Open Journal of Medical Microbiology
Vol.3 No.4(2013), Article ID:41014,7 pages DOI:10.4236/ojmm.2013.34034
Screening of Class 1 and Class 2 Integrons in Clinical Isolates of Pseudomonas aeruginosa Collected from Seven Hospitals in Turkey: A Multicenter Study
1Department of Medical Microbiology, Faculty of Medicine, Recep Tayyip Erdogan University, Rize, Turkey
2Department of Biology, Faculty of Arts and Sciences, Artvin Coruh University, Artvin, Turkey
3Department of Genetics and Bioengineering, Faculty of Engineering and Natural Sciences, Gümüşhane University, Gumushane, Turkey
4Clinical Microbiology Laboratory, Kecioren Research and Training Hospital, Ankara, Turkey
5Clinical Microbiology Laboratory, Research and Training Hospital, Kayseri, Turkey
6Clinical Microbiology Laboratory, General State Hospital, Tokat, Turkey
7Department of Medical Microbiology, Faculty of Medicine, Dicle University, Diyarbakir, Turkey
8Clinic of Infectious Diseases and Clinical Microbiology, Private OSM Ortadogu Hospital, Sanliurfa, Turkey
9Clinical Microbiology Laboratory, Ahi Evren Chest and Cardiovascular Surgery Research and Training Hospital, Trabzon, Turkey
10Clinical Microbiology Laboratory, Akcaabat Hackali Baba General State Hospital, Trabzon, Turkey
11Department of Biology, Faculty of Arts and Sciences, Recep Tayyip Erdogan University, Rize, Turkey
Email: *cemal.sandalli@erdogan.edu.tr
Copyright © 2013 Aysegul Copur Cicek et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In accordance of the Creative Commons Attribution License all Copyrights © 2013 are reserved for SCIRP and the owner of the intellectual property Aysegul Copur Cicek et al. All Copyright © 2013 are guarded by low and by SCIRP as a guardian.
Received August 15, 2013; revised September 16, 2013; accepted September 23, 2013
Keywords: Pseudomonas aeruginosa; Antibiotic Resistance; Class 1 Integron; AadA; Multicenter Study
ABSTRACT
Pseudomonas aeruginosa is one of the leading nosocomial pathogens worldwide, and their infections are difficult to treat due to acquired resistance to many antibiotics. This study aimed to detect class 1 and 2 integrons and antibiotic susceptibility of clinical isolates of P. aeruginosa. Two hundred and five P. aeruginosa strains were collected from the seven general state hospitals in Turkey. They were characterized by antimicrobial susceptibility testing, screened for class 1 and class 2 integrons, and evaluated for the association between antibiotic resistance phenotypes and the presence of integrons. intI gene was amplified in 10 isolates (4.87%) by PCR and in seven isolates of them (70%) were found different gene cassettes. The aadA gene integrated into the class 1 integrons was most frequently found and it was followed by aac genes and blaOXA family genes. Sequence analysis of variable regions of the class 1 integrons showed five gene cassette arrays; aadA1(99%), aac(3)-Id(82%)-orf-aac(3”)-Ia(99%), aac(3)-Ie(83%)-blaoxa10(100%)- aadA1 (100%), aadA6(99%, 100%), aac(6’)-I(97%)-orf-aadA2(99%). No class 2 integron was detected. This study is the first multicenter study for class 1 integrons and it indicates the low rate of presence of class 1 gene cassette in P. aeruginosa.
1. Introduction
Pseudomonas is a genus of bacteria known for its metabolic capacity and ability to occupy a wide range of environments. Genomes of these genus members can influence niche and degree of virulence [1]. Pseudomonas aeruginosa (P. aeruginosa) is a ubiquitous, Gram-negative, opportunistic organism. This organism can cause various infections such as hospital-acquired pneumonia and fulminant sepsis especially in immunocompromised hosts [2-4]. P. aeruginosa can also cause urinary tract infections. It survives on minimal nutritional requirements and tolerates a variety of physical conditions so it can remain in both community and hospital settings. In hospitals, P. aeruginosa can be isolated from a variety of sources, including respiratory therapy equipment, antiseptics, soap, sinks, mops, medicines, and physiotherapy and hydrotherapy pools [5]. It grows well on most laboratory media and commonly is isolated on blood agar or eosin-methylthionine blue agar. It is identified on the basis of its Gram morphology, inability to ferment lactose, a positive oxidase reaction, its fruity odor, and its ability to grow at 42˚C [6].
