CellBio
Vol.2 No.3(2013), Article ID:37008,6 pages DOI:10.4236/cellbio.2013.23014
Changes in the Nuclei of Infected Cells at Early Stages of Infection with EMCV
Institute of Molecular Biology of the National Academy of Sciences of the Republic of Armenia, Laboratory of Cell Biology, Yerevan, Armenia
Email: *zkaralyan@yahoo.com
Copyright © 2013 Zaven A. Karalyan et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received July 17, 2013; revised August 5, 2013; accepted August 12, 2013
Keywords: EMCV; Acidic Proteins; RNA; Nucleolus; Nucleus
ABSTRACT
By the methods of quantitative cytophotometry, we have identified the changes in the nucleus and of some intranuclear compartments in the early stages of infection with encephalomyocarditis virus (EMCV). They can be characterized as early 1 - 2 hours post infection (hpi) and temporary increase (duration about 1 hour) in the content of the acidic proteins of the nucleolus, changing their decline to the control values. Then (after 1 - 2 hours) follows an increase in RNA content of nucleoli to 4 hours post infection (the process takes about 2 hours). The increase in RNA content in nucleoli is in approximately the same time (slightly behind) with the activation of PML bodies (2 - 4 hpi). Then, the RNA content in nucleoli decreased to the control values, while simultaneously decreasing activity of PML bodies (ranging from 5 - 6 hpi). The early stages of infection EMCV are also characterized by the tendency to increase in the size of the nuclei of infected cells, and preserve at a later time. Then there is an increase in RNA content in the nucleus, roughly coinciding with the increased content of RNA in the nucleoli.
1. Introduction
The early stages of the replication of picornaviruses represent an extreme interest. This is due to a relatively short period of time that the viruses will start their own replication, but also establish control over a number of important cellular metabolic processes. The result of this phenomenon is a radical restructuring of the entire physiology of the infected cell. Part of the mechanism of this transformation has been clear for a long time, while others have only recently been clarified, but much of it is still unknown.
It is well known that picornaviruses are able to block cap-dependent translation. Since picornaviral translation is cap-independent by virtue of the 5 IRES, many of these viruses have evolved potent mechanisms to inhibit cellular cap-dependent translation during infection, thereby thwarting detrimental antiviral responses. The enteroviruses and aphthoviruses, for example, encode secondary proteases at their 2A and L positions respectively, which target eIF4G [1]. Cardioviruses do not have secondary proteases. Their L and 2A proteins have essential host shut-off roles, but use non-proteolytic mechanisms to achieve them. The EMCV L (67 aa) contributes to the inhibition of cap-dependent translation by triggering disruption of nucleocytoplasmic trafficking during infection. For the disruption of cap-dependent translation of the host cells with EMCV, 2A protein is responsible. As shown by [1], 2A NLS sequence is required for virus shutoff of cap-dependent host protein synthesis.
It has been shown that proteins of EMCV-the 2A, 3B (VPg), 3C (pro) and 3D (pol) can also be found within the nucleoli. The localization of these proteins occurs in the first 2 - 4 hours following infection of cells [2,3].
Therefore, it is important to study the influence of the picornaviruses on the nuclear structure of infected cells. As it is well known, the replication of picornaviruses occurs within the cytoplasm. However, at the earliest stages of viral infection many viral proteins are observed in the nucleus of affected cells. The research reported in this paper was directed towards determining the DNA, RNA and acidic proteins (non histone) as well as PML bodies in nuclei of EMCV infected cells.
2. Materials and Methods
2.1. Cells
SK-N-MC human neuroblastoma cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Sigma) supplemented with 10% heat-inactivated fetal bovine serum (FBS) at 37˚C in 5% CO2.
2.2. Virus
EMCV (Columbia-SK strain) was used at multiplicity of infection 105 TCD50/ml on SK-N-MC. Viral titers were calculated by the method of Kärber. As a control the parallel conducted passages of noninfected cultures were used.
2.3. Image Cytophotometry
In order to quantitative DNA analyze of the received data, the cells preparations were fixed in 96% ethyl alcohol for 30 minutes and painted in fresh Shiffs reactive, by Feulgen (hydrolyze 5N HCL 60 minutes at 22˚C). The content of DNA in a nucleus and nucleolus was defined by computer-equipped microscope-photometer SMP 05 (OPTON). The image cytophotometry of DNA was performed on 575 nm wave [4]. Unstimulated human lymphocytes were used as diploid standards.
For quantification of RNA was used gallocyanin chromalum stain. To obtain reproducible staining results with these large sections, the method of Einarson was adapted to quantitative [5] and image analytical requirements. The image cytophotometry was performed on 610 nm wave. In each case controls were evaluated as 100%.
Fast green FCF staining (for acidic proteins) was used in Deitch modification [6,7]. The image cytophotometry was performed on 434 nm wave. In each case controls were evaluated as 100%.
