Advances in Microbiology
Vol.3 No.6(2013), Article ID:38163,6 pages DOI:10.4236/aim.2013.36066
Incidence of Methicillin Resistant Staphylococcus aureus in Burn Patients Admitted to Burn Unit, Dhaka Medical College Hospital, Bangladesh
1Department of Microbiology and Biotechnology, Jagannath University, Dhaka, Bangladesh
2Department of Microbiology, Jahangirnagar University, Savar, Dhaka, Bangladesh
3Department of Microbiology, University of Dhaka, Dhaka, Bangladesh
Email: *sabita_rahman@hotmail.com
Copyright © 2013 Md. Shariful Islam et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received August 23, 2013; revised September 12, 2013; accepted September 20, 2013
Keywords: Methicillin Resistant Staphylococcus aureus (MRSA); Multidrug Resistant (MDR); Minimum Inhibitory Concentration (MIC)
ABSTRACT
Staphylococcus aureus infection remains an increasing problem for higher morbidity and mortality in burn patients. We sought to determine the frequency of methicillin-resistant S. aureus (MRSA) in burn wound patients and study their drug resistance genes. Samples were collected (August 2010 to October 2011) from burn unit of Dhaka Medical College Hospital (DMCH), Bangladesh. MRSA was identified by conventional culture based methods. S. aureus was confirmed in 44.44% burn wound samples and 22.5% of the isolates were oxacillin resistant. All the S. aureus isolates were resistant to commonly used drugs like amoxicillin, azactam, erythromycin, azithromycin etc. and were sensitive to imipenem. The isolates were devoid of plasmid and the gene mecA, femA and IS431 were detected in their chromosomal DNA. Computational analysis of mecA gene sequence showed homology to S. aureus “penicillin binding protein 2a” (PBP-2a). The higher association of MRSA is in burn unit of DMCH, Bangladesh is alarming and with inappropriate antibiotic use, and the situation gets even complicated to treat. Therefore, the detection system and control practices for MRSA in DMCH should be improved in the hospital settings.
1. Introduction
Burn related diseases were among the top ten diseases according to the number of patients admitted in the Dhaka Medical College Hospital during 2011 in Bangladesh [1]. Burn patients become susceptible to increased rate of infection due to the loss of protective skin barrier and decreased cellular and humoral immunity [2,3]. More than 70% deaths of burn patients occur due to infection with various microorganisms [4,5]. Though the infection may be initiated by the skin flora, after entry of burn patients in a hospital nosocomial infection may initiate.
Methicillin-resistant Staphylococcus aureus (MRSA) is also called oxacillin resistant S. aureus (ORSA) which has long been associated as a major agent of nosocomial infection and is a prominent reason for higher morbidity and mortality in burn patients causing a variety of infections such as bacteremia, pneumonia, septic arthritis, endocarditis, and surgical site infections [6-8]. The spread of MRSA has an enormous impact on patients as well as in the country because it dramatically increases the healthcare expenditure [9].
Higher virulence of MRSA compared to methicillinsusceptible S. aureus (MSSA) is a controversial issue [10,11]. In 2004, Centers for Disease Control and prevention (CDC) reported that MRSA was responsible for more than 50% of all health care associated S. aureus infections in the United States [12]. Data from the sentry antimicrobial surveillance program showed increasing rates of MRSA among S. aureus isolated from intensive care unit patients worldwide [13]. Phenotypic expression of antimicrobial resistance to methicillin, oxacillin and other drugs may vary depending on growth conditions, therefore may provide improper result [14]. The genome of MRSA bears a 21 to 67 kb staphylococcal cassette chromosome mec (SCCmec) containing the mecA gene
(confers methicillin resistance by encoding PBP-2a) and other resistance determinants [15,16]. IS431, a staphylococcal genetic element can transfer gene(s) for antimicrobial resistance [17] and femA is unique to S. aureus [16]. Therefore, detection of the femA, mecA and IS431 genes in the same organism can entail its species and drug resistance phenotype.
Burn unit in Dhaka Medical College Hospital is the sole burn management centre in Bangladesh but ironically the hospital hardly performs any test for the determination of MRSA in burn patients. This study was undertaken to investigate the frequency of MRSA in burn wound patients and investigate the pattern of antimicrobial susceptibility together with molecular basis of methicillin/oxacillin resistance among the isolates.
2. Methods
2.1. Sample Collection
Samples were collected during August, 2010 to October, 2011 from burn wound patients admitted to the burn unit of Dhaka Medical College Hospital (DMCH), Dhaka- 1000, Bangladesh.
2.2. Isolation and Identification
Sterile cotton-swab sticks were used for sampling and were immediately transported to the laboratory in transport media. The swabs were streaked on the blood agar and MacConkey agar and Mannitol salt agar (MSA) plates (Fluka, USA). Blood agar and MacConkey agar plates were incubated at 37˚C and MSA plate was incubated at 33˚C - 35˚C for 24 hour. S. aureus and MRSA was confirmed by different biochemical tests.
2.3. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was performed in vitro by the disc diffusion assay, which is a modification of Kirby-Baur method. Commercially available antimicrobial discs (Oxoid Ltd. Hampshire, UK) of Amoxicillin (30 μg/ml), Cloxacillin (1 μg/ml), Oxacillin (1 μg/ml), Erythromycin (10 μg/ml), Azithromycin (30 μg/ml), Nalidixic acid (30 μg/ml), Ciprofloxacin (10 μg/ml), Tetracycline (30 μg/ml), Levofloxacillin (5 μg/ml), Cefixime (5 μg/ml), Gentamicin (10 μg/ml), Cefepime (30 μg/ml), Azactam (30 μg/ml) Doxycycline (30 μg/ml) and Imipenem (10 μg/ml) were used for this test. E. coli ATCC 25922 was used as control strain for susceptibility testing. Oxacillin resistant strains were later tested for Cefoxitin (30 μg/ml) susceptibility.