Infections caused by this bacterium are difficult to root up because it has innate antimicrobial resistance due to low outer membrane permeability and an extensive efflux pump system [7]. However, genes make the bacteria more resistant to antibiotics such as β-lactams, aminoglycosides and fluoroquinolones acquired newly by this bacterium [8]. Intrinsic and acquired resistance mechanisms of P. aeruginosa cause the evolution of numerous multidrug-resistant (MDR) P. aeruginosa outbreaks in clinical settings [9]. Dissemination of antibiotic resistance genes by horizontal transfer results in the rapid emergence of antibiotic resistance among clinical bacterial isolates [10]. The gene acquisition by horizontal gene transfer is mediated by mobile genetic elements such as plasmids, transposons and integrons. Integrons are one of the main types of mobile elements currently known to be the natural gene capture systems in bacteria that are considered responsible for multi-drug resistance [11]. These mobile elements have two conserved regions (integrase and recombination site) divided by a variable region that includes integrated gene cassettes, which frequently include antibiotic resistance genes [12]. Gene cassettes can be mobile and contain only a single gene and a specific recombination site 59 base element [13,14].
Five classes of multi-resistance integrons are known [15]. Class 1 and class 2 integrons are the most frequent ones in Gram-negative bacteria [16,17]. Class 1 integrons contain 5’ and 3’ conserved segments. While 5’ conserved region of class 1 integron includes class 1 integrase gene encoding a site-specific integrase (intI) and recombination site, 3’ conserved region includes sulI and qacEΔ1 genes [18]. Class 1 integrons have been reported in P. aeruginosa isolates [8,19-27]. Organization of class 2 integrons is similar to the class 1 integrons and they are inserted in the Tn7 transposons family. They have been reported from P. aeruginosa in 2009 [28,29].
Selection of the most appropriate antibiotic is difficult for treatment diseases caused by drug-resistant P. aeruginosa. Drug-resistant P. aeruginosa is associated with significant increases in morbidity, mortality, need for surgical intervention, length of hospital stay and chronic care, and overall cost of treating the infection [5].
In the present study, we investigated the class 1 and class 2 integrons from clinical strains of P. aeruginosa. This is the first multi-center study of investigation of two classes of integrons in P. aeruginosa isolated from Turkey.
2. Materials and Methods
2.1. Bacterial Strains
Two hundred and five clinical strains of P. aeruginosa were collected from the clinical samples of the patients from various hospitals of six different cities, Ankara (n = 46), Diyarbakir (n = 43), Kayseri (n = 36), Trabzon (n = 38), Sanliurfa (n = 33) and Tokat (n = 9) in Turkey between 2011 and 2012. Clinical samples were from blood, urine, body fluids. Identification of the strains was confirmed to the species level by using the automatic system VITEK 2 (bioMerieux, Durham, NC, USA) according to the manufacturer’s instructions.
2.2. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility tests were performed by the disk diffusion method, and the results were interpreted according to the CLSI guidelines [30]. The following antibiotic disks (Oxoid, UK) were used: norfloxacin, tobramycin, piperacillin, ceftazidime, piperacillin-tazobactam, amikacin, gentamicin, imipenem, meropenem, ciprofloxacin, levofloxacin and cefepime. P. aeruginosa ATCC 27853 was used as the standard strain.
2.3. DNA Extraction
Genomic DNAs used as template for PCR assays were obtained from bacterial suspensions grown overnight in Luria-Bertani Broth with shaking incubator at 37˚C. Bacterial suspensions were centrifuged at 13.000 rpm for five minutes. Pellet was suspended in 500 µl distilled water and subsequently boiled in a water bath for 10 minutes. Debris was centrifuged at 13.000 rpm for five minutes. A 500 µl of supernatant was obtained and its 5 µl was used as the DNA template for PCR assays [31].
2.4. PCR Assay for Integrons
All isolates were tested for the presence of the conserved regions of class 1 and class 2 integrases. Primers used for detection for intI1: intI1F (5’-ACATGTGATGGCGACGCACGA-3’) and intI1R (5’ATTTCTGTCCTGGCTGGCGA-3’); for intI2: intI2F (5’-CACGGATATGCGACAAAAAGGT-3’) and intI2R (5’-GTAGCAAACGAGTGACGAAATG-3’); for class 1 integron variable region: 5’CS
(5’-GGCATCCAAGCAGCAAG-3’) and 3’CS (5’-AAGCAGACTTACCTGA-3’). PCRs were performed in a final volume of 50 µl. PCR mix component was as follows; 5 µl of 10X PCR buffer (100 mM Tris-HCl [pH 8.8], 500 mM KCl, 1% Triton X-100), 3 µl of 25 mM MgCl2, 5 µl of 10X dNTP (2 mM dATP, dCTP, dGTP and dTTP), 2 µl each of primer (25 pmol/μL), 34 µl deionised sterile water, 1 U of Taq DNA polymerase and 5 µl of template DNA. PCR amplification condition was as follows: initial denaturation at 94˚C for 3 min, 94˚C for 45 seconds, 55˚C for 1 minute, 72˚C for 3 min followed by 34 cycles and 5 min at 72˚C with a final extension [32,33].
