Purification Kit, GE Healthcare, USA) and ethanol precipitation tore move any remaining contaminants and PCR artifacts.
The mixed amplicons were subjected to pyrosequencing using a Genome Sequencer FLX system at the Dragon Genomics Center (Mie, Japan), and phylogenetic analysis was conducted using the DDBJ 16S rRNA database (Feb. 5, 2013), the Blast program [15] and the RDP classifier ver. 2.3 [16] . An operational taxonomic unit (OTU) was defined as a unique sequence or a group of sequences with sequence homologies of over 97%. Rarefaction analysis was performed using the Esprit program [17] , and sequence alignment was conducted using ClustalX ver. 2.1 [18] . A phylogenetic tree based on the partial sequences obtained and reference strains were constructed using the neighbor-joining method [19] .
2.4. Quantitative Real-Time PCR
The abundance of Geobacteraceae bacteria in the soil was evaluated via quantitative real-time PCR (qPCR) assay using a primer set targeting 16S rRNA genes of bacteria affiliated with the family Geobacteraceae: Geo494F and Geo825R [20] . The Light Cycler system and a Light Cycler DNA Master SYBR Green I kit (Roche Applied Science) were used. As PCR conditions, after initial denaturation at 95˚C for 10 min, the target DNA was amplified in 40 cycles each consisting of denaturation for 5 s at 95˚C, annealing for 5 s at 65˚C and extension for 20 s at 72˚C. Standard curves were generated using seriallydiluted genomic DNA extracted from Geobacter sulfurreducens. The amount of Geobacteraceae DNA (G. sulfurreducens-equivalent value) in the extracted DNA was calculated from the qPCR results, and the ratio of Geobacteraceae DNA to the total of extracted DNA was estimated using the expression Fold Change (FC) = 2−ΔΔCT. The abundance of Archaea was also evaluated by qPCR assay using a primer set: Ar109f and Ar915r according to the previous study [21] .
2.5. Nucleotide Sequence Accession Numbers
The nucleotide sequence data reported here have been deposited on the NCBI Short Read Archive database under accession number DRA001685.
3.1. Performance of BES Reactor for the Suppression of CH4 Gas Emissions
The BES reactors were allowed to acclimatize for 364 days after electrode installation, as a period of around 200 days is needed for ORP values in the sediment to stabilize [10] . During this time, 4.8 mmol of acetate was periodically injected into the vicinity of the working electrode or anode using a needle. Five injections were made before methane gas emissions were determined, and three were made during the determination period.
The temporal progressions of typical performance in each reactor are shown in Figure 2. Methane gas concentration in the gas phase above the reactor gradually increased after acetate injections, and evolution ceased within around eight days. The behavior response of methane gas evolution showed a correlation with current
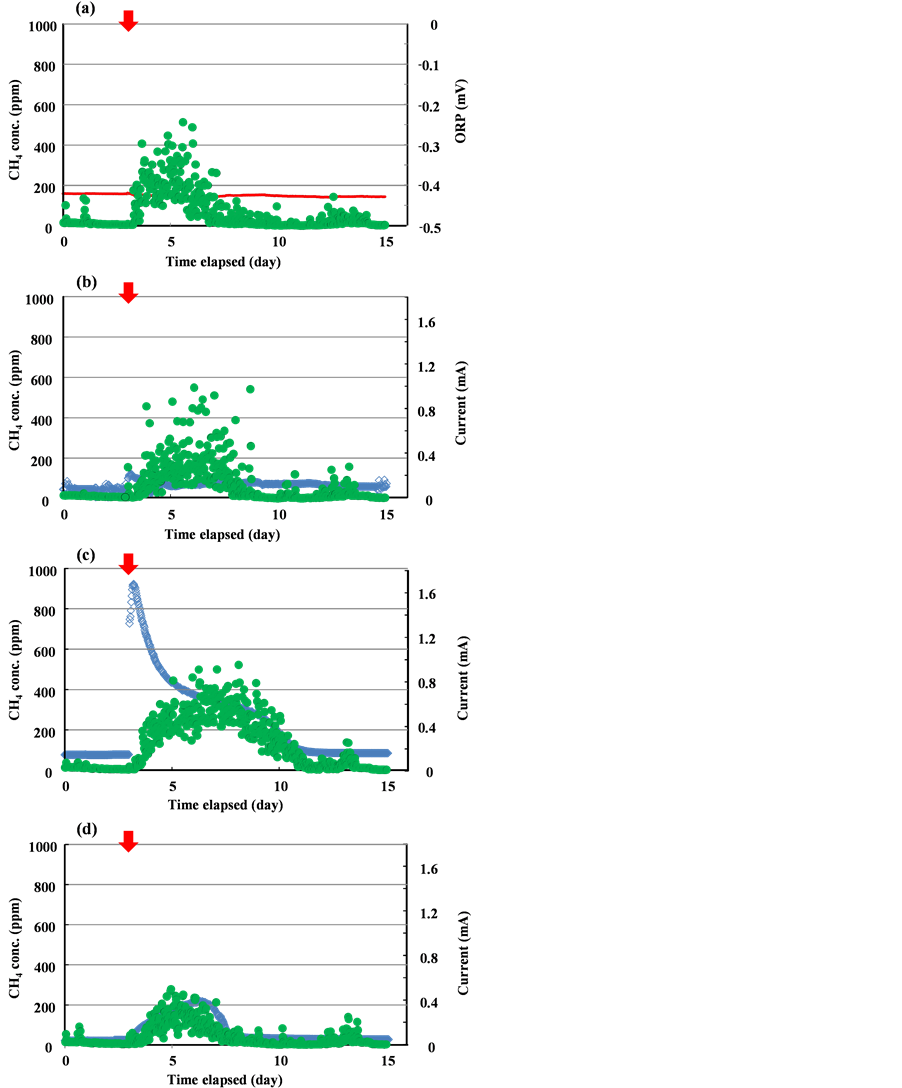

Figure 2. Time courses of current generation and methane gas emissions from BES reactors. Arrows indicate acetate injection. (a) Reactor A; (b) Reactor B; (c) Reactor C; (d) Reactor D.