2.4. Determination of Minimum Inhibitory Concentration (MIC)
MIC is the lowest concentration of drug which inhibits visible growth of bacteria. MIC of the selected isolates was calculated by the standard broth microdilution method [18]. Cell density of S. aureus was set to 5 × 105 CFU/ml and was added as inoculum to 2% Muller Hinton broth [19] for MIC determination. The strains were referred to as Resistant (R) and Susceptible (S) in relation to the MIC of oxacillin, erythromycin and azithromycin tested as shown on the Table 1.
2.5. Plasmid Profiling
Plasmid DNA was extracted according to the simplified alkaline lysis method described by Kado and Liu (1981) [21]. Extracted plasmid DNA was separated by horizontal electrophoresis in 0.8% agarose gels and stained with ethidium bromide prior visualization. The molecular weight of unknown plasmid was determined by comparing their position on the stained gel with that of known molecular weight plasmid markers (500 bp - 10 kb).
2.6. DNA Extraction
Staphylococci were grown on blood agar. Bacterial inoculum was suspended in 100 μL of lysis solution containing 0.05 mg of lysostaphin (Sigma, Germany), 2 mg of lysozyme (Sigma), 2 mg of ribonuclease A (Sigma), 2 μL of 20 mM Tris-HCl, pH 8.0, 2 μL of 2 mM EDTA and 1 μL of Triton X-100. The suspension was incubated at 37˚C for 45 min on a shaker at 300 rpm. Then 10 μL of proteinase K solution and 100 μL of buffer AL (Qiagen, Germany) were added. After incubation at 56˚C for 45 min, the sample was processed for next step.
2.7. PCR Amplification and Agarose Gel Electrophoresis
The primers and PCR condition were set according to Pascal vannuffel et al. [22]. The methicillin resistance
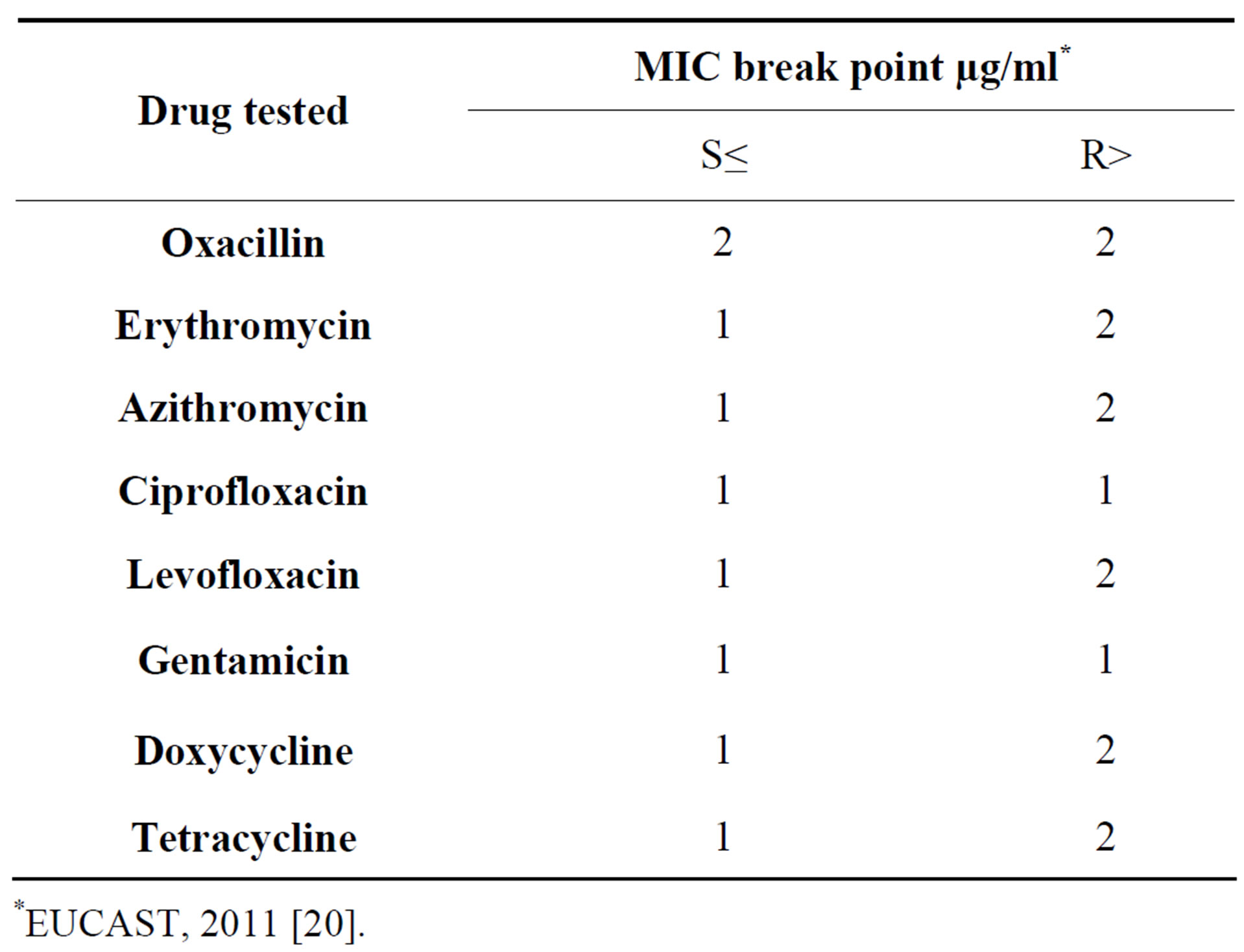
Table 1. Breakpoint for interpretation of MICs for test drugs against MRSA.