2.4. Determination of the PML
Cells grown on glass cover slips were fixed in 4% paraformaldehyde/PBS (pH 7.5) for 5 minutes at room temperature, permeabilized in 0.5% Triton X-100 in PBS for 5 minutes at room temperature. PML protein was visualized with the monoclonal antibody. The determination of the PML was performed using monoclonal antibodies “PML PG-M3, Santa Cruz Biotechnology Inc”-catalogue no. sc-966 FITC [8].
2.5. Statistics
All experiments were conducted in triplicate. The significance of virus-induced changes was evaluated by two-tailed Student’s t-test. p values < 0.05 were considered significant. SPSS version 15.0 software package (SPSS Inc., Chicago, IL, USA) was used for statistical analyses.
3. Results
The lytic EMCV infection was received by virus introduction on 48 h confluences of SK-N-MC culture. At 8 hours post infection (hpi) the virus titer reached 3.0 lg TCD50/ml, at 12-4.0, at 24-4.5, at 24-72-6.5 (Figure 1). The time-course of a single cycle of EMCV reproduction in SK-N-MC cells take place about 8-12 h.
The average content of the nucleolus to a nucleus does not change during the whole period of the experiment with the exception of some minuscule increases of the content of nucleolus in a nucleus (2.5-control, 2.9-at 6 hpi). The cytometry of the nucleolus values revealed the following results: in the whole there are not definite changes, but there may an emphasized tendency to increase the given value up to the 6 hpi. The total area of the nucleoli increases almost by 35%, but due to large variations in individual performance the difference is not reliable.