2.5. DNA Sequencing and Data Analysis
Electrophoresis of PCR products was performed 1% agarose containing 0.5 μg/ml ethidium bromide, and subsequently visualized under UV light. Class 1 integron positive PCR products were ligated into pGEM-T easy vector (Promega) at 16˚C for 16 h by T4 DNA ligase. Ligation mixture were then transformed into E. coli JM101 strain prepared according to Sambrook et al. (1989) [34] and spread in ampicillin plates (50 μg/ml) containing X-gal (40 μg/ml). We collected the white colonies and plasmids were purified from these colonies by using Promega Plasmid Purification Kits. Recombinant plasmids carrying amplicons of class 1 integrons were sent to Macrogen Inc., Seoul, Korea for sequencing by using the universal oligonucleotide primers, T7 and SP6. Sequencing results were analysed using alignment search tool, BLAST (http://www. ncbi.nlm.nih.gov/BLAST) [35] and the multiple sequence alignment program, CLUSTALW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
3. Results
All isolates were tested for susceptibility to twelve antimicrobials by using disk diffusion method. The frequency of resistance of two hundred and five clinical isolates to each antibiotics tested was determined. Resistance to norfloxacin (13.2%), tobramycin (10.2%), piperacillin (23.9%), ceftazidime (15.6%), piperacillin-tazobactam (18.5%), amikacin (11.2%), gentamicin (16.1%), imipenem (22.9%), meropenem (21.5%), ciprofloxacin (19%), levofloxacin (18%) and cefepime (15.1%) was observed. The lowest resistance rates were for tobramycin (10.2%) and amikacin (11.2%). The highest resistance rates were observed for piperacillin (23.9%), imipenem (22.9%) and meropenem (21.5%). Susceptibility testing of 205 P. aeruginosa isolates showed that all isolates were resistant to at least one of the antibiotics and thirty six (18%) of the isolates were multi-resistant. Freqeuncies of resistance of strains against twelve antibiotics were shown in Figure 1.
All 205 P. aeruginosa isolates tested for susceptibility to antibiotics were screened for the presence of class 1 and 2 integrases. Ten isolates (4.8%) were identified as being positive for class 1 integrase. Class 2 integrase was not detected. Seven distinct variable regions were detected among seven isolates with sizes ranging from 1070 bp to 2104 kb. The rest 1.4% of isolates harboured class 1 integron elements without any gene cassette attached. Integron-bearing strains were only detected in the strains from three cities (Trabzon, Sanliurfa and Diyar bakir) (Table 1).
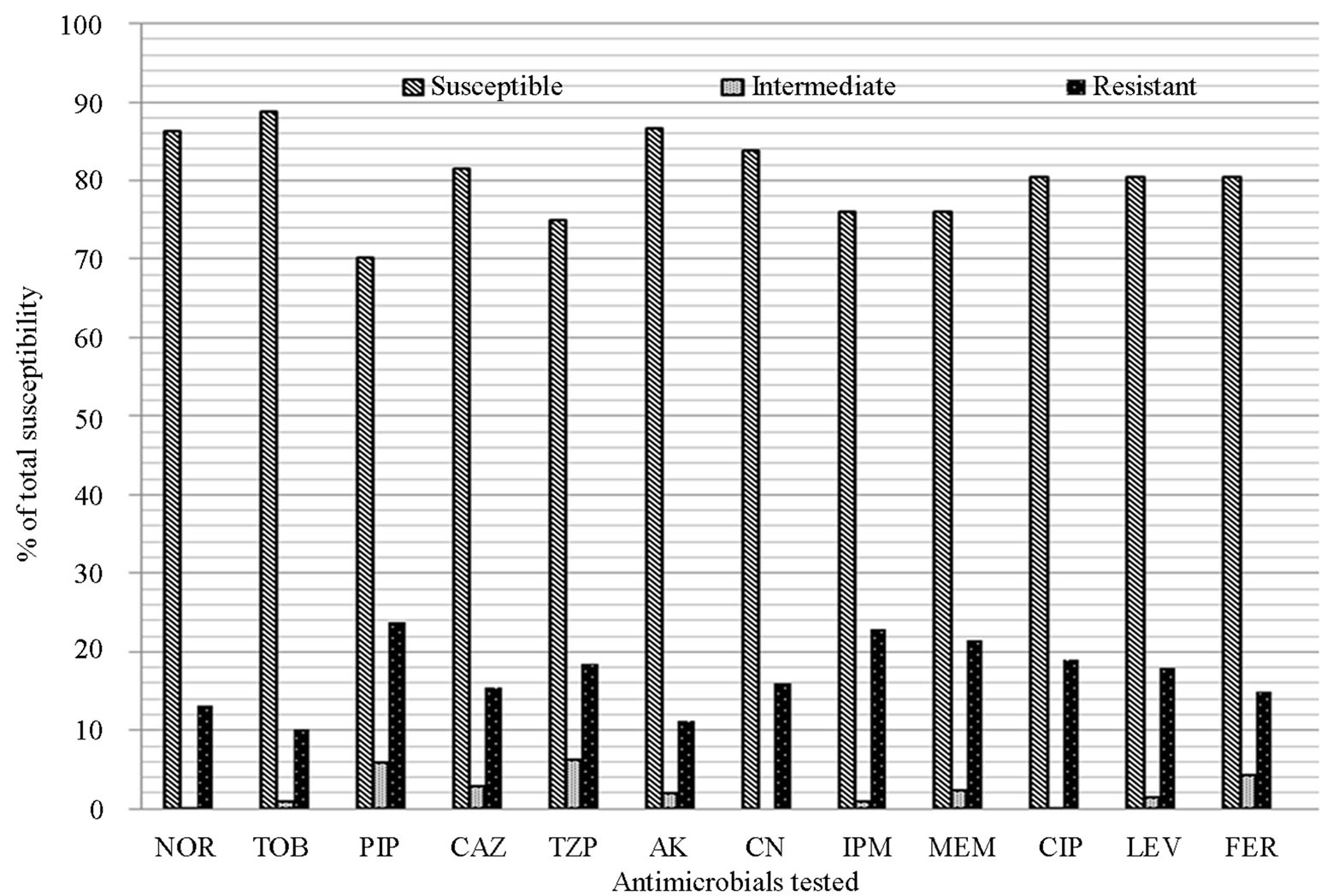
Figure 1. Resistance frequency of the strains against twelve antibiotics.