generation. The concentrations of the methane gas evolved differed in each reactor tested; the value was lower in Reactor D, in which a potential of +300 mV (vs Ag/AgCl) as set at the working electrode, while no reduction of concentration was observed in Reactor B (SMFC mode). Acetate was not detected in the reactor water phases after the end of the 14-day period following acetate injections, when both methane gas evolution and current generation ceased. This implies that the injected acetate had completely decomposed by this time. The methane gas emissions of Reactor D were 36% lower than those of the reactor without electrode installation (Reactor A). Meanwhile, methane gas concentration drastically increased when the electrode potential was set at −200 mV (vs Ag/AgCl) (Reactor C). These results suggest that electrode installation in the sediment and positive potential setting reduced methane gas concentration in the gas phase in the reactor.
The highest current generation was observed in Reactor C, while that seen in SMFC mode (Reactor B) was lower than previously reported values in MFCs, yielding a coulumbic efficiency level of 1% (Table 1). This may have been due to mass transfer limitation and/or corrosion of the electrode used, as the present study continued for around year after electrode installation. Electron recovery in the experiments was calculated from the amount of acetate injected, the amount of current generated and the amount of methane gas evolved (Table 1). In each reactor, most of the electrons donated from acetate were recovered except for those taken up by methane and current, indicating that oxygen respiration and other electron-accepting reactions were predominant in the present experimental system, although Fe3+, Mn4+ and NO3− are also electron acceptor candidates under anaerobic conditions. Electron recovery for the current was approximately 1% in Reactor B, where electrode potential was not artificially set. Current generation was extremely low in SMFC mode in this experimental system.
3.2. Pyrotag Sequencing
Although the number varied slightly, approximately 4000 - 8000 reads were obtained for each sample. Around 10,000 representative examples were chosen from the sequences, accounting for more than 97% of identities. Sediment from before the experiment was also analyzed to provide control data.
The results of rarefaction analysis are shown in Figure 3. The read number was not saturated for each sample analyzed, implying that a much more diversified population cloud existed within the samples. The number of representative sequences represents the richness of microbial diversity in the samples, and the most diversified population was observed in the original sediment analyzed before the experiment to provide control data. A secondary diversified population was observed in the non-electrode sample injected with acetate during the same experimental period (Reactor A). The read numbers were lower in the samples from BES reactors B, C and D than those of the control sample and Reactor A, implying that the presence of specific microorganisms on the electrode surface was enriched by electrode installation and potential setting. The levels of enrichment differed among the BES reactors, while richness in diversity showed no correlation with the setting potential. The microbial population was the most accumulated in Reactor C, where potential of −200 mV was applied.