Thus, when the infection with picornaviruses occurs the activation of the nucleolar values is detected. It has a temporary nature—first of all increases in the synthesis of the acidic proteins and after that an increase in the content of the RNA and the area of the nucleoli occurs.
The quantitative indicators of DNA of the nucleus do not differ from the control values throughout the experiment, nor in the DNA content and neither in the distribution of DNA in ploidy classes.
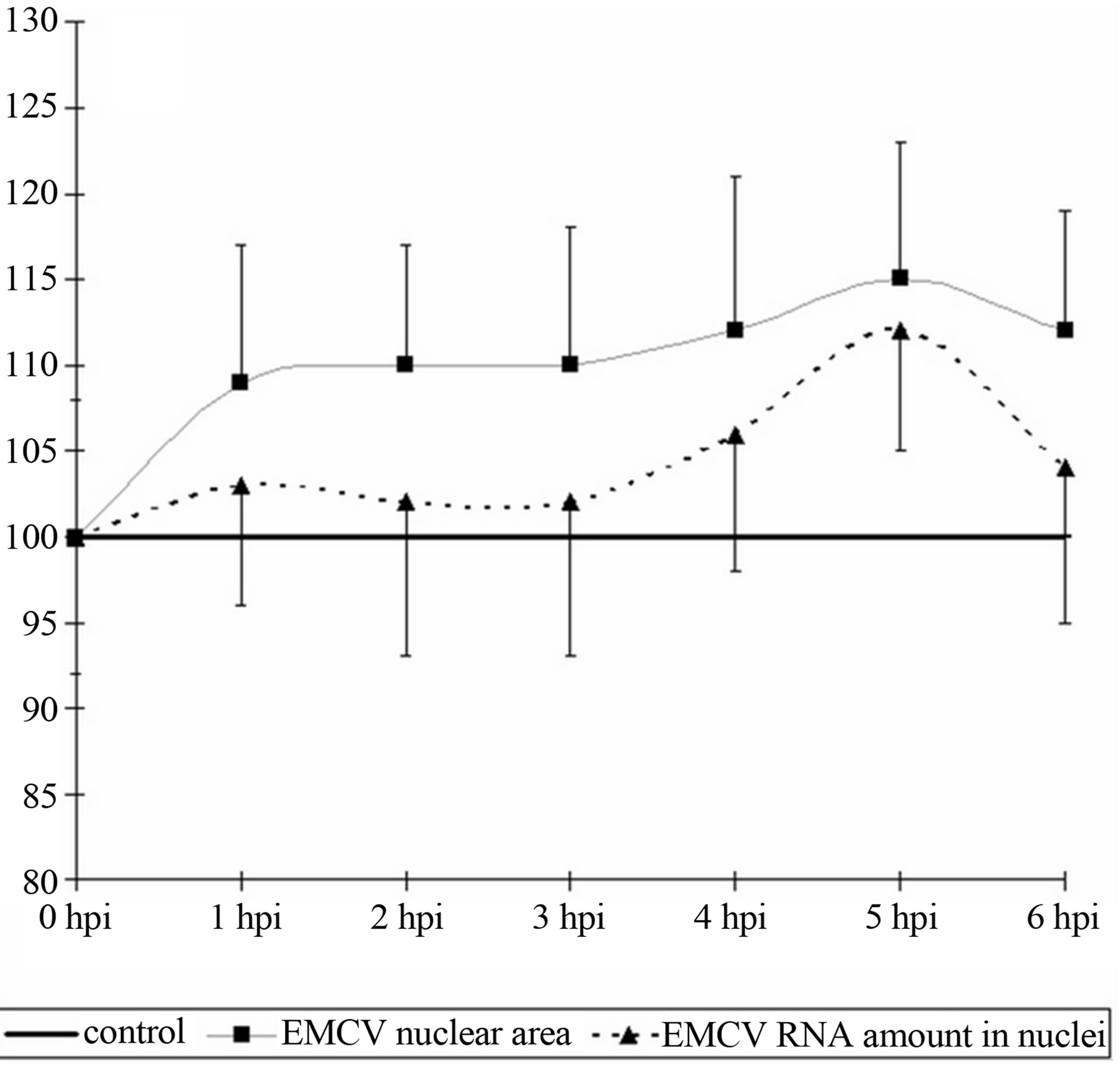
The size of the nucleus tends to increase in comparison with the control (p < 0.1), by 1 hpi and remain so until the end of the observation period (6 hpi) (Figure 2(a)). In addition the increase in the size of the nucleus is small and varies from 8% - 11% from the baseline. It is important to note that the increase in the size of the nuclei of infected cells is not accompanied by an increase in the RNA or acidic proteins of the nucleus. However, if the content of acidic proteins of the nucleus remained in a relatively stable index throughout the study, the RNA has a tendency (t = 1.8, p < 0.1) increased by 5hpi, and coin-
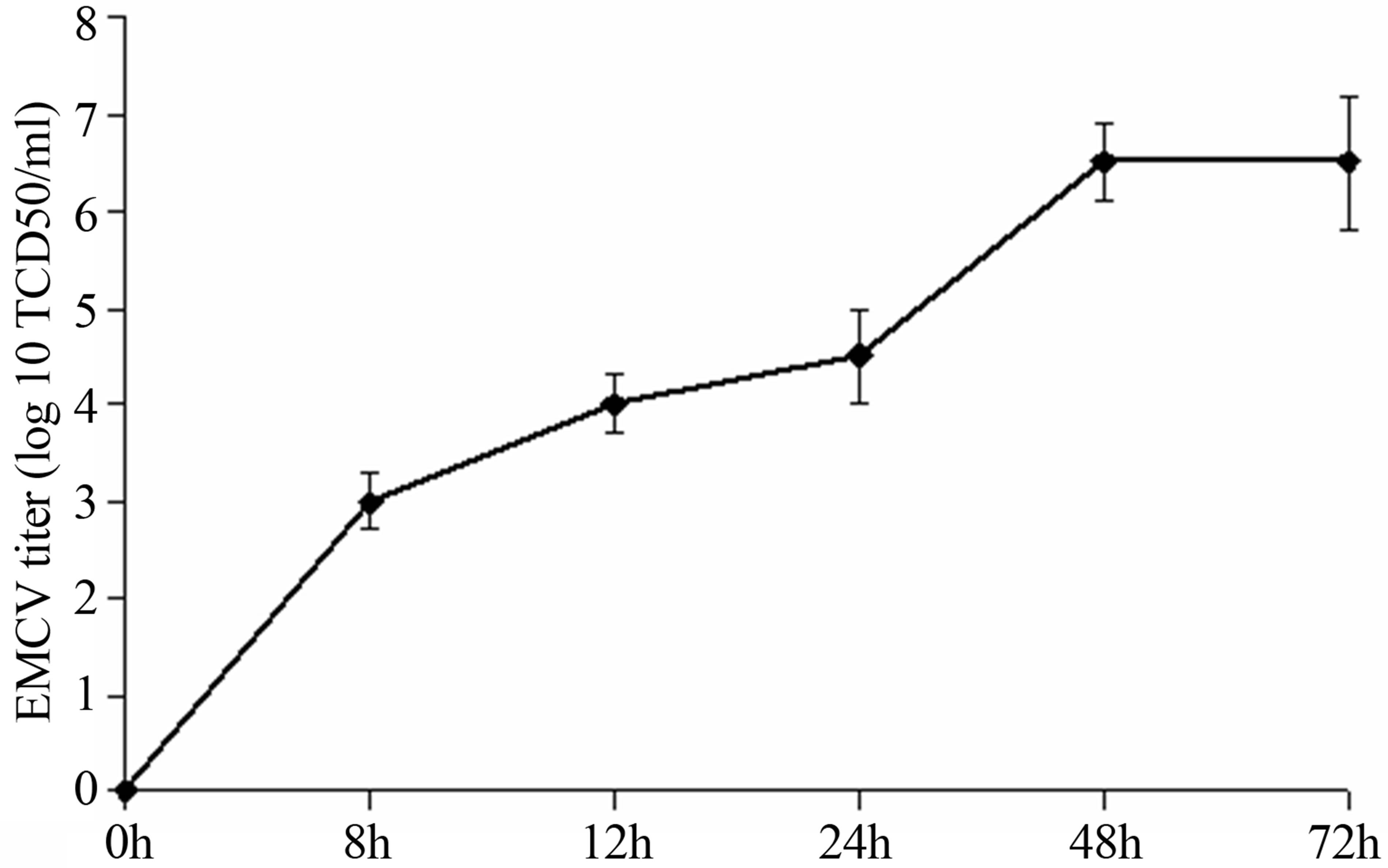
Figure 1. The titer of EMCV calculated on SK-B-MC cells.
