Advances in Microbiology
Vol. 3 No. 3 (2013) , Article ID: 33773 , 10 pages DOI:10.4236/aim.2013.33037
Bacteria on Meat Abattoir Process Surfaces after Sanitation: Characterisation of Survival Properties of Listeria monocytogenes and the Commensal Bacterial Flora
Nofima, Norwegian Institute of Food, Fishery and Aquaculture, Aas, Norway
Email: *even.heir@nofima.no
Copyright © 2013 Trond Møretrø et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received May 10, 2013; revised June 10, 2013; accepted June 25, 2013
Keywords: Listeria monocytogenes; Meat Bacteria; Desiccation; Biofilm; Disinfection
ABSTRACT
Contamination of food with spoilage bacteria and pathogens from food processing environment remains a challenge for the food industry. Bacteria able to persist in such environments over time must survive several hygienic hurdles. The aim of this study was to identify bacteria surviving practical disinfection and compare their survival abilities with representative isolates of the pathogen Listeria monocytogenes. Bacteria isolated from processing surfaces after cleaning and disinfection in a meat abattoir were identified. Selected isolates of the most frequently isolated bacterial genera along with eight meat associated L. monocytogenes were further characterized with regard to biofilm formation abilities at 12˚C and 20˚C, tolerance to desiccation (stainless steel at 70% RH at 12˚C) and bactericidal effects of recommended in-use-concentrations of four commercial disinfectants on stainless steel surface. The most dominating bacterial genera based on counts on non-selective agar were Aerococcus, Acinetobacter, Pseudomonas, Serratia and Staphylococcus. Isolates of Citrobacter. Enterobacter and Serratia dominated on agar plates selective for Enterobacteriaceae. In general, Gram negative bacteria formed more biofilm than Gram positives, especially at 12˚C with the best biofilm formers being Acinetobacter, Citrobacter and Pseudomonas. Listeria monocytogenes were poor biofilm formers. Gram positives survived better air drying than Gram negatives. Strains of L. monocytogenes were more sensitive to desiccation than the other Gram positives; Aerococcus, Kocuria and Staphylococcus. Two disinfectants containing peracetic acid and a disinfectant containing alkylaminoacetate had limited or no antibacterial effect against bacteria dried on stainless steel. A quaternary ammonium compound-based disinfectant provided >2 log reductions of Aerococcus, Acinetobacter and Listeria. Only 0.5 log reductions were obtained against Staphylococcus and no bactericidal effect against Serratia. In this study the dominating flora in a meat abattoir was isolated and identified. Several of these bacteria were better biofilm formers and more resistant to desiccation and disinfection than L. monocytogenes. The disinfectants tested had limited bactericidal activity against surface associated bacteria.
1. Introduction
The food industry has a strong focus on hygiene in order to produce safe food with high quality. Though cleaning and disinfection are performed daily, few surfaces or equipment are sterile. Bacteria present on surfaces may cross-contaminate the food during processing. In meat production there is a focus on potential faecal pathogens like Salmonella and Escherichia coli, and considerable resources are used to sample the environments for these potential pathogenic bacteria, often with high numbers of negative samples. It has been suggested that survival of E. coli in these environments is not connected to enhance survival properties, but that the bacterium is associated with certain raw materials and niches [1]. For L. monocytogenes, it has been proposed that persistence is connected to survival abilities of the bacterium itself [2,3]. Several studies have been conducted to describe important properties of L. mononcytogenes associated with survival in food production environments [4-8], but much less is known concerning other commensal bacteria. It may be hypothesized that bacteria surviving cleaning and disinfection is likely to have improved abilities to survive and also to form persistent bacterial populations in food production environments. It is therefore of specific interest to identify bacteria isolated after cleaning and disinfection and characterise their surviving properties. There is limited information about the prevalence of the commensal bacterial flora in meat processing environments. In a study where the bacterial composition was studied on conveyor belts from a lamb boning room, Sphingomonas dominated among non-cultivable bacteria while Pseudomonas, Serratia, Alcaligenes and Microbacterium were identified by culture-dependent techniques [9]. In another study Pseudomonas and Staphylococcus dominated on the floor in a ground meat processing facility [10]. A comparison of qualitative determination of bacteria from different types of food production environments revealed that Pseudomonas, Staphylococcus, Acinetobacter, Bacillus, lactic acid bacteria and coryneforms commonly dominated [9- 15]. The role of the commensal bacterial flora on food safety is not completely understood, but it may be involved in food spoilage, and affect pathogenic bacteria present in the food producing environment [16-19]. Strains of L. monocytogenes may also frequently be isolated after sanitation and still remain the most challenging microbial threat to many parts of the food industry, including meat processing industry.
In the present study, survival characteristics of meat associated L. monocytogenes were compared to isolates of the bacterial genera dominating in a meat abattoir. The flora isolates were collected from processing surfaces after cleaning and disinfection. The bacteria were identified and characterized with regards to factors anticipated as important for growth and persistence in the food production environment.
2. Materials and Methods
2.1. Isolation and Identification of Bacterial Isolates
A total of 20 surfaces of equipment, floors, doors, knives, saws, handling panels from the slaughter area (from flaying to evisceration, splitting and cooling) in an abattoir for bovine slaughter were swabbed. The swabbing was performed approximately 6 h after cleaning and disinfection and prior to slaughter activities in the abattoir. In each site an area of approx. 100 cm2 was swabbed (3M Swab-Sampler—Letheen Broth; 3 M, St. Paul, USA). In certain sites reduced sampling areas were available for sampling. The samples were cooled and transported to the laboratory for plating within 10 h after sampling. Direct plating of the samples from the separate surfaces was performed by adding 0.1 ml of whirl mixed samples and surface plate spreading using a sterile loop on agar media (Plate count agar (PCA, Oxoid, Basingstoke, UK), Blood agar (Oxoid), Pseudomonas agar base with CFC selective supplement (Oxoid), Violet red bile glucose agar (VRBGA, Oxoid)), Chromogenic E. coli/coliform agar (Chrom, Oxoid) and incubated at 15˚C and 30˚C (PCA), 25˚C (Pseudomonas agar) and 37˚C (Blood agar, VRBGA, Chrom) for 48 - 96 h. Colony morphology on the separate plates was inspected. Quantitatively dominating colonies from the sampling sites were picked, plated and incubated to obtain pure cultures before being stored at −80˚C in 15% glycerol. Bacterial isolates were identified by 16S rDNA sequencing after automatic DNA extraction on colony material and subsequent 16S rDNA PCR using primers 8F and 1492R according to protocol of Schirmer et al. [20]. The 16S rDNA sequencing were performed using the Big Dye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) using the 534r primer [20]. Obtained 16S rDNA sequences were compared to known sequences using BLAST (http://www.ncbi.nlm.nih.gov/BLAST) for DNA sequence homology and isolate identification.