*EUCAST, 2011 [20].
determinant gene mecA specific primers (F: 5’-TGGCTATCGTGTCACAATCG-3’, R: 5’-CTGGAACTTGTTGAGCAGAG-3’), S. aureus species identification gene femA specific primers (F: 5’-CTTACTTACTGGCTGTACCTG-3’, R: 5’-ATGTCGCTTGTTATGTGC- 3’) and internal negative control gene IS431 specific primers (F: 5’-AGGATGTTATCACTGTAGCC-3’, R: 5’-GATGTACAATGACAGTCAGG-3’) were used for PCR assay. For mecA and IS431 multiplex PCR was performed. After amplification of the target genes, 15 µl of the PCR sample was loaded on a 2% (wt/vol) agarose gel for horizontal transfer. Electrophoresis was performed in 0.1 M Tris HCl (pH 8.6)-80 mM boric acid-1 mM EDTA containing 0.5 mg of ethidium bromide/ml. DNA fragments were visualized at 300 nm on a UV transilluminator.
2.8. Sequencing and Comparison
The PCR products of mecA gene was purified using PCR product purification kit (QIA Quick50, Qiagen, Germany) and the purified products were sequenced in forward direction by DNA sequencer (Macrogen DNA sequencer, Korea). Sequence obtained were analyzed using Chromas 2.33 software, Bio-edit 7.1.3 for comparing the sequences with GenBank, EMBL and DDBJ databases using BLAST and sequence alignment was carried out using Clustal 1.8.
3. Results
Out of 180 samples collected from burn wound infected hospitalized patients, 80 were S. aureus positive. Antibiotic susceptibility pattern revealed that 22.5% of the isolates were resistant to oxacillin (2.5% isolates were border line resistant to oxacillin), 90% to amoxicillin, 82.5% to azactam, 57.5% to erythromycin, 55% to azithromycine, 55% to cefixime, 52.5% to ciprofloxacin, 40% to levofloxacin, 30% to tetracycline, 15% to nalidixic acid, 12.5% to cefepime and 7.5% to gentamycin. However, all the isolates (n = 80) were truly (100%) sensitive to imipenem and to a greater extent to cloxacillin (97.5%), gentamycin (92.5%), and nalidixic acid (85%). Figure 1 shows pattern of drug sensitivity of the selected isolates of this study. All the oxacillin resistant isolates were resistant to Cefoxitin as well.
About 70% of the S. aureus isolates (n = 80) were resistant to more than two antibiotics. However, isolates with oxacillin resistance and borderline resistance (n = 20) exhibited non-susceptibility to higher number of drugs. All the oxacillin resistant S. aureus (ORSA) were completely resistant to β-lactam and macrolides groups, 10% of the isolates were resistant to aminoglycosides, and 65% were resistant to fluoroquinolone. Drug resistance phenotype among the S. aureus isolates as shown
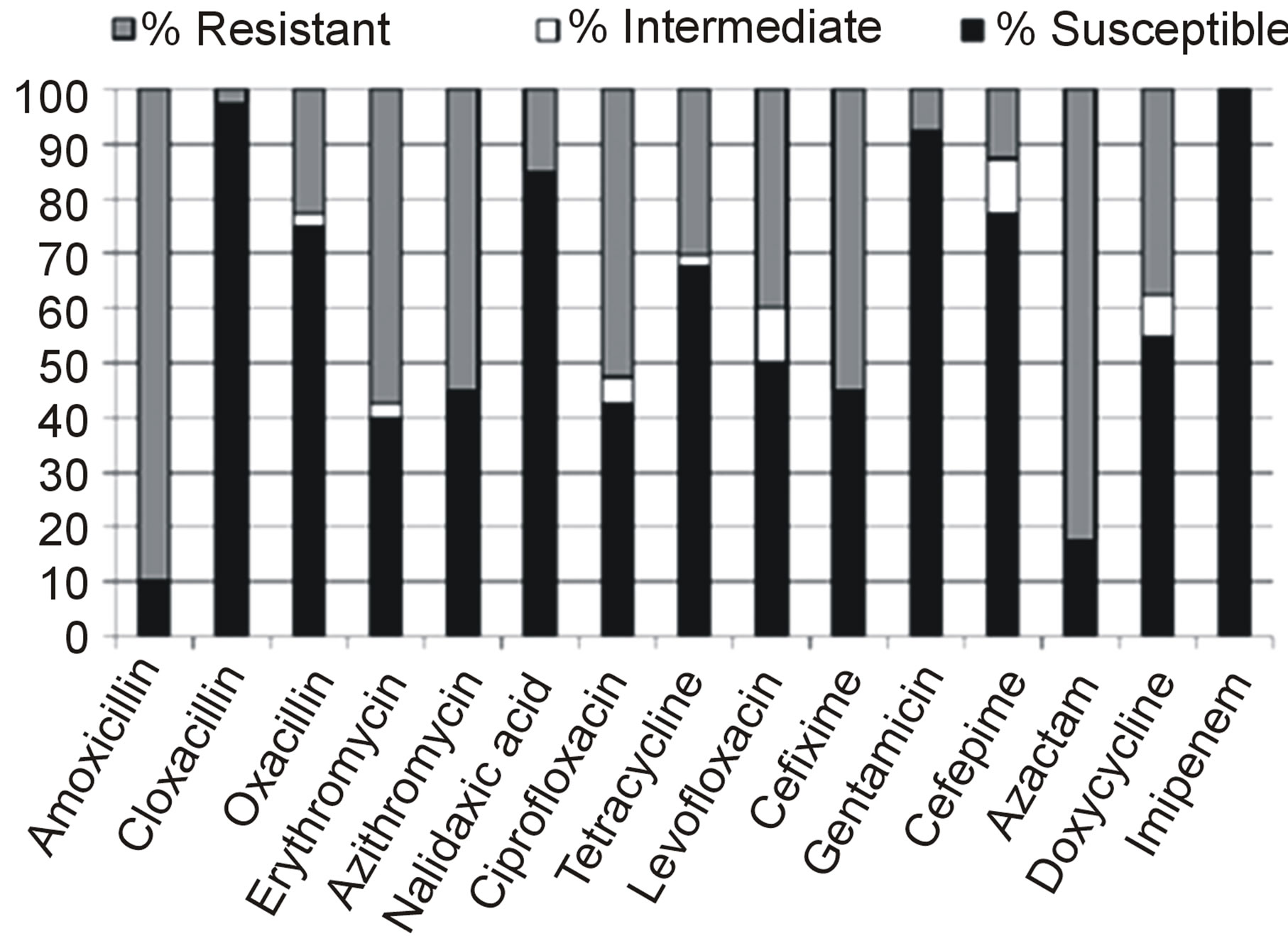
Figure 1. Antimicrobial susceptibility patterns of S. aureus isolated from burn wound infected patients in Dhaka Medical College Hospital, Bangladesh.