 (a)
(a)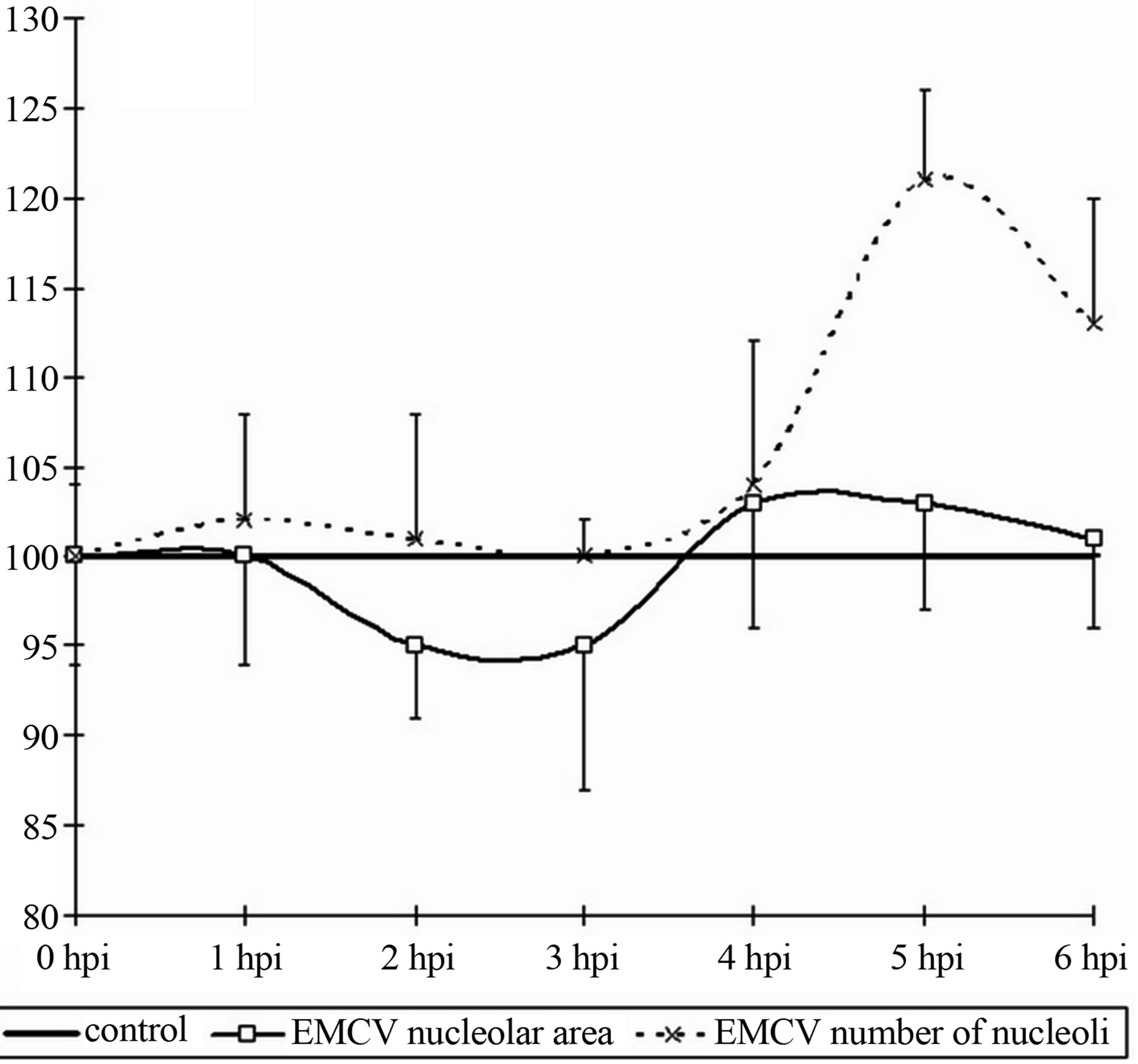 (b)
(b)
Figure 2. Dynamics of nuclear (a) and nucleolar (b) indices under the influence of EMCV infection. Data show percentage of control levels (average of 300 cells = 100%).
cides with an increase in RNA in the nucleoli (Figure 2(b)).
As it follows from the Figure 3, in the early stages of the infection, there is an increase in the content (amount, concentration) of the nucleolar acidic proteins (with a 1- 2 hpi) (t = 3.07, p < 0.05). The elevated level of acidic proteins has a short-period character, and after 1 hour (to 3 hpi) its levels in the nucleolus does not differ from the benchmarks. The increased levels of the acidic proteins vary between 20% - 25% of the initial content.
