Open Journal of Animal Sciences
Vol.3 No.1(2013), Article ID:27074,9 pages DOI:10.4236/ojas.2013.31004
Association between microsatellite markers and bovine tuberculosis in Chadian Zebu cattle
![]()
1ReproGen—Animal Bioscience Group, Faculty of Veterinary Science, University of Sydney, Sydney, Australia
2CSIRO Animal, Food and Health Sciences, Australian Tropical Science and Innovation Precinct, James Cook University, Townsville, Australia
3Division of Genetics and Bioinformatics, Department of Veterinary Clinical and Animal Sciences, Faculty of Health and Medical Sciences, University of Copenhagen, Frederiksberg, Denmark; *Corresponding Author: hajak@sund.ku.dk
Received 13 September 2012; revised 14 October 2012; accepted 21 October 2012
Keywords: Bovine Tuberculosis; Breeding Disease-Resistant Cattle; Genetic Markers; Mycobaterium bovis; African Cattle
ABSTRACT
Bovine tuberculosis (BTB) is a considerable threat to livestock keepers and public health in many developing and underdeveloped countries. We investigated associations between 20 microsatellite markers and three phenotypes for BTB in a sample of the Chadian cattle population. The phenotypes measured were: 1) single intra-dermal comparative cervical tuberculin test (SICCT) performed on live animals, 2) presence of lesion post-slaughter, 3) a bacteriological tissue culture test for Mycobacterium bovis using the samples with observed lesions and 4) a predicted Bayesian model (BM) estimate of a true BTB disease status using all tested animals. All traits were recorded in binary form and as either 1 = presence or 0 = absence. A total of 224 animals for SICCT, lesion and BM traits and 96 animals with bacteriological culture test were genotyped. Generalised linear models were fitted to the binary BTB phenotypes that consisted of age (covariate), sex (2 levels), breed (2 levels) and markers (alleles: 5 - 14 levels) as explanatory variables and implemented in R using glm with a logit link function. The model was fitted for each marker, separately. Six out of 20 markers tested were significantly associated with at least one trait considered; these were ILSTS005, ILSTS006, TGLA227, BM2113 and CSRM66. Genomic regions around these markers may serve as a basis for further functional investigations. This is the first study to report association of microsatellite markers with bovine tuberculosis traits in African or Chadian cattle population.
1. INTRODUCTION
Mycobacterium bovis causes bovine tuberculosis (BTB), an important disease of domesticated cattle that has a major economic and health impact throughout the world [1]. Worldwide, agricultural losses due to BTB are estimated at around $3 billion annually [2]. BTB has a zoonotic potential and may be transmitted from animals to humans through consumption of contaminated, unpasteurised milk, poorly heat-treated meat or close contact with infected animals. Tuberculosis in humans causes almost two million deaths annually [3] and tuberculosis incidence is rapidly increasing in sub-Saharan African countries, fuelled by the concurrent HIV epidemic [4].
The incidence of BTB has been significantly reduced or eradicated from domestic cattle in many developed countries by state compensation of a test and cull policy that removes infected cattle [5]. However, in Africa, due to cost implication, lack of capacity and infrastructure, the test and cull policy is not possible [6]. Consequently, farmers face extreme pressure from disease episodes. A study on two main Chadian zebu cattle (Arab and Mbororo) populations estimated the incidence of BTB to be 11% [7].
The traditional animal breeding strategies (i.e. animal performance and pedigree recording for genetic evaluations of animals) for developing resistant strains of livestock species is also impractical due to organisational, operational, technical and infrastructural difficulties. However, the use of genetic markers for the genetic improvement of resistance of the host is a critical component of effective disease control. Better knowledge on host genetic mechanisms of susceptibility and/or resistance are prerequisites for the development of animal breeding tools which may open ways for possible effective and sustainable methods of BTB control. One approach could be to exploit the host genetic variation in response to BTB, as seen in a study conducted in red deer [8] where results of experimental challenged with M. bovis evidenced a wide spectrum of responses and a high heritability of resistance to BTB (0.48 ± 0.096). This result indicates that approximately 48 percent of the between-animal variation seen in response to infection with M. bovis is due to genetics. More recent studies that utilised a large dataset of tuberculin skin tests from Irish and British dairy cows [9,10] estimated heritability were ranged from 0.14 to 0.18 (se: ±0.044). These results suggest that exploitable genetic variation exists and that selection for resistance to BTB is feasible. Interestingly, in cattle it has been demonstrated that differences in susceptibility to BTB is at the level of genus [11], indicating that Bos indicus cattle are more resistant than Bos taurus. Although there is no particular gene known to be responsible for differences in BTB infection susceptibility in cattle, natural resistant associated macrophage protein 1 (NRAMP1) was a known candidate gene; it is associated with natural resistance to infection with Mycobacterium spp., in cattle [12,13]. Recently, a genome-wide association study on Holstein-Friesian herds in Irish cattle that used the bovine 50 k SNP chip has identified three SNPs in a 65 kb genomic region on chromosome 22 associated with BTB susceptibility [14]. These studies provide substantial evidence that polymorphisms in the genome sequence play an essential role in determining resistance/ susceptibility to BTB infection. However, there is a lack of information on the potential genetic markers for BTB resistance in African cattle.