in Table 2 revealed that some of the isolates were resistant to 11 of the 15 test drugs.
MRSA/ORSA isolates (n = 20) revealed higher MIC values for oxacillin, erythromycin and azithromycin when compared to the standard table as shown in Table 1. For oxacillin, erythromycin, and azithromycin antibiotics, two, four, and two isolates respectively showed MIC value of 32 µg/ml concentration.
Plasmid DNA was not present in the MRSA isolates of this experiment (figure not shown). Since, Plasmid profile of the MRSA isolates separated on 0.8% agarose gel revealed band for only chromosomal DNA.
The PCR amplification product of femA gene revealed 686 bp amplicon from all of the S. aureus isolates tested and positive control (S. aureus ATCC 6538) where as the product was absent in negative control (Salmonella typhimurium ATCC 13311) as shown in Figure 2. The presence of femA among the isolates confirms them as S. aureus.
All the oxacillin resistant isolates as shown in the Figure 3 provided PCR amplification products of 310 and 500 bp for mecA and IS431 control fragment respectively while no amplification product was detected for any methicillin susceptible strain (S. aureus ATCC 6538) regardless of the pattern of β-lactam susceptibility and coagulase production. The amplification band of mecA positive strains of this experiment were later compared with that of S. aureus ATCC 49476 strain (positive control for mecA) and found similar pattern of migration on agarose gel.
Sequencing of approximately 145 bp of DNA fragment followed by sequence alignment showed 100% homology of the amplified fragment with the mecA of S. aureus (Accession no. NC 2952.2). Further analysis of the mecA sequence by BLASTX displayed 100% homology with the altered penicillin binding protein (PBP2a) of S. aureus.
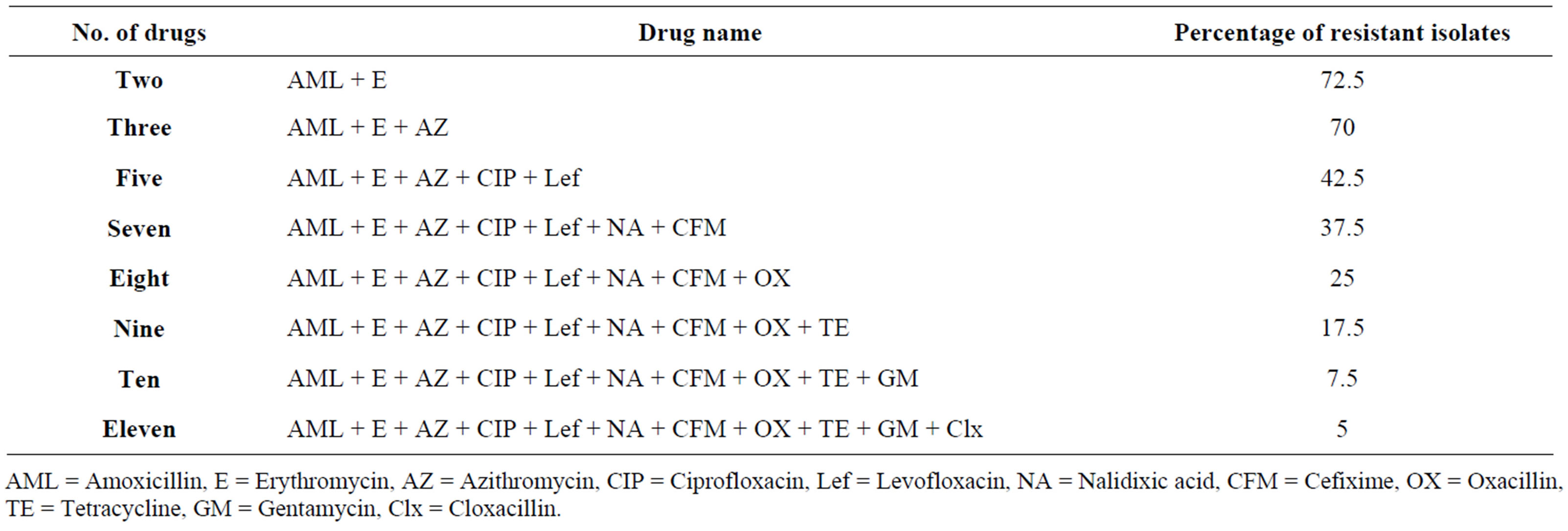
Table 2. Drug resistance phenotype of the S. aureus isolates (n = 80) from burn wound infected patients admitted in the burn unit of Dhaka Medical College Hospital, Dhaka, Bangladesh.