The RNA in the nucleoli of the infected and control cells do not differ from each other, up to 4 hpi, when an
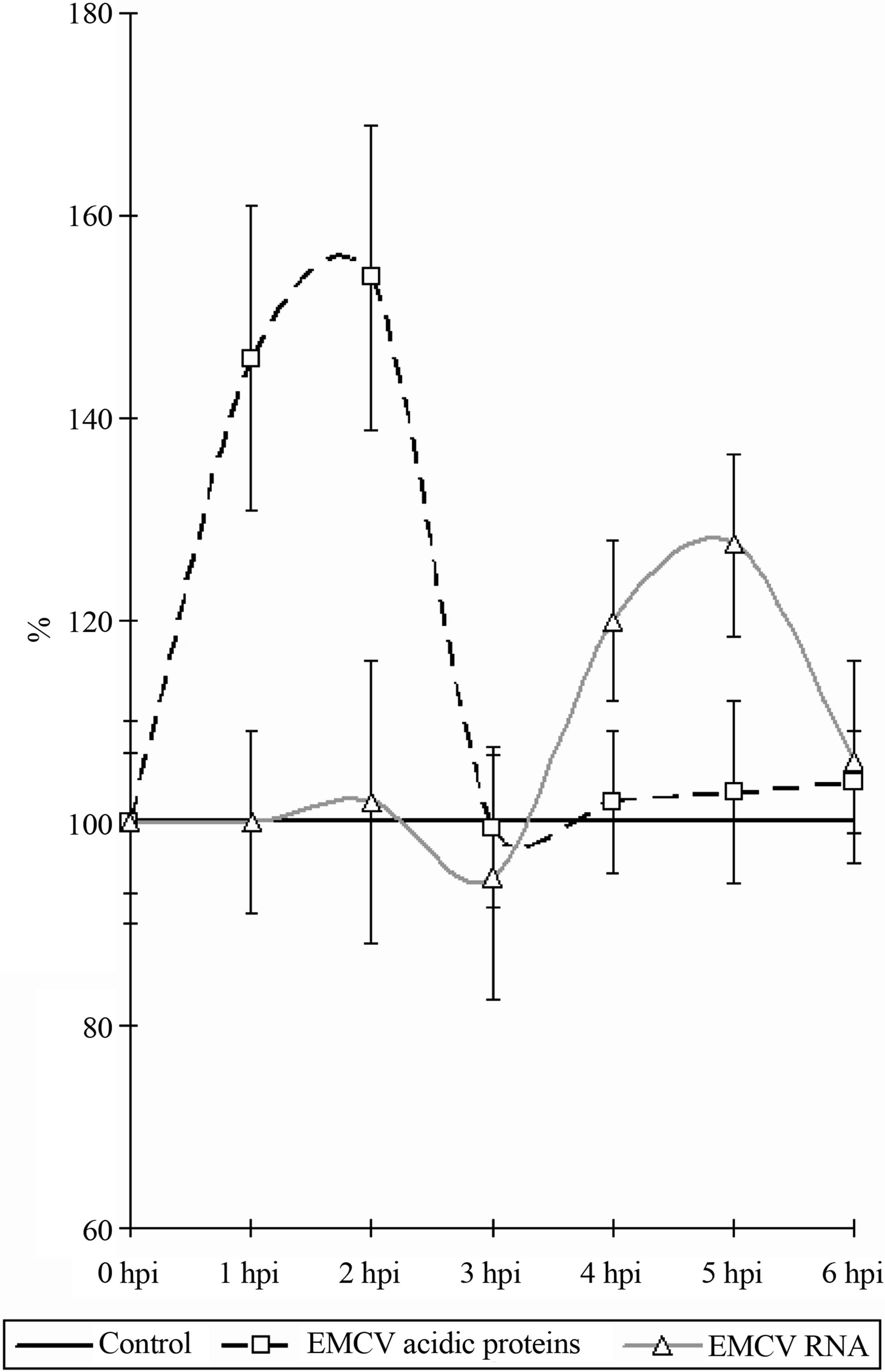
Figure 3. Dynamics of nucleolar RNA and acidic proteins under the influence of EMCV infection. Data show percentage of control levels (average of 300 cells = 100%).
increase in the RNA content in the nucleoli of infected cells begins to be observed (Figure 3) (t = 2.17, p < 0.05). The increase of the RNA in the nucleolus reaches 30% - 35% in comparison with the control values. The increase of the RNA as long with the proteins has a short period character and finishes after 5 hpi, decreasing down to the levels of the control values (6 hpi).
The intensity of the luminescence PML bodies, in the nuclei of the infected cells does not differ from the background values in the range from 1 to 2 hpi. Then there is a sharp increase in the intensity of luminescence, which indicates about the increased activity of PML bodies by 3 hpi. Increased activity of PML bodies completely fading away to a 4 HPI, after which it does not differ from the control values (Figure 4).