The objective of this study was to investigate associations between a panel of 23 microsatellite markers with the following four phenotypes: 1) single intra-dermal comparative cervical tuberculin (SICCT) test outcome, 2) post-slaughter lesion presence, 3) a bacteriological tissue culture test outcome sampled from detected lesion and 4) a predicted true BTB infection status using a Bayesian model for naturally occurring BTB in Chadian zebu cattle.
2. MATERIALS AND METHODS
2.1. Animals
This study was a part of a large Swiss National Science Foundation funded project on the study of molecular epidemiology, host-pathogen interactions in BTB and cattle genetic diversity in Africa. A total of 251 animals were sampled during three intervals of approximately one month between July and November 2005 at abattoirs in Southern Chad, approximately 500 km from N’Djamena, the capital of Chad. Details of the resource population can be found in [15,16]. Briefly, animals belonged to two indigenous cattle breeds known as “Arab” and “Mbororo” of zebu origin. Animals were a random subsample of cattle raised in a long-distance trans-human livestock production system with frequent trans-border movement of herds between the Central African Republic and Chad in search of pasture during the dry season. Thus, there was no information available on the parental population or specific sires. Because regional farming systems focus primarily on milk production, mainly young males and old females are generally sold from the same herd to different traders who, in turn, sell off their animals to different butchers. Therefore, we assumed that the sampled animals can be considered a representative sample from a large number of different herds and an extensive area of southern Chad. Variables recorded were: two breeds (Arab cattle breed, n = 164 and Mbororo cattle breed, n = 87), sex, age in years, body condition status (1 = good, 2 = bad or 3 = very bad condition) based on live body weight and external characteristics and a number of bovine tuberculosis diagnostic tests.
2.2. Phenotypes
The phenotypic data used in this study were the results of three diagnostic tests conducted on animals prior to and after slaughter at abattoirs in southern Chad. Full details of the phenotyping are previously described [12]. Briefly, the SICCT was performed on live animals, three days before slaughter. Animals were injected with an aliquot of 0.1 ml containing 50,000 IU/ml bovine purified protein derivative (PPD-B) and 0.1 ml of 25,000 IU/ml avian purified protein derivative (PPD-A) on two different sites of the previously shaved mid-neck. The PPD-B and PPD-A were supplied by the licensed Instituto Zooprofilattico dell Umbria e delle Marche, Perugia, Italy (http://www.izsum.it/IZSUM/). After 72 hours, the skin thickness at the injection sites was measured. The interpretation of the results was such that when reactions were observed at both injection sites, the difference between the two reaction sizes was considered. An animal was classified as SICCT positive if the increase in skin thickness at the PPD-B injection site was more than 4 mm compared to the reaction shown at the PPD-A site, and considered negative if the skin thickness was equal to or less than 4 mm [17]. The SICCT phenotype was recorded as 1 = skin thickness > 4 mm or 0 = skin thickness ≤ 4 mm. Some animals do not respond to the SICCT test, despite presenting physical signs of BTB infection such as lesion post-slaughter. Therefore, post-slaughter inspection of carcasses and bacteriological culture to isolate M. bovis from tissue are important measures for confirming the SICCT test; hence we also obtained postslaughter lesion and culture information. All experiments were carried out in accordance with Chadian government regulations and as per the standard policies of the Laboratory of Veterinary and Zootechnique Research, Farcha, Chad.
After slaughter, all cattle carcasses were subjected to inspection for the presence of visible lesions in organs and lymph nodes (as seen in Figure 1) indicative of infection by M. bovis. The inspection was conducted by trained local veterinary officers, following the standard procedures as previously described [18]. A total of 108 carcasses had visible lesion across various organs. The lesion trait was recorded as: 1 = if a lesion was observed, 0 = otherwise.