AML = Amoxicillin, E = Erythromycin, AZ = Azithromycin, CIP = Ciprofloxacin, Lef = Levofloxacin, NA = Nalidixic acid, CFM = Cefixime, OX = Oxacillin, TE = Tetracycline, GM = Gentamycin, Clx = Cloxacillin.
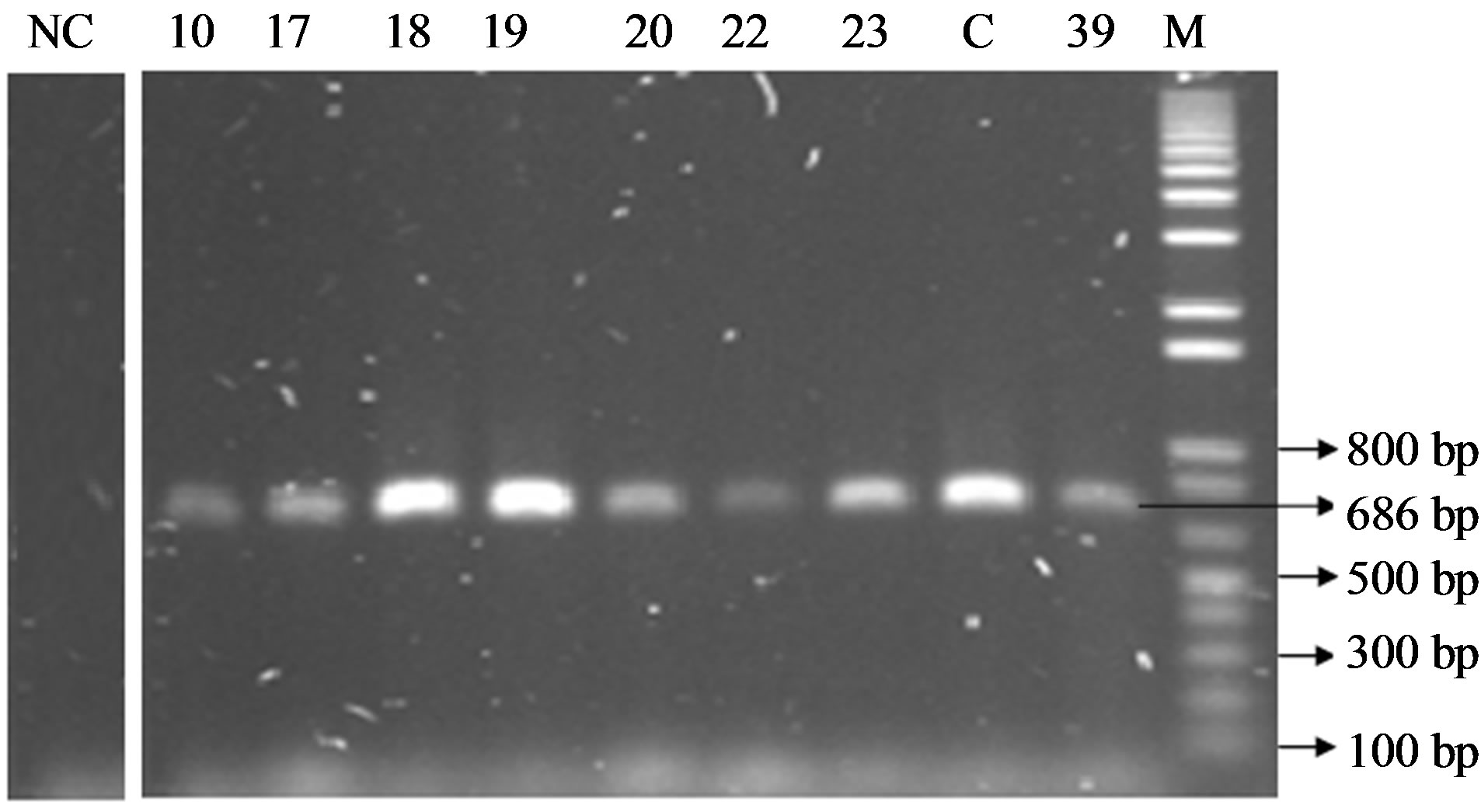
Figure 2. PCR products on 2% agarose gel after amplification of femA gene (686 bp). Lane M indicate marker, lane C indicate positive control, NC indicate negative control and lane 10, 17, 18, 19, 20, 22, 23, 39 indicates sample ID. The lengths of amplified fragments and molecular size markers (in base pairs) are indicated on the right and left, respectively.