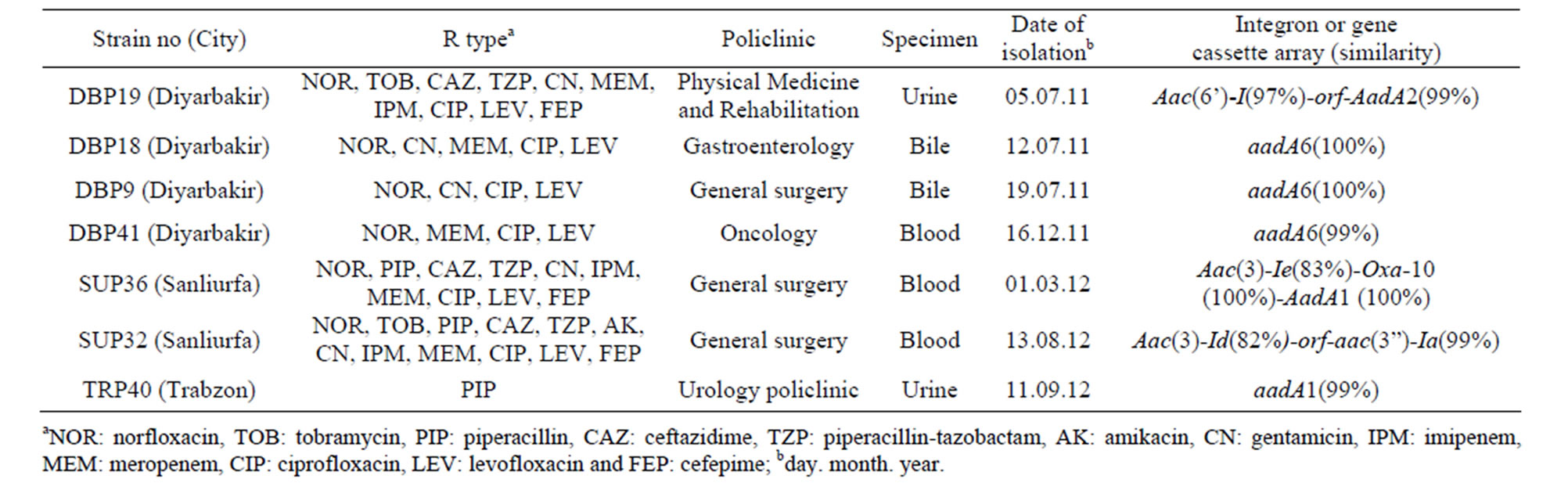
Table 1. Epidemiological data for seven integron-bearing P. aeruginosa strains obtained from six different cities in Turkey.
According to Blast analysis of sequencing results, several genes and gene cassettes were determined in class 1 integrons (Table 1). The genes in integron gene cassettes showed differences in amino acid level from 82% to 100% follows: aadA1 (99%), aac(3)-Id(82%)- orf-aac(3”)-Ia(99%), aac(3)-Ie(83%)-oxa10(100%)-aadA1 (100%), aadA6(100%), aac(6’)-I(97%)-orf-aadA2 (99%).
4. Discussion
Resistance to beta-lactams, quinolones, carbapenems and aminoglycosides are often detected in P. aeruginosa isolates and the rapid spread of antibiotic resistance genes among bacterial isolates is an increasing problem in infectious diseases. Recent studies have shown that resistance genes might have been carried by an integron. Many resistance genes exist as gene cassettes within integrons, which may themselves be located on transmissible plasmids and transposons [36]. Production of an integrinmediated β-lactamases from different molecular classes (carbenicillinases and extended-spectrum β-lactamases belonging to class A, class D oxacillinases and class B carbapenem hydrolysing enzymes) and synthesis of aminoglycoside modifying enzymes (phosphoryl-transferases, acetyltransferases and adenylyltransferases) are some of the resistance mechanisms in P. aeruginosa [37]. In Turkey, integron-borne resistance to various antibiotics is common in P. aeruginosa [38]. However, the prevalence, properties, and roles of integrons and gene cassettes in this species have not yet been studied in a multi-center study.