One hundred and eight carcasses with confirmed gross visible lesions were tissue-sampled and stored at –20˚C prior to processing. For bacteriological culture testing, the specimens were homogenised with sterile saline solution, then inoculated into Middlebrook 7H9 medium flasks containing oleic acid-albumin-dextrose-catalase and PANTA (polymyxin, amphotericin B, nalidixic acid, trimethoprim, azlocillin) and incubated at 37˚C (without CO2) for 8 weeks. The presence of acid fast bacillus, an indicator of Mycobacterium spp., was confirmed by the smear Ziehl-Neelsen staining method. Six out of 108 tissue samples were discarded due to poor quality. Fifty out of 102 samples (49%) that displayed bacterial growth were positively identified for the presence of acid fast bacilli, evidence of Mycobacterium spp. The culture test s were recorded as 1 = presence of acid fast bacilli or 0 = absence of acid fast bacilli.
A predicted true BTB disease status of all animals tested was estimated using a Bayesian model (BM). The model combined the results of both ante-mortem (SICCT) and post-mortem (lesion and culture) diagnostic outcome tests for the entire sampled animal population and was based on the binomial assumption for each phenotype. Under the BM estimate, animals were designated either non-diseased = 0 or diseased = 1. A detailed BM and the mathematical description are provided in [15].
2.3. Genotypes
Blood samples were collected from animals before the SICCT test was performed and the DNA was extracted using the QIAamp® DNA Blood Kit (QIAGEN, Cat. No. 51106) method. The DNA samples were sent to Van Haerigen Laboratory (VHL) Wageningen, The Netherlands for genotyping. Twenty three microsatellite markers were genotyped, namely: BM1818, BM1824, BM2113, CSRM60, CSSM66, ETH10, ETH225, ETH3, ETH152, ETH185, TGLA122, TGLA126, TGLA227, TGLA53, ILSTS005, ILSTS006, HEL5, HAUT27, SPS115, INRA32, INRA35, INRA23, and MM12. These markers were selected from a recommended panel for genetic diversity in cattle populations by the Domestic Animal Diversity (DAD) Information System of the Food and Agriculture Organisation [19]. The FAO markers are polymorphic and segregating independently.
2.4. Statistical Analyses