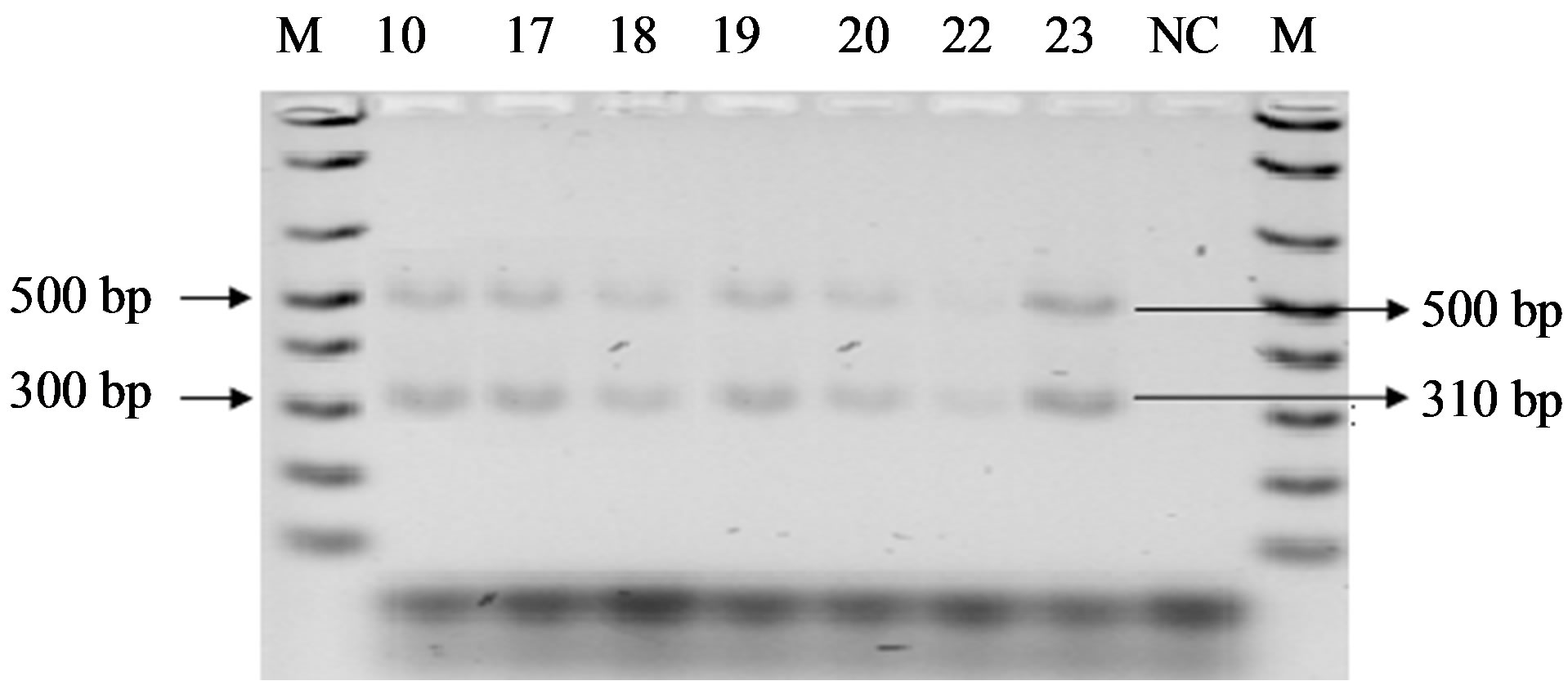
Figure 3. PCR products for the detection of mecA and IS431 on 2% agarose gel. Lane M indicate marker, lane NC indicate negative control and lane 10, 17, 18, 19, 20, 22, 23 indicates sample ID. The lengths of amplified fragments and molecular size markers (in base pairs) are indicated on the right and left, respectively.
4. Discussion
Methicillin resistant S. aureus is a wide spread pathogen both in the community and in hospital settings. Infection of burn wound patients with MRSA and pathogen causes higher morbidity and mortality which may contribute death in about three fourths of the burn wound infected patients [23]. Burn wound infections in Bangladesh become of more concern due to lower socioeconomic status and tropical weather that intensify the occurrence and spread of various pathogens. Due to imprudent uses and suboptimal doses of antibiotics, common pathogenic microorganism develops antimicrobial resistance against commonly used drugs. It increases the burden of patient as well as national cost in the health care sectors. In Bangladesh frequency of MRSA at different hospitals and cities may vary form 32% - 63% [24]. In this study, the isolation rate of MRSA among the clinical specimens was 25% which was in agreement with the findings of Sadia Afroz et al. (2008) as they reported 28% frequency of MRSA in hospitalized burn patients. Methicillin resistant S. aureus can spread from one patient to another patient during their nursing, and the hospital personnel may promote the transmission [25].
A large proportion of Bangladeshi people live below the poverty line and they are unable to complete the treatment course, where more expensive third-line antibiotics may not be available or affordable. On the other hand due to arbitrary use of antibiotics, common pathogen develops resistance against frequently used drugs. Negligence of infection worsens the condition and makes treatment complicated to unaffordable. In most cases, patients initially take antibiotic without consulting an expert. As a result microorganisms get suboptimal pressure at which they are not killed conversely, they acquire their ability to withstand the antibiotic pressure and emerge as antibiotic resistant strain. This study reports higher rate of resistance to amoxicillin and azactam in the MRSA isolates. Resistance to erythromycin, azithromycin and cefixime was noted in more than 50% of the isolates. However the isolates were sensitive to imipenem, cloxacillin and gentamycin. Methicillin resistance in S. aureus is a gold standard for multidrug resistance, mainly to penicillin family (such as cephalosporins, ampicillin, amoxicillin), ticarcillin-clavulanic acid, piperacillin-tazobactam, and the carbapenems regardless of the susceptibility testing results [25,26] and this study also supports the same. The drug resistance pattern of MRSA isolates of this study coincides with the findings of Chambers et al. (2009) [27]. Such higher degree of drug resistance complicates the treatment process [27]. Plasmid DNA was not found in MRSA isolates of this study, however, evidence for both plasmid free and plasmid borne methicillin resistance among S. aureus has been reported by Gelmi et al. (1987) [28] and Coia et al. (1988) [29]. PCR is a rapid and sensitive technique for detecting various genes in genetic materials over conventional sucseptibility testing methods. Presence of mecA and femA in the methicillin resistant isolates of this study confirms the isolates to be MRSA. Further sequencing of mecA product showed 100% homology with the “penicillin binding protein 2a” (PBP2a) sequence of S. aureus (Accession no NC 002952.2). PBP2a binds to altered penicillin binding site therefore methicillin fails to inhibit synthesis of bacterial cell wall and the cell becomes resistant to drug.