Class 1 integrons were demonstrated in P. aeruginosa worldwide, and the frequency of class 1 integrons harboured in P. aeruginosa displayed diversity. One study performed from Brazil and the frequency of class 1 integrons was determined 41.5% (44/106). Another study was performed from United Kingdom and, the frequency of class 1 integrons was determined about 60% in A. baumannii isolates. The frequency of class 1 integrons from different studies were determined 43% in Europe (gram-negative isolates), >50% in The Netherlands (Enterobacteriaceae), 59% in France (Enterobacteriaceae), and 52% in Taiwan (Escherichia coli) [36].
Class 1 integrons are the most prevalent among P. aeruginosa isolates from different hospitals of six provinces in Turkey. The screening for integrons in clinical P. aeruginosa isolates (n = 205) from the Turkey showed that 10/205 (4.8%) isolates carried class 1 integrases. However, the analysis of the integron variable region revealed that 3/10 (30%) were lacking in gene cassettes, indicating a low occurrence of empty class 1 integrons among these strains. In comparison, this proportion is highly low to that found in other studies [8,36]. The absence of class 2 integrons among our isolates established the restricted distribution of these two genetic elements among P. aeruginosa isolates.
Genes causing resistance to aminoglycosides and β-lactams are often seen in integrons from Pseudomonas and the family members of Enterobacteriaceae, and aad and aac families are the most common aminoglycoside resistance mechanisms [8]. The most frequent resistance gene found in our isolates was from the aadA family, aacA family followed by blaOXA. OXA β-lactamase family has been found in bacterial isolates from some European countries and Turkey [15]. According to obtained results, aadA1, aac(3)-Id-orf-aac(3”)-Ia, aac(3)-Ie -blaoxa10-aadA1, aadA6, aac(6’)-I-orf-aadA2 genes or gene cassette arrays are seen common in clinical P. aeruginosa isolates in Turkey. To our knowledge, this is the first description of aadA1 and aadA6 gene cassette arrays in P. aeruginosa from Turkey. AadA1 AadA2 and AadA6 proteins are produced by AadA1, AadA2 and AadA6 genes and each proteins confer resistant to streptomycin and spectinomycin [39,40]. In five isolates (TRP40, DBP19, DBP9, DBP18 and DBP14), the presence of class 1 integron located aadA1, aadA2 and aadA6 gene is the reason for antibiotic resistance to streptomycin and spectinomycin [41]. The AAC(3)-I enzymes confer resistance to gentamicin, sisomicin, and fortimicin
(astromicin) and are widespread among Enterobacteriaceae and nonfastidious gram-negative nonfermenters [42]. Classical OXA enzymes confer resistance to carboxypenicillins and ureidopenicillins (Piperacillin) [43]. Piperacillin and gentamicin resistance in SUP36 is associated with class 1 integron gene cassettes. AAC(6’)-I, an enzyme that confers resistance to tobramycin, netilmicin, amikacin, dibekacin, and kanamycin [44]. Tobramycin resistance in DBP19 is associated with Aac-(6’)-I gene cassette.
Sanliurfa and Diyarbakir are located in Southeast Anatolia while Trabzon is located in the Black Sea region. Class 1 integrons found in strains collected from Diyarbakir carried only aadA genes. blaOXA-10 was found only in one strain from Sanliurfa. These findings indicate that selective pressure in different geographic regions causes variations in integron gene content. Each isolate had reflected the same or highly similar gene cassette arrays from geographically distant P. earuginosa. This finding implies that contact with mobile genetic elements such as integrons contributes to the development of this pathogen as multi-drug resistant. This study is the first nationwide multicenter study for class 1 integrons and it indicates the low rate of presence of class 1 integrons in P. aeruginosa in Turkey.
5. Acknowledgements
This work was supported by Recep Tayyip Erdogan University Research Fund Grants BAP-2013.102.03.12 and BAP-2013.102.03.13.
REFERENCES
- G. L. Winsor, D. K. W. Lam, L. Fleming, R. Lo, M. D. Whiteside, N. Y. Yu, R. E. W. Hancock and F. S. L. Brinkman, “Pseudomonas Genome Database: Improved Comparative Analysis and Population Genomics Capability for Pseudomonas Genomes,” Nucleic Acids Research, Vol. 39, 2011, pp. 596-600.