We have identified the changes in the nucleus and of some intranuclear compartments in the early stages of infection EMCV.
They can be characterized as early (1 - 2 hpi) and temporary increase (duration about 1 hour) in the content of the acidic proteins of the nucleolus, changing their
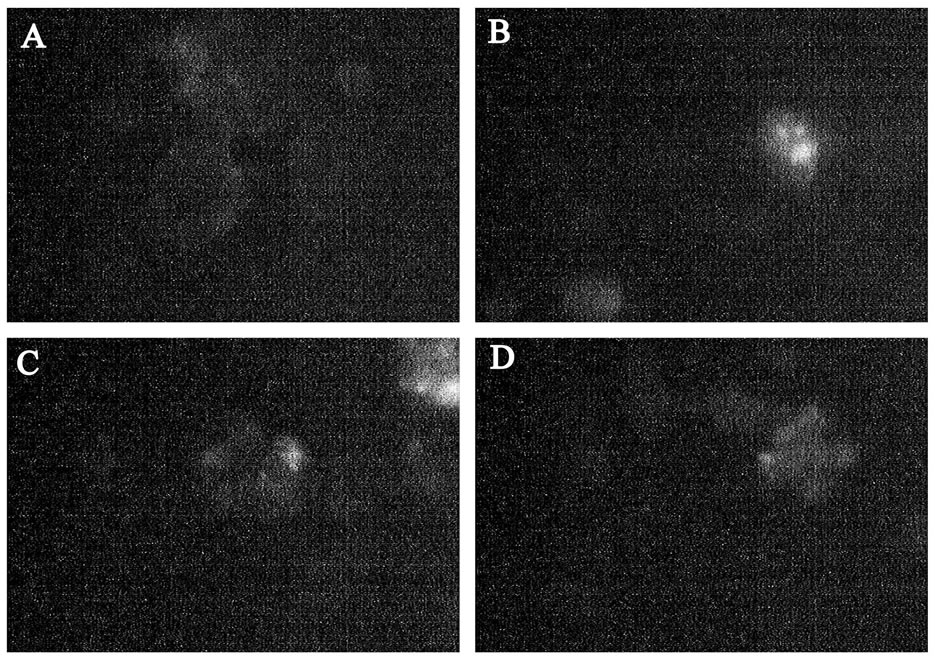
Figure 4. PML bodies in EMCV infected SK-N-MC cells. (A) PML bodies in control nuclei of SK-N-MC cells; (B) PML bodies in infected nuclei of SK-N-MC cells (3 h.p.i). (C) PML bodies in infected nuclei of SK-N-MC cells (6 h.p.i. EMCV); (D) PML bodies in infected nuclei of SK-N-MC cells (9 h.p.i. EMCV) (×400).
decline to the control values. Then (after 1 - 2 hours) follows an increase in RNA content of nucleoli to 4 hpi (the process takes about 2 hours). The increase in RNA content in nucleoli in approximately the same time (slightly behind) with the activation of PML bodies (2 - 4 hpi). Then, the RNA content in nucleoli decreased to the control values, while simultaneously decreasing activity of PML bodies (ranging from 5 - 6 hpi). The early stages of infection EMCV is also characterized by the tendency to increase in the size of the nuclei of infected cells, and preserves at a later time. Then there is an increase in RNA content in the nucleus, roughly coinciding with the increased content of RNA in the nucleoli.
The total content of acidic proteins of the nucleus is a relatively stable index, changing a little both in the control and in the early stages of infection EMCV.
4. Discussion
For EMCV is found the involvement of the viral proteins in the nuclear processes. Already in the early stages of infection (2 - 3 hpi) cardiovirus protein 2A, as well as proteins 3BVpg, 3Cpro, 3Dpolas a single precursor 3BCD are located in the nucleoli of the cells, where the synthesis of rRNA and ribosome assembly take place (Aminev et al., 2003a, Aminev et al., 2003b). The function of the cardiovirus proteins in the nucleus of infected cells have not been studied sufficiently. Protease 3Spro presumably responsible for the inhibition of synthesis of cellular mRNA, whereas the work of RNA polymerases I and III in cardiovirus infection are not inhibited [3]. The function performed by the protein 2A of cardiovirus in the nucleoli of the cells is not completely clear. Amines and colleagues suggest that the protein 2A in the nucleoli embedded in the mature ribosomal subunits, resulting in formation of modified ribosomes engaged mainly in the cytoplasm of virus-specific protein synthesis [2]. As shown by us, at this stage is a series of important structural and functional changes in the nucleoli of infected cells.