In Bangladesh, more than 250,000 people get injured due to burn and of them more than 3000 die each year [30]. Since burn unit in Dhaka Medical College Hospital is the only burn management centre in Bangladesh, therefore, the frequency of MRSA occurrence in burn wound and its antimicrobial profile obtained in this study are alarming. Burn patients may acquire the pathogen from hospitals or visitors, however, the source of infection was not documented in this study. Proper arrangement and synchronized endeavor by the concerned person in controlling MRSA distribution in the burn unit of DMCH are necessary to manage the disease successfully. Regular monitoring of the drug resistance profile of the pathogen is necessary and rapid diagnostic method for MRSA detection in the burn unit of DMCH is urgently needed in this regard. This will help the physicians in determining appropriate control strategy for the pathogen in question.
5. Acknowledgements
The authors thank the Ministry of Education, Govt. of Bangladesh for funding this research.
REFERENCES
- Health Bulletin, “Ministry of Health and Family Welfare (MOHFW),” Dhaka Medical College Hospital, 2012, P. 7. [http://app.dghs.gov.bd/lhb/reportbyadmin.php?yid=2012&lvl=3&om=dmch@hospi.dghs.gov.bd&oti=&organame=Dhaka%20Medical%20College%20Hospital]
- R. Kaushik, S. Kumar, R. Sharma and P. Lal, “Bacteriology of Burn Wounds—The First Three Years in a New Burn Unit at the Medical College Chandigarh,” Burns, Vol. 27, No. 6, 2001, pp. 595-597. http://dx.doi.org/10.1016/S0305-4179(01)00023-7
- T. H. Wong, B. H. Tan, M. L. Ling and C. Song, “Multi-Resistant Acinetobacter baumannii on a Burns Unit—Clinical Risk Factors and Prognosis,” Burns, Vol. 28, No. 4, 2002, pp. 349-357. http://dx.doi.org/10.1016/S0305-4179(02)00012-8
- S. Nasser, A. Mabrouk and A. Maher, “Colonization of Burn Wounds in Ain Shams University Burn Unit,” Burns, Vol. 29, No. 3, 2003, pp. 229-233. http://dx.doi.org/10.1016/S0305-4179(02)00285-1
- N. P. Singh, R. Goyal, V. Manchanda, S. Das, I. Kaur and V. Talwar, “Changing Trends in Bacteriology of Burns in the Burns Unit, Delhi, India,” Burns, Vol. 29, No. 2, 2003, pp. 129-132. http://dx.doi.org/10.1016/S0305-4179(02)00249-8
- M. C. Enright, D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann and B. G. Spratt, “The Evolutionary History of Methicillin-Resistant Staphylococcus aureus (MRSA),” Proceedings of the National Academy of Sciences of the United States of America, Vol. 99, No. 11, 2002, pp. 7687-7692. http://dx.doi.org/10.1073/pnas.122108599
- F. D. Lowy, “Staphylococcus aureus Infections,” The New England Journal of Medicine, Vol. 339, No. 8, 1998, pp. 520-532. http://dx.doi.org/10.1056/NEJM199808203390806
- G. J. Moran, A. Krishnadasan, R. J. Gorwitz, G. E. Fosheim, L. K. McDougal, R. B. Carey and D. A. Talan, “Methicillin-Resistant S. aureus Infections among Patients in the Emergency Department,” The New England Journal of Medicine, Vol. 355, No. 7, 2006, pp. 666-674. http://dx.doi.org/10.1056/NEJMoa055356
- B. Capitano, O. A. Leshem, C. H. Nightingale and D. P. Nicolau, “Cost Effect of Managing Methicillin-Resistant Staphylococcus aureus in a Long-Term Care Facility,” Journal of the American Geriatrics Society, Vol. 51, No. 1, 2003, pp. 10-16. http://dx.doi.org/10.1034/j.1601-5215.2002.51003.x
- L. A. Selvey, M. Whitby and B. Johnson, “Nosocomial Methicillin-Resistant Staphylococcus aureus bacteremia: Is It Any Worse than Nosocomial Methicillin-Sensitive Staphylococcus aureus Bacteremia?” Infection Control and Hospital Epidemiology, Vol. 21, No. 10, 2000, pp. 645-648. http://dx.doi.org/10.1086/501707
- S. I. Blot, K. H. Vandewoude, E. A. Hoste and F. A. Colardyn, “Outcome and Attributable Mortality in Critically Ill Patients with Bacteremia Involving Methicillin-Susceptible and Methicillin-Resistant Staphylococcus aureus,” Archives of Internal Medicine, Vol. 162, No. 19, 2002, pp. 2229-2235. http://dx.doi.org/10.1001/archinte.162.19.2229
- National Nosocomial Infections Surveillance (NNIS) “System Report, Data Summary from January 1992 through June 2004, Issued October 2004,” American Journal of Infection Control, Vol. 32, No. 8, 2004, pp. 470-485. http://dx.doi.org/10.1016/j.ajic.2004.10.001
- K. B. Anand, P. Agrawal, S. Kumar and K. Kapila, “Comparison of Cefoxitin Disc Diffusion Test, Oxacillin Screen Agar, and PCR for mecA Gene for Detection of MRSA,” Indian Journal of Medical Microbiology, Vol. 27, No. 1, 2009, pp. 27-29.