In order to ensure robust association testing, both animals and genotyped markers were checked to identify and exclude individuals and genotyped markers with high missing information, an indicator of poor genotyping. Individuals or/and markers with missing information of greater than 5% and markers with minor allele frequency (MAF) less than 1% were excluded from further analyses. Twenty-seven animals and three markers (TGLA53, HEL5 and HAUT27) were excluded from association analyses. A total of 224 animals and 20 markers with complete genotype information were available for SICCT test, lesion and BM true BTB status, whereas for the culture test trait, 96 animals and 20 markers with complete genotype were used in the association analyses.

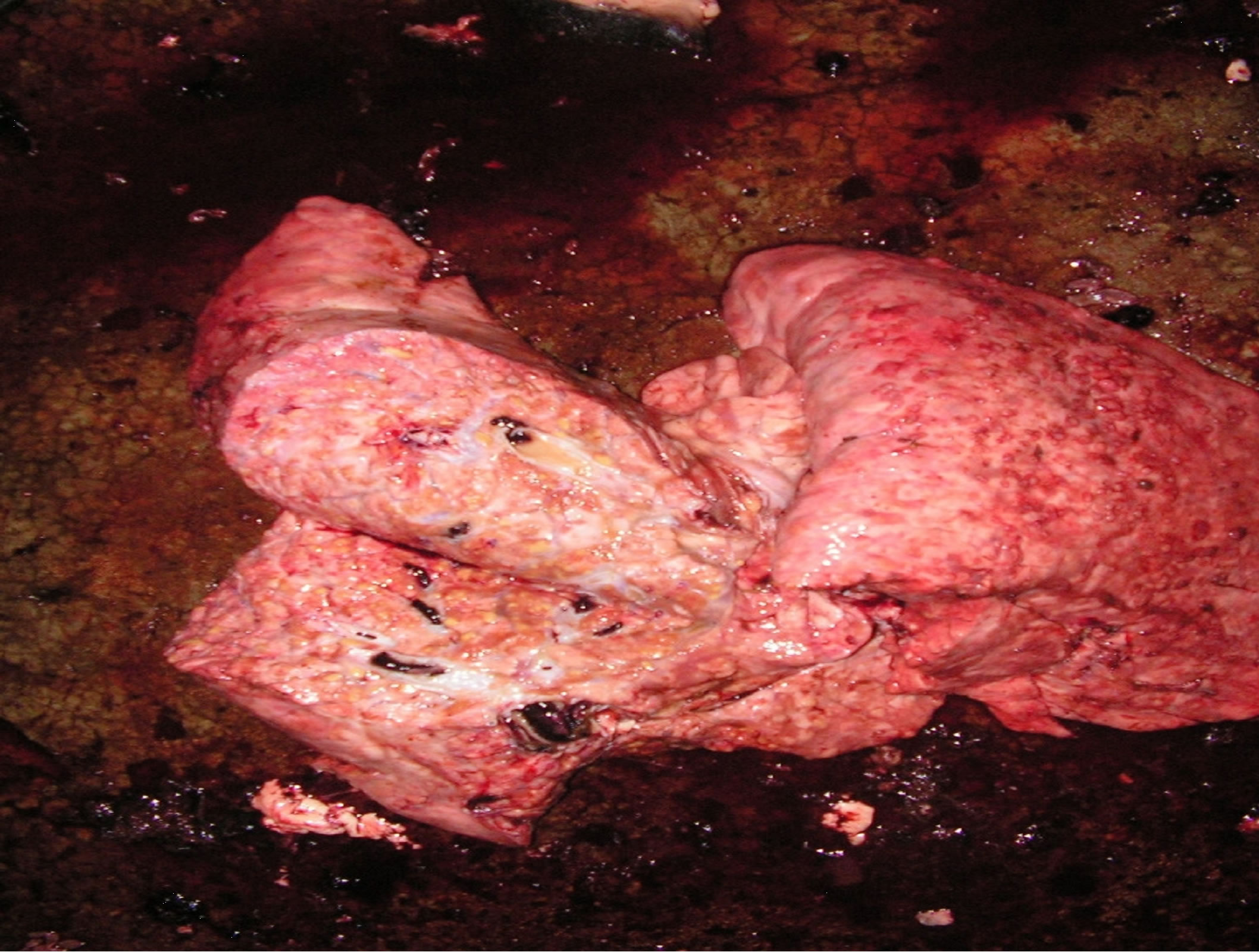
Figure 1. Left picture: a cattle lung showing presence of visible lesions in the post-mortem examination. Right picture: Arab cattle (white coat) and Mbororo cattle (dark coat) at the holding facility of abattoirs in southern Chad (photo republished with permission from [12], Animal Genetics 42: 656-658).
Empirical proportions of the number of animals exhibiting positive status for SICCT, lesion, BM and tissue culture were obtained. Univariate and multivariate logistic regression models were fitted to the BTB phenotypic data to assess the effect of age (covariate), sex and breed, as these are considered common potential risk factors. The models were then extended to include the effect of a marker, fitting each marker in a separate model. Each fitted model took the following form:
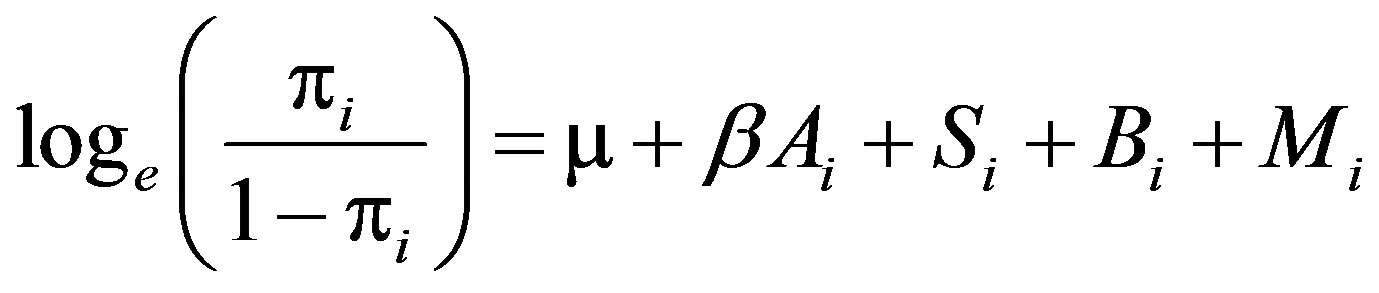
where  is the probability of a phenotype being scored positive (Y = 1) for animal i, μ is overall constant, Ai is age (covariate, 1 to 12 years), Si is sex (female, male), Bi is breed (Arab, Mbororo), and Mi is a marker (alleles: 5 to 14 levels). We assumed no relationships between individuals. The effect of each allele on a trait was evaluated based on effect size and associated P-value where a threshold P-value ≤ 0.05 was considered significant. Significance of the model was assessed by a likelihood ratio test comparing the full model to a reduced model (a model without the marker being tested). The statistical concepts including likelihood functions behind a fitting logistic regressison model in marker-trait association studies involving binary traits can be found in [20]. Model fitting was conducted using the glm function in R [21].