The L. monocytogenes strains used in this study are described in Table 1. The strains were meat/meat industry related, in addition to the Scott A and EGDe strains which are commonly used as reference strains in scientific studies of L. monocytogenes.
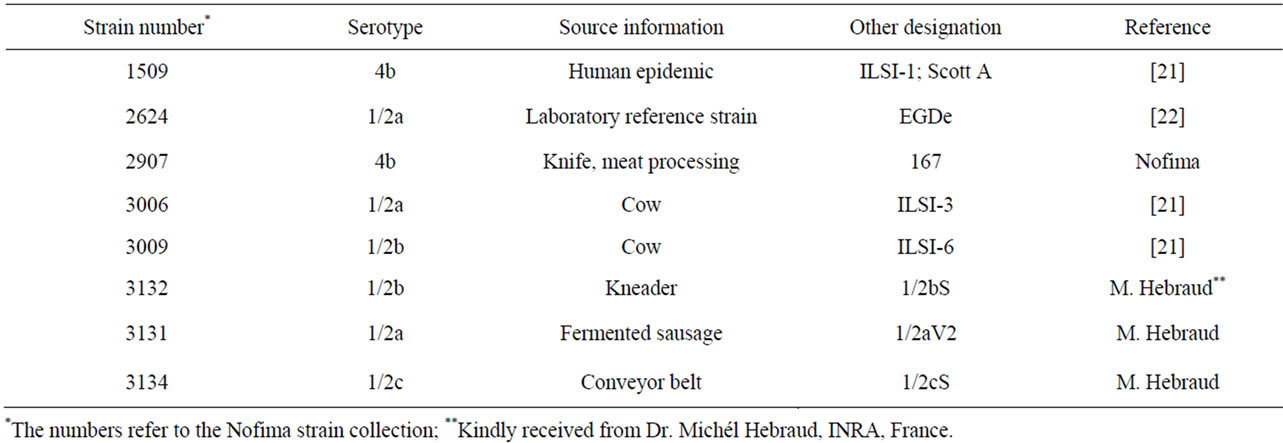
Table 1. Listeria monocytogenes strains used in this study.
2.2. Logging of Temperature and Humidity in the Abattoir
The temperature and relative humidity was monitored in the abattoir for one week using an automatic logging device (EL-USB-2, Lascar electronics Ltd., Salisbury, UK).
2.3. Biofilm Formation
Biofilm-forming ability was measured by staining of polystyrene-attached bacteria with crystal violet (CV). Cultures of L. monocytogenes and isolated slaughter house bacteria were inoculated from freezer stocks at −80˚C, and grown individually in Brain Heart Infusion broth (BHI; Oxoid) in two cultivation steps at 25˚C for 48 h. Bacterial cultures were used as inoculum to obtain approx. 106 CFU/ml in each well of 96-well polystyrene plates, U-bottom (Bibby Sterilin; Bibby Scientific, Staffordshire, UK) containing a total of 150 µl bacterial suspensions in BHI. Four parallel wells were used for each strain and cultivation condition. Negative control wells contained BHI only. Biofilm formation was tested after incubation at 12˚C and 20˚C for 48 h and seven days. Total cell mass was measured as absorbance at 600 nm (TitertekMultiskan RC plate reader; Labsystems, Helsinki, Finland). Biofilm formation was quantified according to the following procedure: Bacterial suspensions were pipetted off and the remaining biofilm washed twice with 300 µl distilled water (dH2O), using a semiautomatic microtiter plate washer (Wellwash AC, Thermo Electron Corporation, Waltham, Massachusetts, USA). Surface-attached bacteria were dried at 30˚C for 15 min and thereafter stained with 200 µl 0.1% CV for 5 - 10 min. After two washes with 300 µl dH2O, surface-bound CV was extracted by addition of 200 µl 33% acetic acid and incubation for 5 min. A volume of 100 µl was transferred to a new microtiter plate and absorbance was measured at 600 nm. Absorbance measurements were subtracted the absorbance values from wells containing BHI only. Each strain was tested in three to five independent experiments per cultivation condition.
2.4. Tolerance to Drying
Bacterial survival after air-drying on stainless steel was studied in a model system, as described previously [23]. An overnight culture (16 - 18 h) in BHI-broth incubated at 30˚C, inoculated from BHI-agar (Oxoid), was diluted ten times in fresh BHI. From the resulting suspension, four drops of 20 µl each was applied to a stainless steel (AISI 304, 2B, NorskStål AS, Nesbru, Norway) coupon of 20 × 20 mm, leading to a start concentration of log 6 - 7 cfu per coupon. The coupons were incubated for 1 h at 20˚C in a safety hood before being transferred to a plastic box with lid. A petri dish with a 20 ml saturated solution of lithium acetate was placed in the box, resulting in an atmosphere of approx. 70% relative humidity (RH). After 1, 7 and 14 days incubation at 12˚C, a steel coupon was transferred to a tube with 6 ml peptone water. To release cells from the coupon, the tube was sonicated for 15 min in a sonication bath (40˚C, 45 kHz/100 W, Bransonic 3510, Bransonic Ultrasonic B. V., Soest, The Netherlands). The number of surviving bacteria was determined after plating to BHI-agar and incubation at 30˚C. The strains were tested in two to five independent experiments.