With regard of the above said the increased content of nucleolar acidic proteins in early stages of infection EMCV could be explained by two reasons. The first is the accumulation of the viral proteins and particularly proteins 2A, 3BVpg, 3Cpro, 3Dpol and its predecessor 3BCD. The second - the accumulation of nucleolar proteins with enzymatic activity (discussed below).We have identified the data which allow us to assert that in the early stages of infection (2 - 4 hpi) significant changes occur in the nucleoli, coinciding with the localization of viral proteins in them. These effects disappear in the later stages of infection (6 hpi and later). This suggests a short-term increase in the functional activity of the nucleolus [9] under the influence of infection EMCV. In addition to changes in the nucleoli, this process is accompanied by an increase in the PML bodies. PML bodies-small spherical domains are present in the nucleus of cells, they undergo morphological changes during the cell cycle. Their number is intensely variable, depending on the physiological state of the cells, cell cycle stage in viral infections and so on. The PML bodies are destroyed during some viral infections.
Incubation of the cells with IFN induces the synthesis of the PML-protein and inhibits the multiplication of the viruses. These cells are destroyed during a viral infection and for the replication of adenovirus DNA their destructtion is a necessary step that underlines the possible involvement of the corpuscles to provide antiviral activity of cells. Incubation of the cells with interferon induces the synthesis of PML-protein and inhibits proliferation of some viruses [10]. Upon infection of cells with the virus EMCV, it is shown a decrease in the number of the PML bodies, under the influence of the viral 3C protease. As the mechanism of reduction of PML bodies, the authors consider a possible direct degradation of the PML bodies 3C protease EMCV [11]. Thus, the sharp decrease in emission intensity of monoclonal antibodies to PML, to 5.6 HPI, is explained by the influence of non-structural viral proteins (3Cprotease), and aimed at suppressing IFN-mediated protection of the infected cell.
We have also investigated the acidic proteins of chromatin and the nucleolus in a control experiment and under the influence of the virus. Acidic proteins of the chromatin play an important role in the regulation of the genetic activity. It was shown that in proliferating tissues contain more acidic proteins than that of resting, and these proteins are richer with euchromatin than with heterochromatin. The acidic proteins restore the DNA-dependent RNA synthesis, inhibited by histone, which enhance the transcription of chromatin in vitro and the activation of genes. Acidic proteins, in contrast to the histones are tissue specific and bind to DNA only within the tissue from which they are allocated [12,13]. Taking into account of our data it should be concluded that the activation of the transcription processes are in the range of 2 - 4 HPI.
Nucleolar acidic proteins—are a large group of proteins associated with the functional activity of the nucleolar proteins and the equivalent argentophilic proteins [14]. The most important argentophilic proteins: RNA polymerase I, transcription factor UBF, nucleolin (C-23), nucleophosmin (nyumatrin or B-23), etc. In contrast to the widespread methods of silver protein, the quantitative cytochemical determination of acid proteins, allows us more accurate identify variations in protein content, and hence the functional activity of the nucleolus [15-17]. The role of these proteins in the replication of EMCV is unquestionable, since it is next to nucleophosmin (B-23) are localized EMCV proteins-the 2A, 3B (VPg), 3C (pro) and 3D (pol).
It should be noted that the EMCV does not need nuclear structures sensitive cells, as it is able to replicate in a cell-free medium containing the individual components of the damaged cells in the lysates [18]. Consequently, for successful replication the virus does not require modification of nucleolar apparatus.
We can therefore conclude that any changes in the nucleoli are the result of the reaction of cells to viral infection or virus modifies the activity of the nucleolus to the successful suppression of cellular antiviral mechanisms.
In our experiments, an elevated level of the acidic proteins in the nucleoli was preceded by the increased content of RNA. The time gaps were approximately 1 - 2 hours. RNA synthesis in the nucleoli is directly related to the formation of the nucleoli. It should be noted that in the temporal boundaries the biosynthesis of the nucleoli fits into a space equivalent to about 1 hour. 7 - 10 minutes is the transcription of the rDNA, the synthesis of the small subunit of the ribosome—15 - 30 min, the synthesis of the large subunit of the ribosome—20 - 40 minutes [19]. It is possible that identified by us the time difference in the content of the acidic proteins and RNA in the nucleolus is associated with the time of the biosynthesis of ribosomes.
REFERENCES
- R. Groppo, B. A. Brown and A. C. Palmenberg, “Mutational Analysis of the EMCV 2A Protein Identifies a Nuclear Localization Signal and an eIF4E Binding Site,” Virology, Vol. 410, No. 1, 2011, pp. 257-267. doi:10.1016/j.virol.2010.11.002
- A. G. Aminev, S. P. Amineva and A. C. Palmenberg, “Encephalomyocarditis Viral Protein 2A Localizes to Nucleoli and Inhibits Cap-Dependent mRNA Translation,” Virus Research, Vol. 95, No. 1-2, 2003, pp. 45-57. doi:10.1016/S0168-1702(03)00162-X
- A. G. Aminev, S. P. Amineva and A. C. Palmenberg, “Encephalomyocarditis Virus (EMCV) Proteins 2A and 3BCD Localize to Nuclei and Inhibit Cellular mRNA Transcription but Not rRNA Transcription,” Virus Research, Vol. 95, No. 1-2, 2003, pp. 59-73. doi:10.1016/S0168-1702(03)00163-1
- A. D. Deich, “Introduction to Quantitative Cytochemistry,” Academic Press New York/London, 1966, pp. 65-67.