- R. J. Gillis, K. G. White, K. H. Choi, V. E. Wagner, H. P. Schweizer and B. H. Iglewski, “Molecular Basis of Azithromycin-Resistant Pseudomonas aeruginosa Biofilms,” Antimicrobial Agents Chemotherapy, Vol. 49, No. 9, 2005, pp. 3858-3867. http://dx.doi.org/10.1128/AAC.49.9.3858-3867.2005
- J. Patzer, M. A. Toleman, L. M. Deshpande, W. Kaminska, D. Dzierzanowska, P. M. Bennett, R. N. Jones and T. R. Walsh, “Pseudomonas aeruginosa Strains Harbouring an Unusual blaVIM-4 Gene cassette Isolated from Hospitalized Children in Poland (1998-2001),” Journal Antimicrobial Chemotherapy, Vol. 53, No. 3, 2004, pp. 451- 456. http://dx.doi.org/10.1093/jac/dkh095
- J. Sarlangue, O. Brissaud and C. Labreze, “Clinical Features of Pseudomonas aeruginosa Infections,” Archives de Pediatrie, Vol. 13, No. 1, 2006, pp. 13-16.
- P. D. Lister, D. J. Wolter and N. D. Hanson, “Antibacterial-Resistant Pseudomonas aeruginosa: Clinical Impact and Complex Regulation of Chromosomally Encoded Resistance Mechanisms,” Clinical Microbiology Reviews, Vol. 22, No. 4, 2009, pp. 582-610. http://dx.doi.org/10.1128/CMR.00040-09
- http://textbookofbacteriology.net/themicrobialworld/Pseudomonas.html
- J. K. Lutz and J. Lee, “Prevalence and AntimicrobialResistance of Pseudomonas aeruginosa in Swimming Pools and Hot Tubs,” International Journal of Environmental Research and Public Health, Vol. 8, No. 2, 2011, pp. 554-564. http://dx.doi.org/10.3390/ijerph8020554
- E. L. Fonseca, V. V. Vieira, R. Cipriano and A. C. Vicente, “Class 1 Integrons in Pseudomonas aeruginosa Isolates from Clinical Settings in Amazon Region, Brazil,” FEMS Immunology and Medical Microbiology, Vol. 44, No. 3, 2005, pp. 303-309. http://dx.doi.org/10.1016/j.femsim.2005.01.004
- F. Budak, M. Kasap, F. Kolaylı, A. Karadenizli and M. H. Vahaboğlu, “Integron-Associated Resistance Genes among Multidrug-Resistant Pseudomonas aeruginosa Isolated from Clinical Specimens,” Turkish Journal of Medical Science, Vol. 42, No. 1, 2012, pp. 149-156.
- Y. Khosravi, S. T. Tay and J. Vadivelu, “Analysis of Integrons and Associated Gene Cassettes of Metallo- β-Lactamase-Positive Pseudomonas aeruginosa in Malaysia,” Journal of Medical Microbiology, Vol. 60, No. 7, 2011, pp. 988-994. http://dx.doi.org/10.1099/jmm.0.029868-0
- D. Mazel, “Integrons: Agents of Bacterial Evolution,” Nature Reviews. Microbiology, Vol.4, No. 8, 2006, pp. 608-620.
- G. D. Recchia and R. M. Hall, “Gene Cassettes: A New Class of Mobile Element,” Microbiology, Vol. 141, No. 12, 1995, pp. 3015-3027. http://dx.doi.org/10.1099/13500872-141-12-3015
- C. M. Collis and R. M. Hall, “Gene Cassettes from the Insert Region of Integrons Are Excised as Covalently Closed Circles,” Molecular Microbiology, Vol. 6, No. 19, 1992, pp. 2875-2885. http://dx.doi.org/10.1111/j.1365-2958.1992.tb01467.x
- R. M. Hall, D. E. Brookes and H. W. Stokes, “Site-Specific Insertion of Genes into Integrons: Role of the 59- Base Element and Determination of the Recombination Cross-Over Point,” Molecular Microbiology, Vol. 5, No. 8, 1991, pp. 1941-1959. http://dx.doi.org/10.1111/j.1365-2958.1991.tb00817.x
- A. C. Fluit and F. J. Schmitz, “Resistance Integrons and Super-Integrons,” Clinical Microbiology and Infection, Vol. 10, No. 4, 2004, pp. 272-288. http://dx.doi.org/10.1111/j.1198-743X.2004.00858.x
- C. Goldstein, M. D. Lee, S. Sanchez, C. Hudson, B. Phillips, B. Register, M. Grady, C. Liebert, A. O. Summers, D. G. White and J. J. Maurer, “Incidence of Class 1 and 2 Integrases in Clinical and Commensal Bacteria from Livestock, Companion Animals, and Exotics,” Antimicrobial Agents and Chemotherapy, Vol. 45, No. 3, 2001, pp. 723-726. http://dx.doi.org/10.1128/AAC.45.3.723-726.2001
- D. A. Rowe-Magnus, A. M. Guerout and D. Mazel, “Bacterial Resistance Evolution by Recruitment of SuperIntegron Gene Cassettes,” Molecular Microbiology, Vol. 43, No. 6, 2002, pp. 1657-1669. http://dx.doi.org/10.1046/j.1365-2958.2002.02861.x