2.5. Disinfection Test
The effect of disinfectants against bacteria dried on stainless steel was tested by the European surface test EN13697 with a few modifications [24,25]. The lowest recommended user-concentration of four commercial disinfectants was used (Table 2). The abattoir where the strains were isolated from used TP-99 for daily disinfection and Topactive DES for disinfection three times a year. 50 µl of an overnight culture grown in BHI at 25˚C was applied in one drop to a stainless steel coupon of 20 × 20 mm (AISI 304, 2B, Norskstål, Nesbru, Norway). The coupon was allowed to dry for 1 h in a safety hood at 20˚C. The disinfectant to be tested was diluted and added bovine serum albumin (0.3%) immediately before the test. The bacteria were exposed to the disinfectant for 5 min at 20˚C. 100 µl of the diluted disinfectant was added to the coupon to cover the area where the bacterial suspension was added. After 5 min exposure the coupon was transferred to a tube with 6 ml Dey/Engley Neutralizing broth (Difco Laboratories, Detroit, MI, USA). The bacteria were released from the surface by sonication as de-

Table 2. Disinfectants used in this study.
scribed above. The number of surviving bacteria was determined after serial dilution and plating on BHI-agar. As a control, deionized water was used instead of disinfectants. The different strain/disinfectant combinations were tested in three to four independent experiments.
2.6. Statistics
Statistical differences between different treatments or genera were calculated using analysis of variance (Anova, Minitab v16, Minitab Ltd, Coventry, UK) and difference between means by Tukeys test (Minitab). Calculations on desiccation and disinfection data were performed on log transformed data.
3. Results
3.1. Temperature and Humidity in the Meat Abattoir
The automatic monitoring of air temperature and humidity was performed in the same slaughter area as swab sampling. During one production week, temperatures were in the range 14˚C - 25˚C with temperatures below 20˚C during production and in the weekend. Relative humidity (RH) measurements varied between 35% - 90% RH. The highest temperatures and RH were obtained during cleaning and disinfection. The lowest RH was recorded during the weekend with no production while RH during production was in the range 50% - 70%. As an example, data from the flaying area is shown in Figure 1. Logging of temperatures and RH were also performed in the cutting and packaging departments with temperatures during production ranging between 13˚C and 18˚C.
3.2. Bacteria Isolated from Abattoir
After direct plating (PCA, 15˚C) bacterial colonies were detected in 19 of 20 sampled surfaces after cleaning and

Figure 1. Temperature and humidity in flaying department during a five day period.
disinfection. A semi-quantitative interpretation of total counts after direct plating showed variable bacterial levels in different sites. Highest bacterial loads were on console-joystick surfaces for slaughter process control. Lowest bacterial levels were observed on surfaces not regularly contaminated during the slaughter process (door, curtain for area separation, water hose at splitting saw). Aerococcus was identified as the most dominant bacterial genus, isolated from eight of the swabbed surfaces. Other bacteria identified after PCA plating (15˚C) were Pseudomonas, Serratia, Acinetobacter and Kocuria. All isolates showed 99% - 100% identity according to 16S rDNA BLAST sequence homology results.
Pseudomonas spp. and Staphylococci were isolated from Pseudomonas agar and Blood agar, respectively. Bacteria of the genera Citrobacter, Enterobacter, Acinetobacter, Pseudomonas and Serratia were isolated on VRBGA from two or more of the sample sites. The presence and survival of presumptive E. coli isolates were tested by plating swab samples on E. coli/coliform agar. No E. coli isolates were found.
Based on bacterial identification, we selected bacterial isolates for studies on surface survival and biofilm formation (Table 3). Isolates of the genera, Acinetobacter, Aerococcus, Citrobacter, Pseudomonas, Serratia and Staphylococcus were chosen as they dominated on two or more of the sampling sites. In addition, we also included
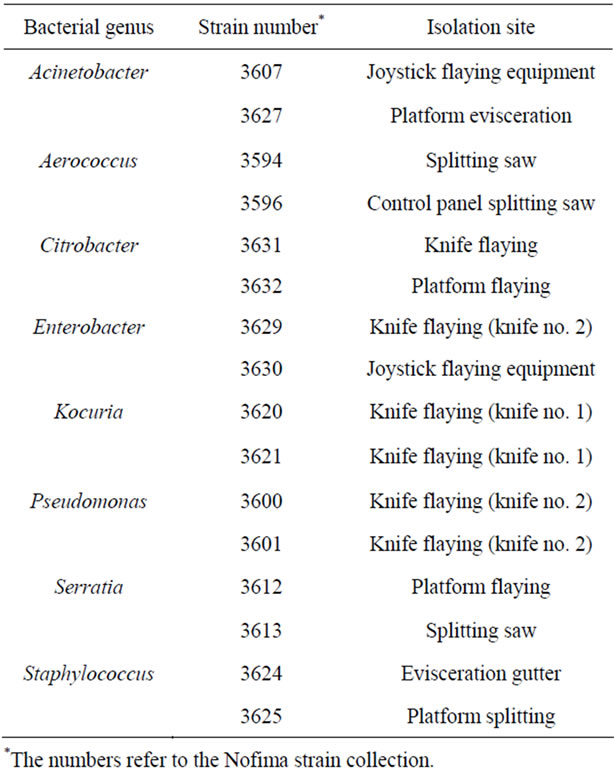
Table 3. Identity and site of isolation for bacteria used in this study.
two isolates of Kocuria based on their reported effect on biofilm formation in bacterial co-cultures [18].