- W. Sandritter, G. Kifer and W. Rik, “Gallocyaninchromalum Stain,” In: G. L. Wied, Ed., Introduction to Quantitative Cytochemistry, Academic Press, New York and London, 1966, pp. 153-170.
- A. C. Dhar and C. K. Shah, “Cytochemical Method to Localize Acidic Nuclear Proteins,” Stain Technology, Vol. 57, No. 3, 1982, pp. 151-155.
- B. R. Zirkin, “A cytochemical Study of the Nonhistone Protein Content of Condensed and Extended Chromatin,” Experimental Cell Research, Vol. 78, No. 2, 1973, pp. 394-398. doi:10.1016/0014-4827(73)90084-0
- C. C. Yin, A. B. Glassman, P. Lin, J. R. Valbuena, D. Jones, R. Luthra and L. J. Medeiros, “Morphologic, Cytogenetic, and Molecular Abnormalities in TherapyRelated Acute Promyelocytic Leukemia,” American Journal of Clinical Pathology, Vol. 123, No. 6, 2005, pp. 840- 848. doi:10.1309/TJFFK819RPCLFKJ0
- C. C. Morton, J. A. Brown, W. M. Holmes, W. E. Nance and B. Wolf, “Stain Intensity of Human Nucleolus Organizer Region Reflects Incorporation of Uridine in to Mature Ribosomal RNA,” Experimental Cell Research, Vol. 145, No. 2, 1983, pp. 405-413. doi:10.1016/0014-4827(83)90019-8
- H. Zakaryan and T. Stamminger, “Nuclear Remodelling during Viral Infections,” Cellular Microbiology, Vol. 13, No. 6, 2011, pp. 806-813. doi:10.1111/j.1462-5822.2011.01596.x
- B. El McHichi, T. Regad, M. A. Maroui, M. S. Rodriguez, A. Aminev, S. Gerbaud, N. Escriou, L. Dianoux and M. K. Chelbi-Alix, “SUMOylation Promotes PML Degradation during Encephalomyocarditis Virus Infection,” Journal of Virology, Vol. 84, No. 22, 2010, pp. 11634-11645. doi:10.1128/JVI.01321-10
- R. Baserga and G. Stein, “Nuclear Acidic Proteins and Cell Proliferation,” Federation Proceedings, Vol. 30, No. 6, 1971, pp. 1752-1759.
- G. Stein and R. Baserga, “Cytoplasmic Synthesis of Acidic Chromosomal Proteins,” Biochemical and Biophysical Research Communications, Vol. 44, No. 1, 1971, pp. 218-223.
- F. Thiebaut, J. P. Rigaut and A. Reith, “Improvement in the Specificity of the Silver Staining Technique for AgNORAssociated Acidic Proteins in Paraffin Sections,” Stain Technology, Vol. 59, No. 3, 1984, pp. 181-188.
- M. Okuwaki, “The Structure and Functions of NPM1/ Nucleophsmin/B23, a Multifunctional Nucleolar Acidic Protein,” The Journal of Biochemistry, Vol. 143, No. 4, 2008, pp. 441-448. doi:10.1093/jb/mvm222
- E. A. Sorokina, J. A. Wesson and J. G. Kleinman, “An Acidic Peptide Sequence of Nucleolin-Related Protein Can Mediate the Attachment of Calcium Oxalate to Renal Tubule Cells,” Journal of the American Society of Nephrology, Vol. 15, No. 8, 2004, pp. 2057-2065. doi:10.1097/01.ASN.0000133024.83256.C8
- R. Voit, A. Kuhn, E. E. Sander and I. Grummt, “Activation of Mammalian Ribosomal Gene Transcription Requires Phosphorylation of the Nucleolar Transcription Factor UBF,” Nucleic Acids Research, Vol. 23, No. 14, 1995, pp. 2593-2599. doi:10.1093/nar/23.14.2593
- Y. V. Svitkin and N. Sonenberg, “Cell-Free Synthesis of Encephalomyocarditis Virus,” Journal of Virology, Vol. 77, No. 11, 2003, pp. 6551-6555. doi:10.1128/JVI.77.11.6551-6555.2003
- A. S. Stoykova, M. D. Dabeva, R. N. Dimova and A. A. Hadjiolov, “Ribosome Biogenesis and Nucleolar Ultrastructure in Neuronal and Oligodendroglial Rat Brain Cells,” Journal of Neurochemistry, Vol. 45, No. 6, 1985, pp. 1667-1676. doi:10.1111/j.1471-4159.1985.tb10521.x
NOTES
*Corresponding author.