The proportion of clones affiliated to unknown species was high in each sample and particularly high in the control sample, suggesting microbial richness in diversity within the natural sediment used (Figure 4). On the contrary, the microbial population changed drastically depending on experimental conditions including temperature, nutrient (acetate) addition and electrode installation with or without potential setting. These cultivation biases brought about an increase in the proportion of the genera Geobacter and Longilinea. The proportion of Longilinea was 2% - 5% among all BES reactors, while that of Geobacter increased in reactors with buried electrodes (B, C and D), accounting for 37%, 59% and 30% of the total, respectively. The proportion of Geobacter increased to 6% of the total in Reactor A, indicating that acetate addition enriched this genus in the sediment used even without the presence of a buried electrode.
Table 1. Summary of reactor performance for suppression of methane gas emissions using sediment-based BES.
Data were obtained during 15 days of operation after acetate injection (4.8 mmol). aCalculated from CH4 concentration in the gas phase in the hood chamber and the volume of gas continuously sampled for determination; bEvaluated with comparison to Reactor A; cCurrent values are converted to coulomb equivalents.
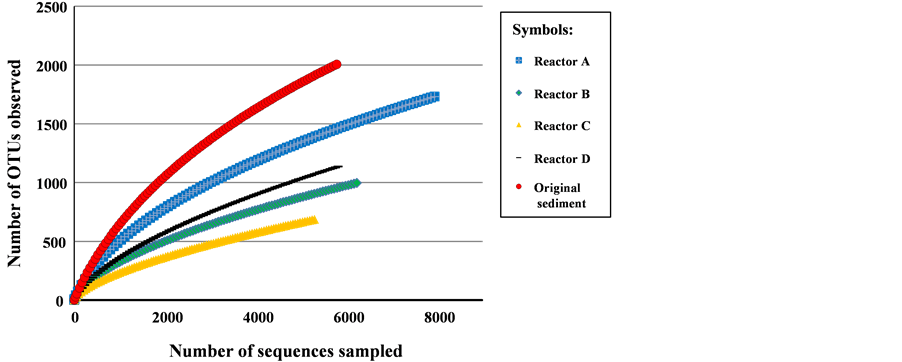
Figure 3. Rarefaction curves based on data obtained from pyrotag-sequencing analysis of 16S rRNA genes in sediment samples (similarity cut off: 97%).
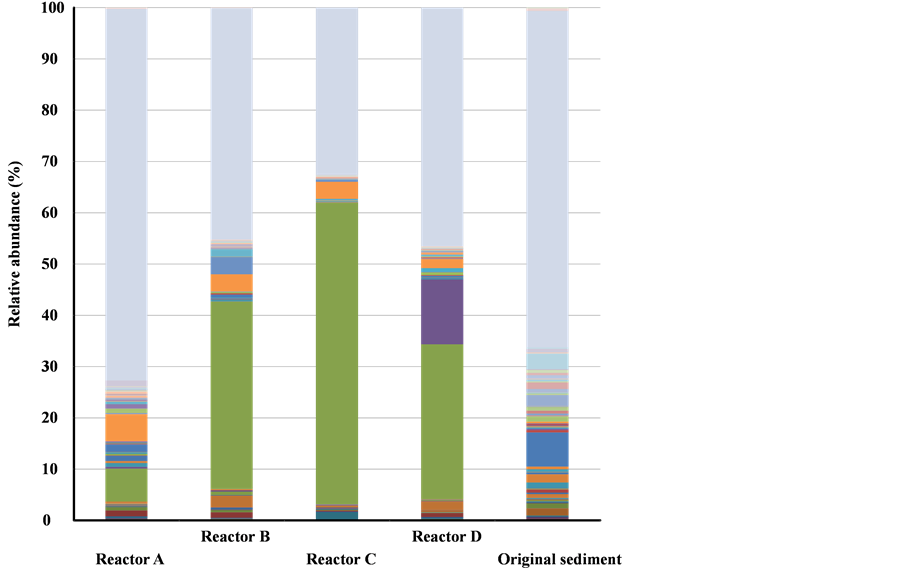

Figure 4. Relative abundance of major genus-level microbial groups obtained in pyrotag-sequencing analysis of 16S rRNA genes in sediment samples. More than 100 reads of detected sequences are shown in the figure. Detailed data are shown in Table S1. Data for the microbial communities in the sediment used as the inoculum are also shown for comparison.