3.3. Biofilm Formation
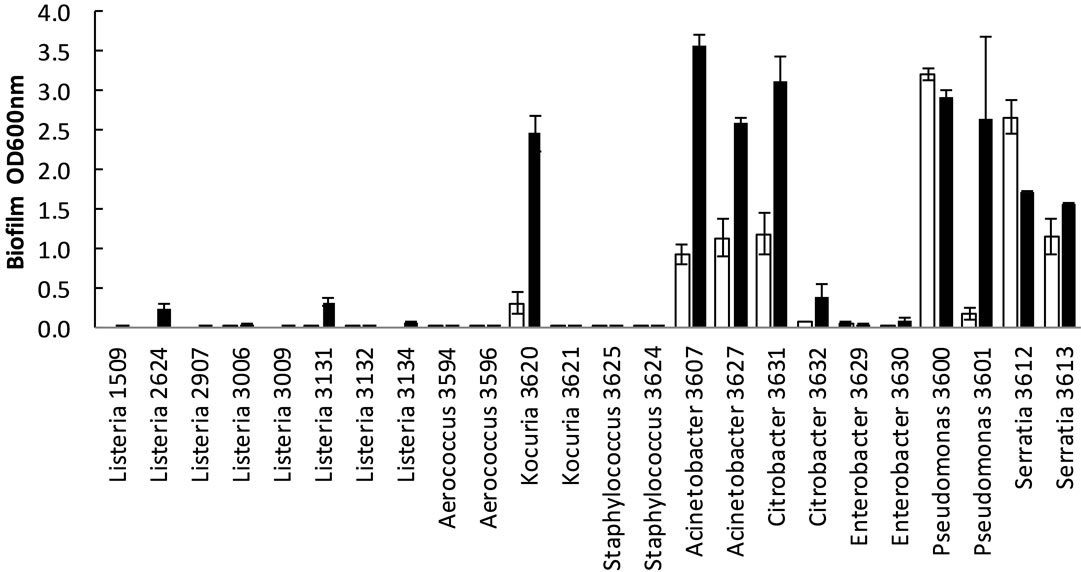
In general Gram negative bacteria formed more biofilm (p < 0.05, all four time/temperature combinations tested) than Gram positive bacteria including L. monocytogenes. The highest biofilm formation was observed for Acinetobacter, Citrobacter and Pseudomonas (Figure 2). Enterobacter formed little biofilm compared to the other Gram negative bacteria. There were no statistical differences in biofilm formation between L. monocytogenes and the other Gram positive bacteria for the four time/
temperature combinations (p = 0.23 − 0.98). The L. monocytogenes isolates 2624 (EGDe) and 3131 formed more (p < 0.05) biofilm than the other L. monocytogenes isolates after 2 d at 20˚C and 7 d at 12˚C. Especially at 12˚C there was very poor biofilm formation among Gram positive bacteria, except for Kocuria isolate 3620. The staphylococci and Kocuria 3621 showed very poor growth at 12˚C, the OD600nm < 0.2 (measured before removal of culture medium). Also at 20˚C these strains had lower growth (OD600nm) than the other strains. To check whether the differences in biofilm formation could be explained by differences in growth, we compared the biofilm/growth ratio between the strains. For the strains
 (a)
(a)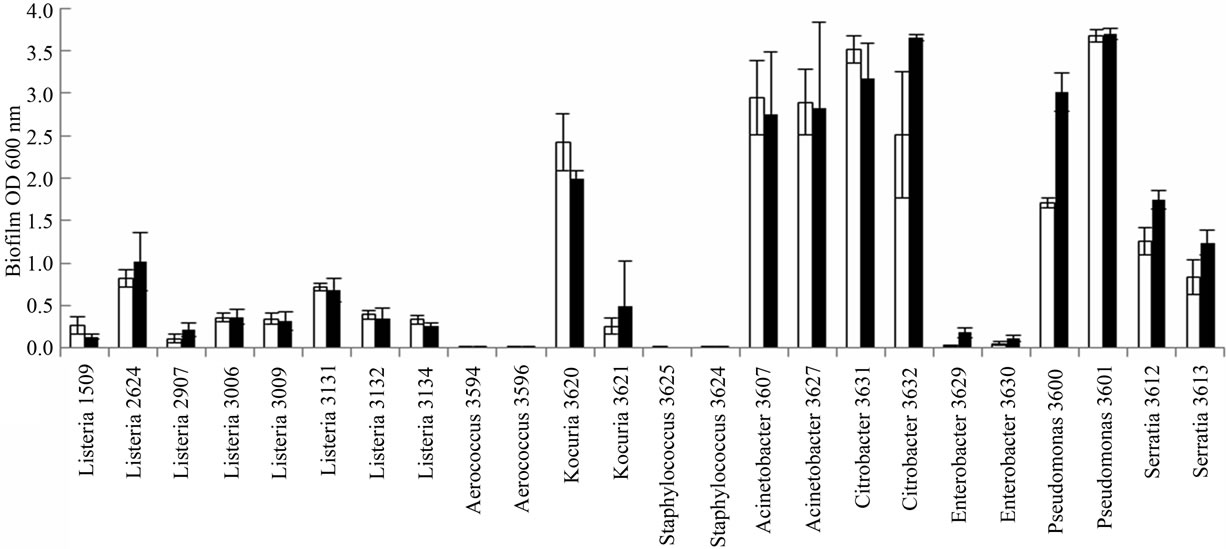 (b)
(b)
Figure 2. Biofilm formation (OD600nm) of bacteria in microtiter plates after incubation for 2 days (¨) and 7 days (n). Means of three to five replicates with standard errors in bars are shown. (a) 12˚C; (b) 20˚C.
with very low growth at 12˚C (Staphylococcus and Kocuria 3621), the denominator in the calculations was very low, which lead to high uncertainty in the ratio, thus we omitted these strains from the comparisons. For the other strains, when comparing biofilm/growth, the highest values were found among the Gram negative bacteria.
3.4. Survival Air-Drying on Stainless Steel
Gram positive bacteria were more tolerant to air-drying than Gram negative bacteria, both after 7 and 14 days (p < 0.001, both incubation times; Figure 3). The L. monocytogenes strains were less tolerant (p < 0.001; d7, p < 0.01; d14) than the other Gram positive bacteria, and more tolerant (p < 0.05) than the Gram negative bacteria after 14 d air-drying. There were no differences in tolerance to air-drying (p = 0.44; d7, p = 0.74; d14) between the L. monocytogenes isolates.