- A. Carattoli, “Importance of Integrons in the Diffusion of Resistance,” Veterinary Research, Vol. 32, No. 3-4, 2001, pp. 243-259. http://dx.doi.org/10.1051/vetres:2001122
- J. Chen, Z. Su, Y. Liu, S. Wang, X. Dai, Y. Li, S. Peng, Q. Shao , H. Zhang , P. Wen , J. Yu , X. Huang and H. Xu, “Identification and Characterization of Class 1 Integrons among Pseudomonas aeruginosa Isolates from Patients in Zhenjiang, China,” International Journal of Infectious Diseases, Vol. 13, No. 6, 2009, pp. 717-721. http://dx.doi.org/10.1016/j.ijid.2008.11.014
- D. Girlich, T. Naas, A. Leelaporn, L. Poirel, M. Fennewald and P. Nordmann, “Nosocomial Spread of the Integron-Located Veb-1-Like Cassette Encoding an Extended-Spectrum Beta-Lactamase in Pseudomonas aeruginosa in Thailand,” Clinical Infectious Diseases, Vol. 34, No. 5, 2002, pp. 603-611. http://dx.doi.org/10.1086/338786
- K. Lee, J. B. Lim, J. H. Yum, D. Yong, Y. Chong, J. M. Kim and D. M. Livermore, “Bla(VIM-2) Cassette Containing Novel Integrons in Metallo-Beta-Lactamase-Producing Pseudomonas aeruginosa and Pseudomonas putida Isolates Disseminated in a Korean Hospital, “ Antimicrobial Agents and Chemotherapy, Vol. 46, No. 4, 2002, pp. 1053-1058. http://dx.doi.org/10.1128/AAC.46.4.1053-1058.2002
- L. Pallecchi, M. L. Riccio, J. D. Docquier, R. Fontana and G. M. Rossolini, “Molecular Heterogeneity of blaVIM-2- Containing Integrons from Pseudomonas aeruginosa Plasmids Encoding the VIM-2 Metallo-Beta-Lactamase,” FEMS Microbiology Letters, Vol. 195, No. 2, 200, pp. 145-150.
- L. Poirel, T. Lambert, S. Türkoğlu, E. Ronco, J. Gaillard and P. Nordmann, “Characterization of Class 1 Integrons from Pseudomonas aeruginosa that Contain the Blavim-2 Carbapenem-Hydrolyzing β-Lactamase Gene and of Two Novel Aminoglycoside Resistance Gene Cassettes,” Antimicrobial Agents and Chemotherapy, Vol. 45, No. 2, 2001, pp. 546-552. http://dx.doi.org/10.1128/AAC.45.2.546-552.2001
- E. L. Schnabel and A. L. Jones, “Distribution of Tetracycline Resistance Genes and Transposons among Phylloplane Bacteria in Michigan Apple Orchards,” Applied and Environmental Microbiology, Vol. 65, No. 11, 1999, pp. 4898-4907.
- P. Severino and V. D. Magalhaes, “The Role of Integrons in the Dissemination of Antibiotic Resistance among Clinical Isolates of Pseudomonas aeruginosa from an Intensive Care unit in Brazil,” Research in Microbiology, Vol. 153, No. 4, 2002, pp. 221-226. http://dx.doi.org/10.1016/S0923-2508(02)01310-4
- H. Yan, L. Shi, S. Yamasaki, X. Li, Y. Cao, L. Li, L. Yang and S. Miyoshi, “A Plasmidic Class 1 Integron from Five Pseudomonas aeruginosa Clinical Strains Harbored aacA4 and Nonsense-Mutated cmlA1 Gene Cassettes,” Journal of Health Science, Vol. 53, No. 6, 2007, pp. 750-755. http://dx.doi.org/10.1248/jhs.53.750
- S. Yousefi, M. R. Nahaei, S. Farajnia, M. Ghojazadeh, M. T. Akhi, Y. Sharifi, M. Milani and R. Ghostaslou, “Class 1 Integron and Imipenem Resistance in Clinical Isolates of Pseudomonas aeruginosa: Prevalence and Antibiotic Susceptibility,” Iranian Journal of Microbiology, Vol. 2, No. 3, 2010, pp. 113-119.
- K. Hansson, L. Sundström, A. Pelletier and P. H. Roy, “IntI2 Integron Integrase in Tn7,” Journal of Bacteriology, Vol. 184, No. 6, 2002, pp. 1712-1721.
- Z. Xu, L. Li, M. E. Shirtliff, M. J. Alam, S. Yamasaki and L. Shi, “Occurrence and Characteristics of Class 1 and 2 Integrons in Pseudomonas aeruginosa Isolates from Patients in Southern China,” Journal of Clinical Microbiology, Vol. 47, No.1, 2009, pp. 230-234. http://dx.doi.org/10.1128/JCM.02027-08
- National Committee for Clinical Laboratory Standards, “Performance Standards for Antimicrobial Susceptibility testing” NCCLS Antim-Icrobial Susceptibility Testing Standards M100-S22, 2012.