Longilinea, an obligate anaerobe belonging to phylum Chloroflexi, is known as a protein decomposer in methanogenic bioreactors [22] . Although it seems to function in granular formation, the function remained unclear in this experimental system. A significant increase in the proportion of Geotoga was observed in the Reactor I sample (13% of the total). The function of this organism was also unclear in the experimental system, as the genus Geotoga is known as a moderate thermophilic carbohydrate-fermenting bacterium [23] . The proportion of methanogenic archaea belonging to the genera Methanobacterium and Methanosarcina was approximately 2% of the total read number.
Sequences affiliated with the family Geobacteraceae contained several major OTUs (more than 100 reads of the total). The phylogenetic relationships linking these OTUs and the reference strains are shown in Figure 5. Although overgrowing Geobacter in the sample of Reactor A (OTU-JBKXN) was affiliated with a distinct cluster comprised of G. grbiciae and G. metallireducens, all other clones were positioned in different clusters of the reference strains. Relatives of G. sulfurreducens, which is a well-characterized EAB with high electricity-generatingcapacity in the presence of acetate [24] , are frequently detected in acetate-fed bioelectrochemical systems [25] -[27] . The Geobacter relatives observed in the present experimental system with freshwater sediment as a microbial source may have been slightly different from species so far reported.
3.3. Proportion of Archaea and Geobacteraceae in BES Reactors
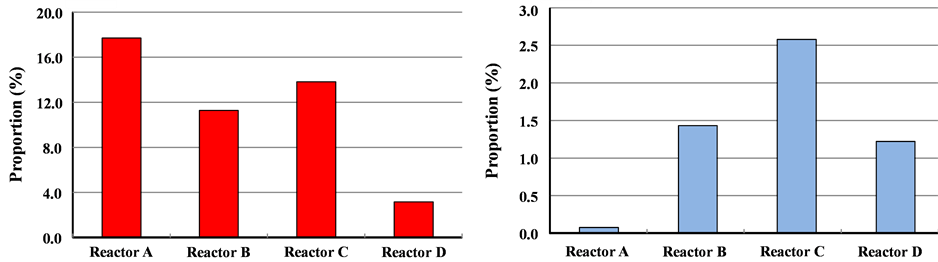
The relative abundances of archaea and Geobacteraceae DNA as a proportion of the total DNA were determined using sediment samples taken from the vicinity of the buried electrodes. It was confirmed that the proportion of archaea was lower in the samples from the BES reactors (Figure 6(a)), with the population decreasing to approximately 1/6 for the electrode set at +300 mV.