3.5. Disinfection
The disinfectants TP-99, Topactive DES and Oxydes had only low or no bactericidal effects against the bacteria tested in the surface disinfection tests (Figure 4). The disinfectant Aco Hygiene Ultra DES had a bactericidal effect of 2 - 3.5 log reductions against Acinetobacter, Aerococcus and the L. monocytogenes isolates. In general Staphylococcus and Serratia were more tolerant than the other bacteria as no significant (p > 0.05) bactericidal reduction was observed for any of the disinfectants. The most sensitive bacteria were Pseudomonas and Acinetobacter, where significant bacterial reductions (p < 0.05)
were observed for all four and three of the four disinfectants, respectively.
4. Discussion
Control of bacteria in the meat processing industry is a key element to assure meat safety and quality. However, bacterial elimination and control strategies in meat manufacture are difficult tasks. Cleaning and disinfection do not eliminate all bacteria present and bacteria surviving the sanitation process may be a source for crosscontamination and bacterial persistence; hazards potentially affecting both food safety and quality. In the food industry, bacteria will also encounter other stressful conditions, e.g. mechanical shear forces, desiccation, low and high temperature and starvation. It is likely that bacteria dominating after cleaning and disinfection have certain characteristics enabling them to survive in the food processing environments. The bacterial genera dominating in the abattoir after cleaning and disinfection were Aerococcus, Acinetobacter, Pseudomonas, Staphylococcus and Serratia. Several studies have reported the latter four genera to be common in the production environment of many types of food [9-15]. Aerococcus has been found in fish [12] and pastry production environments [10]. The semi-quantitative determination of bacterial loads indicated that easy-to-clean sampling sites not in direct product contact had the lowest bacterial counts. Highest bacterial loads were observed on consoles and joysticks for process control which included design not suitable for effective sanitation (rubber with
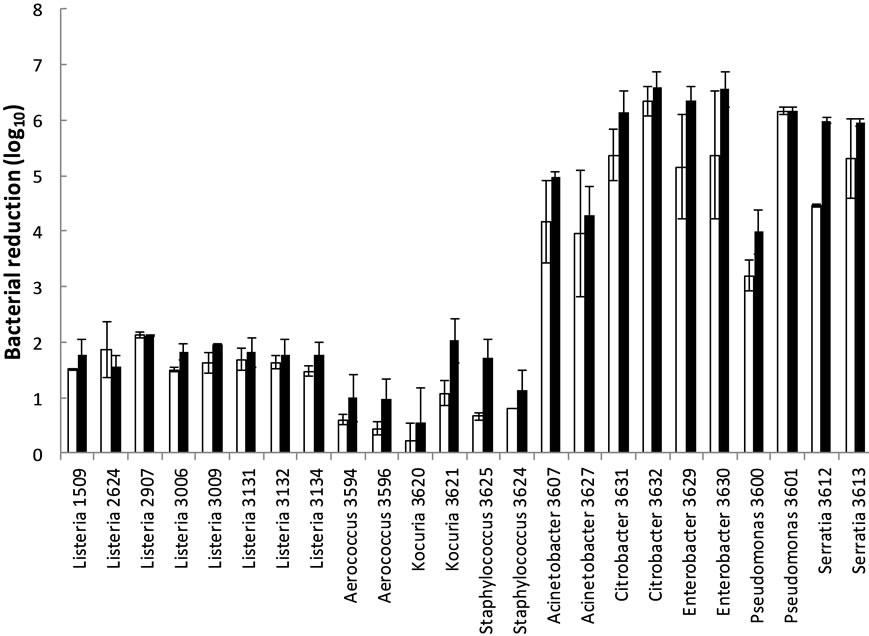
Figure 3. Bacterial reduction during air drying of bacteria at stainless steel at 70% RH at 12˚C for seven (¨) and 14 days (n) incubation. Means of two to four replicates with standard errors in bars are shown.

Figure 4. Effects of disinfectants against bacteria dried on stainless steel. Means and standard errors of three or four independent experiments are shown. Asterisk indicates significant bacterial reduction (p < 0.05).
folds). The results indicated lack of a high selective pressure as the diversity was higher than one would expect for example after an efficient disinfection procedure or long periods of drying. However, since a similar microbiota has been reported from production of a range of foods, food processes and countries it is likely that these genera or strains/species within these genera have attributes that enhance survival and growth under conditions common for a range of food processes. Examples of such conditions could be periods with high nutrient availability combined with low temperature, periods of drying and finally cleaning and disinfection. Therefore these scenarios were investigated further. Listeria monocytogenes (mostly meat associated strains) was included in the study as it is regarded among the most troublesome bacteria in several food industries, with numerous reports on their ability to survive and persist in food industry premises [3,26].
The results indicated large differences in the biofilm forming abilities of different genera and also strains of the same genus. The large differences in biofilm formation observed thus indicate that this property alone cannot explain survival of all bacteria isolated. Biofilm formation has been suggested as an important mechanism for L. monocytogenes persistence, but this hypothesis is disputed [3,26]. In this study L. monocytogenes was found to be a relatively poor biofilm former compared to Gram negative bacteria. The results are in accordance with other studies showing that L. monocytogenes form less biofilm than Pseudomonas spp. [27,28]. The L. monocytogenes isolates producing most biofilms belonged to serovar 1/2a (phylogenetic Division II) while low biofilm producers were serovar 4b isolates (Division I). Differences in L. monocytogenes adherence/biofilm production between persistent and non-persistent strains and between phylogenetic divisions have been reported [4,6, 29]. The strains used in the present study are not reported as persistent, and testing persistent strains of Listeria or other bacteria could have resulted in different results. Other researchers have reported that the type of growth medium can influence biofilm formation of L. monocytogenes [29-31]. In addition to BHI, we also tested TSB and LB (with and without NaCl), but the biofilm formation was comparable to that obtained in BHI. In conclusion, the results obtained did not confirm that bacterial strains surviving in the food production environment share a common property of producing high levels of biofilm at low temperature. However, it cannot be excluded that the results would be different in other biofilm models, e.g. models using materials used in food processing environments.