- H. F. Chambers. “Methicillin Resistance in Staphylococci: Molecular and Biochemical Basis and Clinical Implications,” Clinical Microbiology Reviews, Vol. 10, No. 4, 1997, pp. 781-791.
- S. Boyle-Vavra and R. S. Daum, “Community-Acquired Methicillin Resistant Staphylococcus aureus: The Role of Panton-Valentine Eukocidi-n,” Laboratory Investigation, Vol. 87, No. 1, 2007, pp. 3-9. http://dx.doi.org/10.1038/labinvest.3700501
- S. Ünal, J. Hoskins, J. E. Flokowitsch, C. Y. E. Wu, D. A. Preston and P. L. Skatrud, “Detection of Methicillin-Resistant Staphylococci by Using the Polymerase Chain Reaction,” Journal of Clinical Microbiology, Vol. 30, No. 7, 1992, pp. 1685-1691.
- L. Barberis-Maino, B. Berger-Bachi, H. Weber, W. D. Beck and F. H. Kayser, “IS431, a Staphylococcal Insertion Sequence-Like Element Related to IS26 from Proteus Vulgaris,” Gene, Vol. 59, No. 1, 1987, pp. 107-113. http://dx.doi.org/10.1016/0378-1119(87)90271-X
- L. K. McDougal and C. Thornsberry, “The Role of Beta-Lactamase in Staphylococcal Resistance to Penicillinase-Resistant Penicillins and Cephalosporins,” Journal of Clinical Microbiology, Vol. 23, No. 5, 1986, pp. 832- 839.
- C. Thornsberry and L. K. McDougal, “Successful Use of Broth Microdilution in Susceptibility Tests for Methicillin-Resistant (Heteroresistant) Staphylococci,” Journal of Clinical Microbiology, Vol. 18, No. 5, 1983, pp. 1084- 1091.
- European Committee on Antimicrobial Susceptibility Testing, “Clinical Breakpoint Table v. 1.3,” 2011.
- C. I. Kado and S. T. Liu, “Rapid Procedure for Detection and Isolation of Large and Small Plasmids,” Journal of Bacteriology, Vol. 145, No. 3, 1981, pp. 1365-1373.
- P. Vannuffel, J. Gigi, H. Ezzedine, B. Vandercam, M. Delmee, G. Wauters and J. L. Gala, “Specific Detection of Methicillin-Resistant Staphylococcus Species by Multiplex PCR,” Journal of Clinical Microbiology, Vol. 33, No. 11, 1985, pp. 2864-2867.
- F. C. Tenover, “Mechanisms of Anti Microbial Resistance in Bacteria,” The American Journal of Medicine, Vol. 119, No. 6, 2006, pp. S3-S10. http://dx.doi.org/10.1016/j.amjmed.2006.03.011
- S. Afroz, N. Kobayashi, S. Nagashima, M. M. Alam, A. B. Hossain, M. A. Rahman, M. R. Islam, A. B. Lutfor, N. Muazzam, M. A. Khan, S. K. Paul, A. K. Shamsuzzaman, M. C. Mahmud, A. K. Musa and M. A. Hossain, “Genetic Characterization of Staphylococcus aureus Isolates Carrying Panton-Valentine Leukocidin Genes in Bangladesh,” Japanese Journal of Infectious Diseases, Vol. 61, No. 5, 2008, pp. 393-396.
- J. C. Seguin, R. D. Walker, J. P. Caron, W. E. Kloos, C. G. George, R. J. Hollis, R. N. Jones and M. A. Pfaller, “Methicillin-Resistant Staphylococcus aureus Outbreak in a Veterinary Teaching Hospital: Potential Human-toAnimal Transmission,” Journal of Clinical Microbiology, Vol. 37, No. 5, 1999, pp. 1459-1463.
- J. H. Lee, “Methicillin (Oxacillin)-Resistant Staphylococcus aureus Strains Isolated from Major Food Animals and Their Potential Transmission to Humans,” Applied and Environmental Microbiology, Vol. 69, No. 11, 2003, pp. 6489-6494. http://dx.doi.org/10.1128/AEM.69.11.6489-6494.2003
- H. F. Chambers and F. R. DeLeo, “Waves of Resistance: Staphylococcus aureus in the Antibiotic Era,” Nature Reviews Microbiology, Vol. 7, No. 9, 2009, pp. 629-641. http://dx.doi.org/10.1038/nrmicro2200
- M. Gelmi, I. Foresti, G. Ravizzola, C. Bonfanti, R. Verardi, A. Caruso and A. Turano, “Antibiotic Resistances and Plasmids in Staphylococcus aureus from Italian Hospitals,” Journal of Medical Microbiology, Vol. 23, No. 2, 1987, pp. 111-118. http://dx.doi.org/10.1099/00222615-23-2-111
- J. E. Coia, I. Noor-Hussain and D. J. Platt, “Plasmid Profiles and Restriction Enzyme Fragmentation Patterns of Plasmids of Methicillin-Sensitive and Methicillin-Resistant Isolates of Staphylococcus aureus from Hospital and the Community,” Journal of Medical Microbiology, Vol. 27, No. 4, 1988, pp. 271-276. http://dx.doi.org/10.1099/00222615-27-4-271
- S. R. Mashreky, A. Rahman, S. M. Chowdhury, T. F. Khan, L. Svanstrom and F. Rahman, “Non-Fatal Burn Is a Major Cause of Illness: Findings from the Largest Community-Based National Survey in Bangladesh,” Injury Prevention, Vol. 15, No. 6, 2009, pp. 397-402. http://dx.doi.org/10.1136/ip.2009.022343
NOTES
*Corresponding author.