- F. M. Ausubel, R. Brient, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith and J. Struhl, “Short Protocols in Molecular Biology: 2nd ed.,” John Willey and Sons, New York, 1995.
- S. J. Rosser and H. K. Young, “Identification and Characterization of Class 1 Integrons in Bacteria from an Aquatic Environment,” The Journal of Antimicrobial Chemotherapy, Vol. 44, No. 1, 1999, pp. 11-18. http://dx.doi.org/10.1093/jac/44.1.11
- C. Lévesque, L. Piche, C. Larose and P. H. Roy, “PCR Mapping of Integrons Reveals Several Novel Combinations of Resistance Genes,” Antimicrobial Agents and Chemotherapy, Vol. 39, No. 1, 1995, pp. 185-191. http://dx.doi.org/10.1128/AAC.39.1.185
- J. Sambrook, E. F. Fritsch and T. Maniatis, “Molecular Cloning. A Laboratory Manual,” Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 1989.
- S. F. Altschul, T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller and D. J. Lipman, “Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs,” Nucleic Acids Research, Vol. 25, No. 17, 1997, pp. 3389-3402. http://dx.doi.org/10.1093/nar/25.17.3389
- B. Gu, M. Tong, W. Zhao, G. Liu, M. Ning, S. Pan and W. Zhao, “Prevalence and Characterization of Class I Integrons among Pseudomonas aeruginosa and Acinetobacter baumannii Isolates from Patients in Nanjing, China,” Journal of Clinical Microbiology, Vol. 45, No. 1, 2007, pp. 241-243. http://dx.doi.org/10.1128/JCM.01318-06
- T. Strateva and D. Yordanov, “Pseudomonas aeruginosa —A Phenomenon of Bacterial Resistance,” Journal of Medical Microbiology, Vol. 58, No. 9, 2009, pp. 1133- 1148. http://dx.doi.org/10.1099/jmm.0.009142-0
- O. B. Ozgumus, R. Caylan, I. Tosun, C. Sandalli, K. Aydin and I. Koksal, “Molecular Epidemiology of Clinical Pseudomonas aeruginosa Isolates Carrying the IMP-1 Metallo-β-Lactamase Gene in a University Hospital in Turkey,” Microbial Drug Resistance, Vol. 13, No. 3, 2007, pp. 191-198. http://dx.doi.org/10.1089/mdr.2007.748
- K. Poonsuk, C. Tribuddharat and R. Chuanchuen, “Class 1 Integrons In Pseudomonas Aeruginosa And Acinetobacter Baumannii Isolated from Clinical Isolates,” The Southeast Asian Journal of Tropical Medicine Public Health, Vol. 43, No. 2, 2012, pp. 376-384.
- A. Bito and M. Susani, “Revised Analysis of aadA2 Gene of Plasmid pSa,” Antimicrobial Agents Chemotherapy, Vol. 38, No. 5, 1994, pp. 1172-1175. http://dx.doi.org/10.1128/AAC.38.5.1172
- T. Naas, L. Poirel and P. Nordmann, “Molecular Characterisation of In51, a Class 1 integron Containing a Novel Aminoglycoside Adenylyltransferase Gene Cassette, aadA6, in Pseudomonas aeruginosa,” Biochimica et Biophysica Acta, Vol. 1489, No. 2-3, 1999, pp. 445-451. http://dx.doi.org/10.1016/S0167-4781(99)00202-X
- M. L. Riccio, J. D. Docquier, E. Dell’Amico, F. Luzzaro, G. Amicosante and G. M. Rossolini, “Novel 3-N-Aminoglycoside Acetyltransferase Gene, aac(3)-Ic, from a Pseudomonas aeruginosa Integron,” Antimicrobial Agents and Chemotherapy, Vol. 47, No. 5, 2003, pp. 1746-1748. http://dx.doi.org/10.1128/AAC.47.5.1746-1748.2003
- A. E. Sorour, I. E. Wali and S. K. El-Hodaky, “OXAType-Beta-Lactamases among Extended-Spectrum-Cephalosporin Non-Susceptible Pseudomonas Aeruginosa Isolates Collected from a Large Teaching Hospital in Cairo,” Egyptian Journal of Medical Microbiology, Vol. 17, No. 4, 2008, pp. 565-572.
- G. H. Miller, F. J. Sabatelli, R. S. Hare, Y. Glupczynski, P. Mackey, D. Shlaes, K. Shimizu and K. J. Shaw, “The Most Frequent Aminoglycoside Resistance Mechanisms —Changes with Time and Geographic Area: A Reflection of Aminoglycoside Usage Patterns?” Clinical Infectious Diseases, Vol. 24, No. 1, 1997, pp. 46-62. http://dx.doi.org/10.1093/clinids/24.Supplement_1.S46
NOTES
*Corresponding author.