In many food processing facilities the processing equipment is dry for shorter or longer periods, for example in the weekends or between end of the cleaning period and start of production. Indeed, the temperature recordings showed a very large variation of relative humidity during one production day. The Gram positive bacteria were significantly more tolerant to air drying than Gram negative bacteria. This is in accordance with previous findings. It is believed that the Gram positive cell wall has a protective role during desiccation [32]. L. monocytogenes is often found in humid areas in a production environment. For control of L. monocytogenesit is recommended to keep the production environment as dry as possible, and allow areas to dry up regularly [33]. Interestingly, surfaceassociated L. monocytogenes were less tolerant to desiccation than the other Gram positives, but more tolerant than the Gram negative bacteria. To our knowledge this has not been reported previously, and the mechanisms of lower desiccation tolerance of Listeria than other Gram positives should be subjected to further studies. The presence of a self-produced biofilm matrix, organic matters and food components have been shown to increase the survival of L. monocytogenes and other foodborne bacteria during desiccation [23,34] and the survival during periods of drying may be higher in practice than what was found in this study.
It is important that disinfectants are tested under conditions that resemble the practical usage conditions. It is well known that surface associated bacteria are more tolerant than free living bacteria [25,35,36]. Three (including TP-99, the disinfectant used daily in the abattoir) out of four disinfectants tested had very low efficiency in the surface tests (0 - 1.3 log reductions). Some manufacturers refer to data from suspension tests when marketing the efficiency of their disinfectant and the disinfectants are tested against laboratory strains. Aco Hygiene Ultra DES was more efficient than the other disinfectants. It is not possible to conclude if the differences in susceptibilities are dependent on too low concentrations used in some disinfectants, or if QAC (Aco Hygiene Ultra DES) is more efficient. Surprisingly, the Gram negative bacteria Pseudomonas and Acinetobacter were found to be among the most susceptible to disinfection among the bacteria studied including Gram positive bacteria. This is not in accordance with the majority of previous literature supporting the common opinion that Gram-negative bacteria have higher intrinsic resistance to biocides than Gram positive and among them Pseudomonas being one of the most resistant [37,38]. One may speculate that the desiccation process harmed the cells rendering them more sensitive to subsequent stress like disinfection. Another explanation for discrepancies between other studies and the present study is that commercial disinfectants were used. Often commercial disinfectants are containing several active substances including sequestering agents, such as EDTA, that will disrupt the outer protective membrane of Gram negative bacteria [39]. It cannot be ruled out that the disinfectants tested in the present study contained such agents, although it was not given in the user information. The differences in the bactericidal effects of disinfectants varied between the type of disinfectant. Further studies should be performed to investigate to what extend bactericidal effects of disinfectants is affected by prior bacterial exposure to meat industry relevant stresses. Serratia and Staphylococcus showed highest tolerance to disinfection. Staphylococcus sp. may harbour plasmid encoding for efflux pumps for QAC, which lead to increased tolerance [40,41] and resistance to several biocides has been linked to mucoid growth [42]. Serratia was found to be resistant to the disinfectant containing QAC. Resistance to user-concentrations of amphoteric and cationic tensides has previously been reported for Serratiamarcescens [43,44], and efflux was he proposed resistance mechanism.
As found in the present study and as reported by others, L. monocytogenes is not particularly resistant to environmental stress and not a good biofilm former compared to other bacteria in in vitro tests [27,28]. It may be speculated that the background flora may have a role in protecting L. monocytogenes against stress in the food industry. However, other bacteria have been shown both to promote [18,27] and inhibit [17,45] biofilm formation of L. monocytogenes, and further studies are required to reveal the role of background flora on colonisation and persistence of L. monocytogenes in the food industry.
In conclusion, the dominating bacteria isolated from the meat abattoir were similar as reported for many type of food production environments. The bacteria isolated had different properties believed to be important for survival in the food production environment. Gram negative bacteria were better biofilm formers than Gram positive bacteria (including L. monocytogenes), while the opposite was found for tolerance to desiccation, where L. monocytogenes were more susceptible than other Gram positive bacteria. The disinfectant in daily use in the abattoir, tested at its recommended user-concentration in a surface test, had a low or limited effect against bacteria from the meat abattoir and L. monocytogenes. Our study showed the necessity of applying proper cleaning and disinfection routines in the meat processing industry. Further investigations are needed to understand the factors and mechanisms affecting desiccation tolerance in bacteria and potential cross-tolerance to other stresses. This could have implications for improved intervention strategies for ensuring microbial food safety and quality. Further, isolation of bacteria surviving processing conditions including sanitation routines may be used for designing improved strategies for elimination and control of bacteria in food industry processes.
5. Acknowledgements
This work was in part funded by the Foundation for Research Levy on Agricultural Products, by the Norwegian Research Council, and by the Food Quality and safety, ProSafeBeef Food-CT-2006-36241 research program (part of the EU 6th Framework Program). Dr. Michél Hebraud, INRA, France, is appreciated for providing L. monocytogenes isolates to the study. The authors wish to thank Anette Wold Åsli, Tove Maugesten, Ahmed Abdelgani and Olivier Habimana for excellent technical assistance.
REFERENCES
- J. T. Holah, J. Bird and K. E. Hall, “The Microbial Ecology of High-Risk, Chilled Food Factories; Evidence for Persistent Listeria spp. and Escherichia coli Strains,” Journal of Applied Microbiology, Vol. 97, No. 1, 2004, pp. 68-77. doi:10.1111/j.1365-2672.2004.02272.x
- R. B. Tompkin, “Control of Listeria monocytogenes in the Food-Processing Environment,” Journal of Food Protection, Vol. 65, No. 4, 2002, pp. 709-725.