is the probability of a phenotype being scored positive (Y = 1) for animal i, μ is overall constant, Ai is age (covariate, 1 to 12 years), Si is sex (female, male), Bi is breed (Arab, Mbororo), and Mi is a marker (alleles: 5 to 14 levels). We assumed no relationships between individuals. The effect of each allele on a trait was evaluated based on effect size and associated P-value where a threshold P-value ≤ 0.05 was considered significant. Significance of the model was assessed by a likelihood ratio test comparing the full model to a reduced model (a model without the marker being tested). The statistical concepts including likelihood functions behind a fitting logistic regressison model in marker-trait association studies involving binary traits can be found in [20]. Model fitting was conducted using the glm function in R [21].
3. RESULTS
The animal population sampled consisted mainly of young bulls and old cows, with mean age of 3.64 (SD = 1.03) and 6.16 (SD = 1.42) years for bulls and cows, respectively. Table 1 presents the incidences of various binary BTB traits of animals tested positive. The incidences of BTB phenotypes were calculated as the proportion of animals that were positive. Out of 224 animals used in the analyses, there were 29%, 48% and 28% animals positive for tuberculin skin test, post-slaughter lesion detected and Bayesian estimate for true BTB disease status, respectively, whereas for the 96 tissue culture samples, 52% animals were confirmed for the presence of acid fast bacilli, an indicator of Mycobacterium sp.
The effect of age on various BTB traits considered is shown in Figure 2. Age has a significant positive effect on lesion, Bayesian model estimates for true BTB disease status and culture.
Table 1. Number of animals tested, number of animals tested positive and proportion of animals tested positive for BTB traits in the study.
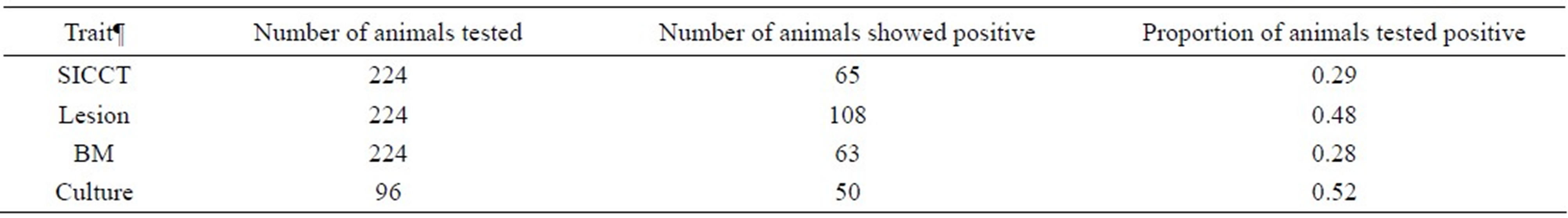
¶SICCT = single intra-dermal comparative cervical tuberculin test; Lesion = presence of visible lesion in organs and lymph nodes; BM = Bayesian model estimate of true BTB disease status; Culture = bacteriological tissue culture test.