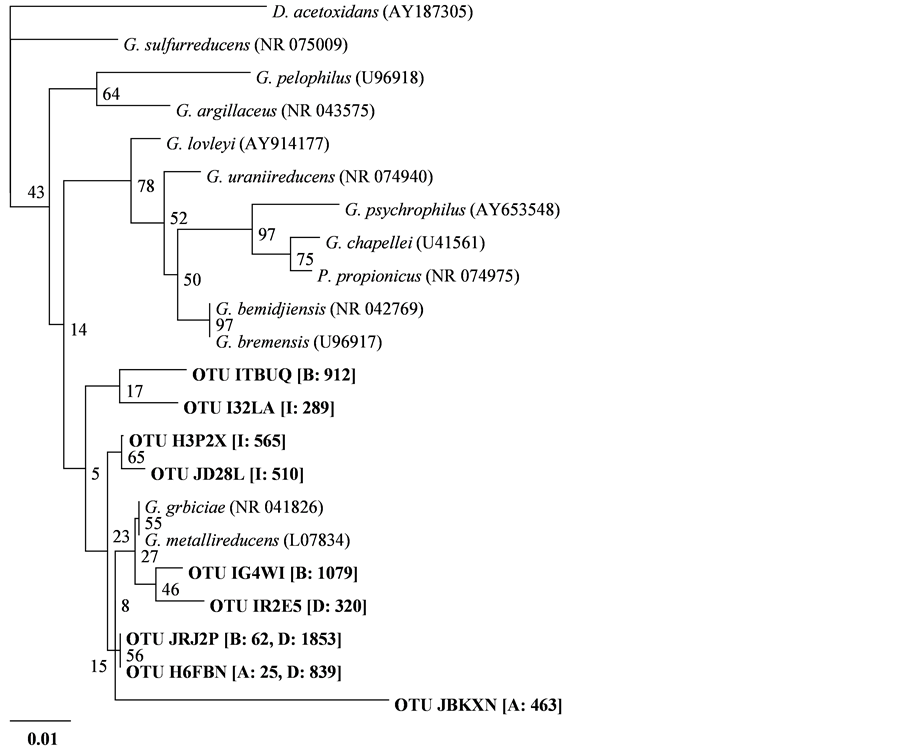
Figure 5. A phylogenetic tree based on partial 16S rRNA gene sequences of representative Geobacter isolates and Geobacteraceae sequences phylotypes retrieved in this study (indicated in bold). The numbers of clones retrieved from different samples are shown in square brackets. The sample names are abbreviated in square brackets as follows: A: Reactor A; B: Reactor B; C: Reactor C; D: Reactor D. Desulfuromonas acetoxidans was used as an out group. Bootstrap values are indicated at branching points. The bar indicates 1% sequence divergence. Accession numbers are shown in parentheses.
 (a) (b)
(a) (b)
Figure 6. Proportion of archaeal (a) and Geobacteraceae (b) DNA in the sediment samples from the electrodes. Values are presented as the means of three independent analysis results.
The relative abundance of Geobacteraceae populations significantly increased in the vicinity of the buried electrodes (Figure 6(b)). The ratio was more than 10 times higher in the reactors in which electrodes were buried than in Reactor A with no electrode. This suggests that electrode installation in the sediment brought about the enrichment of Geobacteraceae, which plays an important role in electricity generation.
A certain degree of suppression for methane gas emissions was observed as a result of setting buried electrodes in sediment, although the effect was not dramatic. Thus, electrode installation and potential control triggered a change in microbial activity in the sediment. To a certain extent, the ORP was raised to a inhibition level for methanogenesis in the vicinity of the buried electrodes (greater than −150 mV vs standard hydrogen electrodes) [7] [8] . The observed lowering of methanogenic activity and reduction of archaeal population in the sediment support this speculation [10] .
Electrons donated from organic compounds should not be used in methanogenesis to reduce methane gas emissions from sediment. If other electron acceptors such as oxygen, Fe3+, Mn4+ or NO3− accept the electrons instead of methanogenesis, methane gas emissions will be reduced. These electron-accepting reactions take place at higher ORPs than methanogenesis. Electrogenesis also competes with methanogenesis [9] , and increased electricity generation will therefore reduce methane gas emissions. However, higher current generation was observed with increased methane gas emissions (Reactor C).
Except in Reactor C, approximately 85% of electrons released from acetate were used for these electron-accepting reactions. In Reactor D, current generation increased with reduced methane gas emissions, while almost the same amounts of electrons were captured by other electron-accepting reactions. This implies that electrons intrinsically used for methanogenesis were recovered in current generation. Meanwhile, methane gas emissions increased with greater current generation, while electron recovery to other reactions was reduced to 60% in Reactor C.
Negative potential control based on electrode installation in a methanogenic reactor brought about efficient methane gas production in a previous study [28] . It was revealed that almost all the methane came from the substrate based on calculation of the current provided and the amount of methane gas evolved, suggesting that effects other than direct methane production from current [29] work toward the efficient operation of the bioelectrochemical reactor. In Reactor C, noequilivalent reduction was provided to the sediment, as the applied potential of −200 mV was more positive than the ORP of the electrode vicinity (data not shown). This may have created milder conditions for microorganisms than the SMFC-mode reactor (Reactor B) and Reactor D, where a positive potential of +300 mV of was applied. It is therefore assumed that this level of potential (−200 mV) provided stable conditions for methanogen enrichment and electricigen activation. The proportion of Geobacter was the highest in samples analyzed either in pyrotag sequencing or in quantitative PCR (Figure 2(b) and Figure 4(b)).