- T. Møretrø and S. Langsrud, “Listeria monocytogenes: Biofilm Formation and Persistence in Food Processing Environments,” Biofilms, Vol. 1, No. 2, 2004, pp. 107-121. doi:10.1017/S1479050504001322
- M. K. Borucki, et al., “Variation in Biofilm Formation among Strains of Listeria monocytogenes,” Applied and Environmental Microbiology, Vol. 69, No. 12, 2003, pp. 7336-7342. doi:10.1128/AEM.69.12.7336-7342.2003
- B. Aase, et al., “Occurrence of and a Possible Mechanism for Resistance to a Quaternary Ammonium Compound in Listeria monocytogenes,” International Journal of Food Microbiology, Vol. 62, No. 1-2, 2000, pp. 57-63. doi:10.1016/S0168-1605(00)00357-3
- J. M. Lunden, et al., “Persistent Listeria monocytogenes Strains Show Enhanced Adherence to Food Contact Surface after Short Contact Times,” Journal of Food Protection, Vol. 63, No. 9, 2000, pp. 1204-1207.
- E. M. Fox, N. Leonard and K. Jordan, “Physiological and Transcriptional Characterization of Persistent and Nonpersistent Listeria monocytogenes Isolates,” Applied and Environmental Microbiology, Vol. 77, No. 18, 2011, pp. 6559-6569. doi:10.1128/AEM.05529-11
- J. Lundén, R. Tolvanen and H. Korkeala, “Acid and Heat Tolerance of Persistent and Nonpersistent Listeria monocytogenes Food Plant Strains,” Letters in Applied Microbiology, Vol. 46, No. 2, 2008, pp. 276-280. doi:10.1111/j.1472-765X.2007.02305.x
- G. Brightwell, et al., “Identifying the Bacterial Community on the Surface of Intralox(TM) Belting in a Meat Boning Room by Culture-Dependent and Culture-Independent 16S rDNA Sequence Analysis,” International Journal of Food Microbiology, Vol. 109, No. 1-2, 2006, pp. 47-53. doi:10.1016/j.ijfoodmicro.2006.01.008
- E. Mettler and B. Carpentier, “Variations over Time of Microbial Load and Physicochemical Properties of Floor Materials after Cleaning in Food Industry Premises,” Journal of Food Protection, Vol. 61, No. 1, 1998, pp. 57-65.
- M. Sharma and S. K. Anand, “Characterization of Constitutive Microflora of Biofilms in Dairy Processing Lines,” Food Microbiology, Vol. 19, No. 6, 2002, pp. 627-636. doi:10.1006/fmic.2002.0472
- D. Bagge-Ravn, et al., “The Microbial Ecology of Processing Equipment in Different Fish Industries—Analysis of the Microflora during Processing and Following Cleaning and Disinfection,” International Journal of Food Microbiology, Vol. 87, No. 3, 2003, pp. 239-250. doi:10.1016/S0168-1605(03)00067-9
- B. Guobjornsdottir, H. Einarsson and G. Thorkelsson, “Microbial Adhesion to Processing Lines for Fish Fillets and Cooked Shrimp: Influence of Stainless Steel Surface Finish and Presence of Gram-Negative Bacteria on the Attachment of Listeria monocytogenes,” Food Technology and Biotechnology, Vol. 43, No. 1, 2005, pp. 55-61.
- S. Langsrud, L. Seifert and T. Moretro, “Characterization of the Microbial Flora in Disinfecting Footbaths with Hypochlorite,” Journal of Food Protection, Vol. 69, No. 9, 2006, pp. 2193-2198.
- T. Møretrø, et al., “Evaluation of the Antimicrobial Effect of a Triclosan-Containing Industrial Floor Used in the Food Industry,” Journal of Food Protection, Vol. 69, No. 3, 2006, pp. 627-633.
- D. K. Jeong and J. F. Frank, “Growth of Listeria monocytogenes at 10˚C in Biofilms with Microorganisms Isolated from Meat and Dairy Processing Environments,” Journal of Food Protection, Vol. 57, No. 7, 1994, pp. 576-586.
- V. Leriche and B. Carpentier, “Limitation of Adhesion and Growth of Listeria monocytogenes on Stainless Steel Surfaces by Staphylococcus sciuri Biofilms,” Journal of Applied Microbiology, Vol. 88, No. 4, 2000, pp. 594-605. doi:10.1046/j.1365-2672.2000.01000.x
- B. Carpentier and D. Chassaing, “Interactions in Biofilms between Listeria monocytogenes and Residant Microorganisms from Food Industry Premises,” International Journal of Food Microbiology, Vol. 97, No. 2, 2004, pp. 111-122. doi:10.1016/j.ijfoodmicro.2004.03.031
- O. Habimana, et al., “Enhanced Surface Colonization by Escherichia coli O157:H7 in Biofilms Formed by an Acinetobacter calcoaceticus Isolate from Meat-Processing Environments,” Applied and Environmental Microbiology, Vol. 76, No. 13, 2010, pp. 4557-4559. doi:10.1128/AEM.02707-09
- B. C. Schirmer, E. Heir and S. Langsrud, “Characterization of the Bacterial Spoilage Flora in Marinated Pork Products,” Journal of Applied Microbiology, Vol. 106, No. 6, 2009, pp. 2106-2116. doi:10.1111/j.1365-2672.2009.04183.x
- E. Fugett, et al., “International Life Sciences Institute North America Listeria monocytogenes Strain Collection: Development of Standard Listeria monocytogenes Strain Sets for Research and Validation Studies,” Journal of Food Protection, Vol. 69, No. 12, 2006, pp. 2929-2938.
- P. Glaser, et al., “Comparative Genomics of Listeria Species,” Science, Vol. 294, No. 5543, 2001, pp. 849-852.
- T. Møretrø, et al., “Factors Affecting Survival of Shigatoxin-Producing Escherichia coli on Abiotic Surfaces,” International Journal of Food Microbiology, Vol. 138, No. 1-2, 2010, pp. 71-77.
- European Committee for standardization, “NS-EN 13697: 2001: Quantitative Non-Porous Surface Test for the Evaluation of Bactericidal and/or Fungicidal Activity of Chemical Disinfectants Used in Food, Industrial, Domestic and Institutional Areas. Test Method and Requirements without Mechanical Action,” 2001.
- T. Møretrø, et al., “Evaluation of Efficacy of Disinfectants against Salmonella from the Feed Industry,” Journal of Applied Microbiology, Vol. 106, No. 3, 2009, pp. 1005-1012. doi:10.1111/j.1365-2672.2008.04067.x
- B. Carpentier and O. Cerf, “Review—Persistence of Listeria monocytogenes in Food Industry Equipment and Premises,” International Journal of Food Microbiology, Vol. 145, No. 1, 2011, pp. 1-8. doi:10.1016/j.ijfoodmicro.2011.01.005
- K. Sasahara and E. Zottola, “Biofilm Formation by Listeria monocytogenes Utilizes a Primary Colonizing Microorganism in Flowing Systems,” Journal of Food Protection, Vol. 56, No. 12, 1993, pp. 1022-1028.
- D. E. Norwood and A. Gilmour, “The Growth and Resistance to Sodium Hypochlorite of Listeria monocytogenes in a Steady-State Multispecies Biofilm,” Journal of Applied Microbiology, Vol. 88, No. 3, 2000, pp. 512-520. doi:10.1046/j.1365-2672.2000.00990.x
- T. Combrouse, et al., “Quantification of the Extracellular Matrix of the Listeria monocytogenes Biofilms of Different Phylogenic Lineages with Optimization of Culture Conditions,” Journal of Applied Microbiology, Vol. 114, No. 4, 2013, pp. 1120-1131. doi:10.1111/jam.12127
- A. Moltz and S. Martin, “Formation of Biofilms by Listeria monocytogenes under Various Growth Conditions,” Journal of Food Protection, Vol. 68, No. 1, 2005, pp. 92-97.
- J. Folsom, G. Siragusa and J. Frank, “Formation of Biofilm at Different Nutrient Levels by Various Genotypes of Listeria monocytogenes,” Journal of Food Protection, Vol. 69, No. 4, 2006, pp. 826-834.
- A. Kramer, I. Schwebke and G. Kampf, “How Long do Nosocomial Pathogens Persist on Inanimate Surfaces? A Systematic Review,” BMC Infectious Diseases, Vol. 6, 2006, p. 130. doi:10.1186/1471-2334-6-130
- R. B. Tompkin, et al., “Guidelines to Prevent Post-Processing Contamination from Listeria monocytogenes,” Dairy, Food and Environmental Sanitation, Vol. 19, No. 8, 1999, pp. 551-562.
- H. Takahashi, et al., “Desiccation Survival of Listeria monocytogenes and Other Potential Foodborne Pathogens on Stainless Steel Surfaces is Affected by Different Food Soils,” Food Control, Vol. 22, No. 3-4, 2011, pp. 633- 637. doi:10.1016/j.foodcont.2010.09.003
- J. W. Costerton, et al., “Microbial Biofilms,” Annual Review of Microbiology, Vol. 49, 1995, pp. 711-742. doi:10.1146/annurev.mi.49.100195.003431
- J. D. Stopforth, et al., “Biofilm Formation by Acid-Adapted and Non-Adapted Listeria monocytogenes in Fresh Beef Decontamination Washings and Its Subsequent Inactivation with Sanitizers,” Journal of Food Protection, Vol. 65, No. 11, 2002, pp. 1717-1727.
- G. McDonnell and A. D. Russell, “Antiseptics and Disinfectants: Activity, Action, and Resistance,” Clinical Microbiology Reviews, Vol. 14, No. 1, 2001, pp. 227-228.
- E. Ortega Morente, et al., “Biocide Tolerance in Bacteria,” International Journal of Food Microbiology, Vol. 162, No. 1, 2013, pp. 13-25. doi:10.1016/j.ijfoodmicro.2012.12.028
- S. Langsrud and G. Sundheim, “Factors Contributing to the Survival of Poultry Associated Pseudomonas spp. Exposed to a Quaternary Ammonium Compound,” Journal of Applied Bacteriology, Vol. 82, No. 6, 1997, pp. 705- 712. doi:10.1046/j.1365-2672.1997.00186.x
- E. Heir, G. Sundheim and A. Holck, “Identification and Characterization of Quaternary Ammonium Compound Resistant Staphylococci from the Food Industry,” International Journal of Food Microbiology, Vol. 48, No. 3, 1999, pp. 211-219. doi:10.1016/S0168-1605(99)00044-6
- E. Heir, G. Sundheim and A. L. Holck, “Resistance to Quaternary Ammonium Compounds in Staphylococcus spp. Isolated from the Food Industry and Nucleotide Sequence of the Resistance Plasmid pST827,” Journal of Applied Bacteriology, Vol. 79, No. 2, 1995, pp. 149-156. doi:10.1111/j.1365-2672.1995.tb00928.x
- D. O. Kolawole, “Resistance Mechanisms of MucoidGrown Staphylococcus aureus to the Antibacterial Action of Some Disinfectants and Antiseptics,” FEMS Microbiology Letters, Vol. 25, No. 2-3, 1984, pp. 205-209. doi:10.1111/j.1574-6968.1984.tb01457.x
- S. Langsrud, T. Møretrø and G. Sundheim, “Characterization of Serratia marcescens Surviving in Disinfecting Footbaths,” Journal of Applied Microbiology, Vol. 95, No. 1, 2003, pp. 186-195. doi:10.1046/j.1365-2672.2003.01968.x
- H. Maseda, et al., “Mutation in the sdeS Gene Promotes Expression of the sdeAB Efflux Pump Genes and Multidrug Resistance in Serratia marcescens,” Antimicrobial Agents and Chemotherapy, Vol. 55, No. 6, 2011, pp. 2922-2926. doi:10.1128/AAC.01755-10
- L. Guillier, et al., “Modelling the Competitive Growth between Listeria monocytogenes and Biofilm Microflora of Smear Cheese Wooden Shelves,” International Journal of Food Microbiology, Vol. 128, No. 1, 2008, pp. 51-57. doi:10.1016/j.ijfoodmicro.2008.06.028
NOTES
*Corresponding author.