As the volume of sediment influenced by one electrode was not optimized in this study, the area and volume affected by electrode potential remains unclear. Since electrode area is known to affect the amount of electricity generation in MFCs [30] , an increased number of electrodes and structural improvement may suppress methane gas emissions even further. Electrode reactivity is also important in evaluating effectiveness. It was demonstrated in a previous study that the chemical profile of sediment was shifted in a zone of 3 cm around an active SMFC anode [31] . Although the area of sediment influenced by potential setting was not determined in the present experiment, a certain amount of sediment was oxidized and the ORP rose to a methanogenesis inhibition level.
However, some studies have demonstrated that a positive soil redox potential does not inhibit the methanogenesis process as long as H2 is sufficientlyavailable [32] . When organic polymers such as carbohydrates and/or proteins are substrates for methanogenesis, molecular hydrogen must be formed. However, hydrogen may be oxidized in the presence of an electrode, leading to a greater presence of appropriate microorganisms [33] .
The results obtained from the present study suggest that BES installation in sediment is applicable to technology intended to suppress GHG emissions from natural environments and to control the decomposition of organic pollutants and environmentally toxic compounds. ORP-dependent accumulation of microorganisms was also demonstrated. Multidisciplinary technologies involving the material properties of electrodes and electrochemistry, confirmation of long-term usage feasibility and prevention of electrode passivation are necessary for the implementation of such systems as an environmental solution in the future.
The effects of bioelectrochemical systems (BESs) for the suppression of methane gas emissions from sediment were examined using a laboratory-scale reactor system. Methane gas emissions from acetate were suppressed by approximately 36% from control based on the installation of a BES in which carbon-graphite electrodes were buried in sediment and arbitrarily set at certain oxidative potentials (+300 mV vs Ag/AgCl) using a potentiostat. Microbial community analysis on the surface of the buried electrodes showed that the genus Geobacter had drastically propagated in a sample from the reactor where the electrodes were buried. Archaeal population had decreased to approximately 1/6 of its original level on the electrode of the BES set at +300 mV. Analysis of electron flux in the experiment revealed that electrons intrinsically used for methanogenesis were recovered via current generation in the sediment where a potential of +300 mV was set for the electrode, although most electrons donated from acetate were captured by oxygen respiration and other electron-accepting reactions. The BES technology can be used as a tool for controlling redox-dependent reactions in natural environments.
The authors thank Ms. Ikuko Sakamoto for her analytical support.
- Rabaey, K. and Rozendal, R.A. (2010) Microbial Electrosynthesis: Revisiting the Electrical Route for Bioproduction. Nature Reviews Microbiology, 8, 706-716. http://dx.doi.org/10.1038/nrmicro2422
- De Schamphelaire, L., Rabaey, K., Boeckx, P., Boon, N. and Verstraete, W. (2008) Outlook for Benefits of Sediment Microbial Fuel Cells with Two Bio-Electrodes. Microbial Biotechnology, 1, 446-462. http://dx.doi.org/10.1111/j.1751-7915.2008.00042.x
- IPCC (2007) Climatechange. Synthesis Report. http://www.ipcc.ch/pdf/assessment-report/ar4/syr/ar4_syr_spm.pdf
- Le Mer, J. and Roger, P. (2001) Production, Oxidation, Emission and Consumption of Methane by Soils: A Review. European Journal of Soil Biology, 37, 25-50. http://dx.doi.org/10.1016/S1164-5563(01)01067-6
- Lovley, D.R. (2006) Bug Juice: Harvesting Electricity with Microorganisms. Nature Reviews, 4, 497-508.
- Devai, I. and Delaune, R.D. (1995) Evidence for Phosphine Production and Emission from Louisiana and Florida Marsh Soils. Organic Geochemistry, 23, 277-279. http://dx.doi.org/10.1016/0146-6380(95)00021-6
- Singh, S.N. (2001) Exploring Correlation between Redox Potential and Other Edaphic Factors in Field and Laboratory Conditions in Relation to Methane Efflux. Environment International, 4, 265-274. http://dx.doi.org/10.1016/S0160-4120(01)00055-1
- Stumm, W. and Morgan, J.J. (1996) Aquatic Chemistry, Chemical Equilibria and Rates in Natural Waters. 3rd Edition, John Wiley & Sons, Inc., New York.
- Ishii, S., Hotta, Y. and Watanabe, K. (2008) Methanogenesis versus Electrogenesis: Morphological and Phylogenetic Comparisons of Microbial Communities. Bioscience, Biotechnology, and Biochemistry, 72, 286-294. http://dx.doi.org/10.1271/bbb.70179
- Ueno, Y. and Kitajima, Y. (2012) Suppression of Greenhouse Gas Emissions from Sediment by Bio Electrochemical System. Environmental Engineering and Management Journal, 11, 1833-1837.
- Tatara, M., Yamazawa, A., Ueno, Y., Fukui, H., Goto, M. and Sode, K. (2005) High-Rate Thermophilic Methane Fermentation on Short-Chain Fatty Acids in a Down-Flow Anaerobic Packed-Bed Reactor. Bioprocess and Biosystems Engineering, 27, 105-113. http://dx.doi.org/10.1007/s00449-004-0387-8
- Logan, B.E., Hamelers, B., Rozendal, R., Schröder, U., Keller, J. and Freguia, S. (2006) Microbial Fuel Cells: Methodology and Technology. Environmental Science & Technology, 40, 5181-5192. http://dx.doi.org/10.1021/es0605016
- Caporaso, J.G., Lauber, C.L., Walters, W.A., Berg-Lyons, D. and Lozupone, C.A. (2011) Global Patterns of 16S rRNA Diversity at a Depth of Millions of Sequences Per Sample. Proceedings of the National Academy of Sciences of the United States of America, 108, 4516-4522. http://dx.doi.org/10.1073/pnas.1000080107
- Dowd, S.E., Callaway, T.R., Wolcott, R.D., Sun, Y. and McKeehan, T. (2008) Evaluation of the Bacterial Diversity in the Feces of Cattle Using 16S rDNA Bacterial Tag-Encoded FLX Ampliconpyro sequencing (bTEFAP). BMC Microbiology, 8, 125. http://dx.doi.org/10.1186/1471-2180-8-125
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J. and Zhang, Z. (1997) Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Research, 25, 3389-3402. http://dx.doi.org/10.1186/1471-2180-8-125
- Wang, Q., Garrity, G.M., Tiedje, J.M. and Cole, J.R. (2007) Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Applied and Environmental Microbiology, 73, 5261-5267. http://dx.doi.org/10.1128/AEM.00062-07
- Sun, Y., Cai, Y., Liu, L., Yu, F. and Farrell, M.L (2009) ESPRIT: Estimating Species Richness Using Large Collections of 16S rRNA Pyrosequences. Nucleic Acids Research, 37, e76. http://dx.doi.org/10.1093/nar/gkp285
- Larkin, M.A., Blackshields, G., Brown, N.P., Chenna, R., McGettigan, P.A., McWilliam, H., Valentin, F., Wallace, I.M., Wilm, A., Lopez, R., Thompson, J.D., Gibson, T.J. and Higgins, D.G. (2007) Clustal W and Clustal X Version 2.0. Bioinformatics, 23, 2947-2948. http://dx.doi.org/10.1093/bioinformatics/btm404
- Saitou, N. and Nei, M. (1987) The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Molecular Biology and Evolution, 4, 406-425.
- Holmes, D.E., Finneran, K.T., O’Neil, R.A. and Lovley, D.R. (2002) Enrichment of Members of the Family Geobacteraceae Associated with Stimulation of Dissimilatory Metal Reduction in Uranium-Contaminated Aquifer Sediments. Applied and Environmental Microbiology, 68, 2300-2306. http://dx.doi.org/10.1128/AEM.68.5.2300-2306.2002
- Lueders, T. and Friedrich, M.W. (2003) Evaluation of PCR Amplification Bias by Terminal Restriction Fragment Length Polymorphism Analysis of Small-Subunit rRNA and mcrA Genes by Using Defined Template Mixtures of Methanogenic Pure Cultures and Soil DNA Extracts. Applied and Environmental Microbiology, 69, 320-326. http://dx.doi.org/10.1128/AEM.69.1.320-326.2003
- Yamada, T., Imachi, H., Ohashi, A., Hrada, H., Harada, S., Kamagata, Y. and Sekiguchi, Y., (2007) Bellilinea caldifistulaegen. nov., sp. nov. and Longilinea arvoryzae gen. nov., sp. nov., Strictly Anaerobic, Filamentous Bacteria of the Phylum Chloroflexi Isolated from Methanogenic Propionate Degrading Consortia. International Journal of Systematic and Evolutionary Microbiology, 57, 2299-2306. http://dx.doi.org/10.1099/ijs.0.65098-0
- Davey, M.E., Wood, W.A., Key, R., Nakamura, K. and Stahl, D. (1993) Isolation of Three Species of Geotoga and Petrotoga: Two New Genera, Representing a New Lineage in the Bacterial Line of Descent Distantly Related to the “Thermotogales”. Systematic and Applied Microbiology, 16, 191-200. http://dx.doi.org/10.1016/S0723-2020(11)80467-4
- Bond, D.R. and Lovley, D.R. (2003) Electricity Production by Geobacter sulfurreducens Attached to Electrodes. Applied and Environmental Microbiology, 69, 1548-1555. http://dx.doi.org/10.1128/AEM.69.3.1548-1555.2003
- Yates, M.D., Kiely, P.D., Call, D.F., Rismani-Yazdi, H. and Bibby, K. (2012) Convergent Development of Anodic Bacterial Communities in Microbial Fuel Cells. The ISME Journal, 6, 2002-2013. http://dx.doi.org/10.1038/ismej.2012.42
- Lee, J., Phung, N.T., Chang, I.S., Kim, B.H. and Sung, H.C. (2003) Use of Acetate for Enrichment of Electrochemically Active Microorganisms and their 16S rDNA Analyses. FEMS Microbiology Letters, 223, 185-191. http://dx.doi.org/10.1016/S0378-1097(03)00356-2
- Kato, S., Kai, F., Nakamura, R., Watanabe, K. and Hashimoto, K. (2010) Respiratory Interactions of Soil Bacteria with (Semi) Conductive Iron-Oxide Minerals. Environmental Microbiology, 12, 3114-3123. http://dx.doi.org/10.1111/j.1462-2920.2010.02284.x
- Sasaki, D., Sasaki, K., Watanabe, A., Morita, M., Igarashi, Y. and Ohmura, N. (2013) Efficient Production of Methane from Artificial Garbage Waste by a Cylindrical Bioelectrochemical Reactor Containing Carbon Fiber Textiles. AMB Express, 3, 17. http://dx.doi.org/10.1186/2191-0855-3-17
- Cheng, S., Xing, D., Call, D.F. and Logan, B.E. (2009) Direct Biological Conversion of Electrical Current into Methane by Electromethanogenesis. Environmental Science & Technology, 43, 3953-3958. http://dx.doi.org/10.1021/es803531g
- Shimoyama, T., Komukai, S., Yamazawa, A., Ueno, Y., Logan, B.E. and Watanabe, K. (2008) Electricity Generation from Model Organic Wastewater in a Cassette-Electrode Microbial Fuel Cell. Applied and Environmental Microbiology, 80, 325-330. http://dx.doi.org/10.1007/s00253-008-1516-0
- Ryckelynck, N., Stecher, H. and Reimers, C.E. (2005) Understanding the Anodic Mechanism of a Seafloor Fuel Cell: Interactions between Geochemistry and Microbial Activity. Biogeochemistry, 76, 113-139. http://dx.doi.org/10.1007/s10533-005-2671-3
- Conrad, R. (2002) Control of Microbial Methane Production in Wetland Rice Fields. Nutrient Cycling in Agroecosystems, 64, 59-69. http://dx.doi.org/10.1023/A:1021178713988
- Rosenbaum, M. (2005) In Situ Electrooxidation of Photobiological Hydrogen in a Photobioelectrochemical Fuel Cell Based on Rhodobacter sphaeroides. Environmental Science & Technology, 39, 6328-6333. http://dx.doi.org/10.1021/es0505447.
Table S1 . Genus-level taxonomic groups detected in the pyrotag-sequencing analysis.

*Corresponding author.