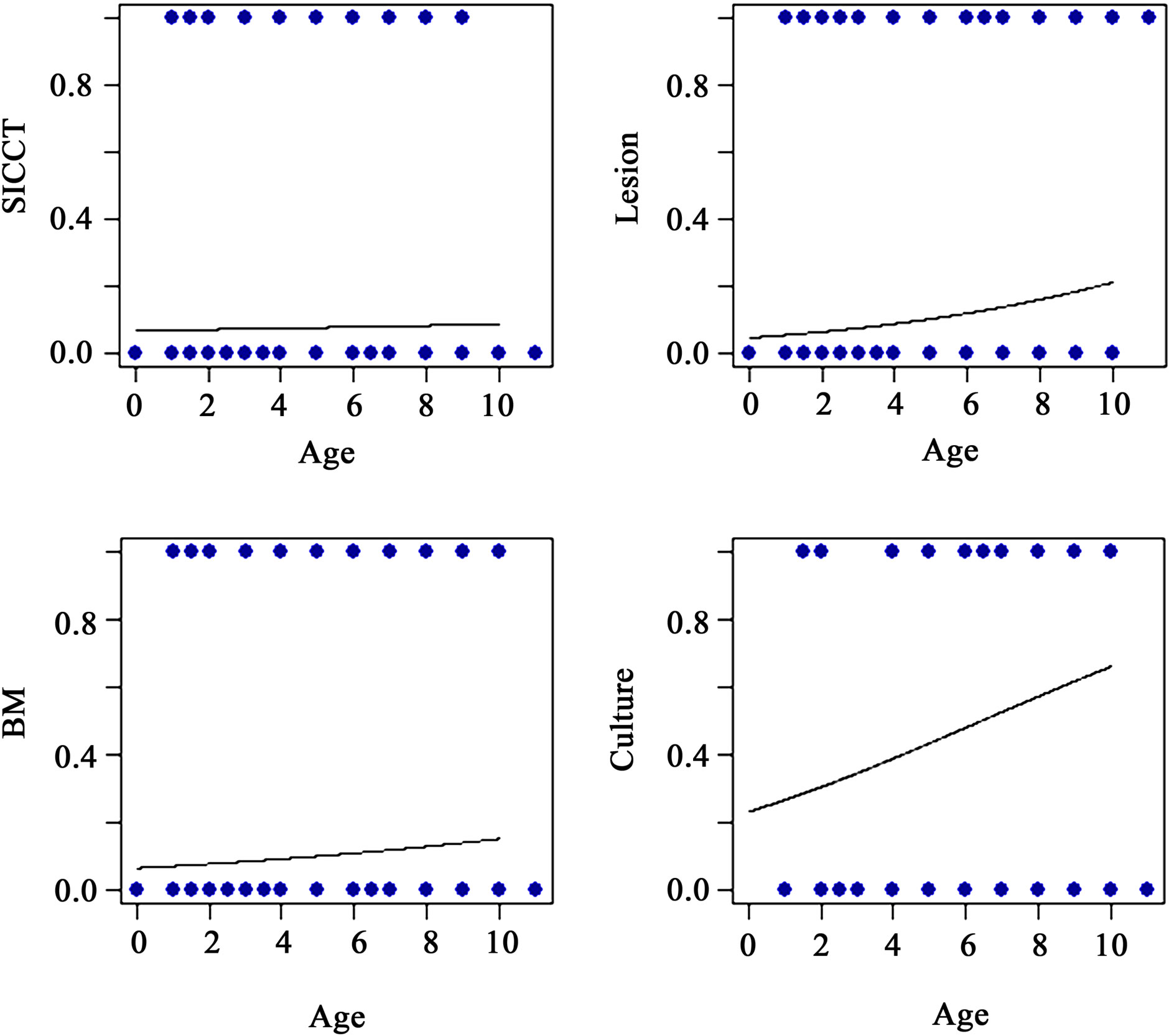
Figure 2. Effect of age on skin tuberculin test (SICCT), lesion, Bayesian model (BM) estimates of true BTB disease status and bacteriological culture test.
Table 2 presents the number of alleles and the number of animals genotyped for each marker across the traits considered. The number of alleles per marker ranged from 5 to 14 with the mean 8.95 (SD = 2.39) alleles, and the average number of animals genotyped was 224 (SD = 13.27) for SICCT, lesion and BM, and 95.55 (SD = 5.52) for culture.
Associations between genetic markers and BTB traits considered are presented in Tables 3-5. Six out of twenty markers reached chromosome-wise significance level at P-value ≤ 0.05 associated with at least one trait considered. Allele frequencies of the markers detected to be significant ranged from 0.0212 to 0.6220. Alleles 181, 187 and 191 in ILSTS005 were significantly associated with increasing SICCT test positive (effect size from 1.911 to 2.49) with significant (P = 0.0180) likelihood ratio (LR) test, whereas alleles 286, 288, 290 and 192 in ILSTS006 marker were significantly associated with decreasing SICCT test (effect size from –3.24 to –1.65) with significant LR test (P = 0.0221). Allele 183 in ILSTS005 and 294 and 296 in ILSTS006 had marginally significant associations with the SICCT test. Allele 87 in TGLA227 was marginally associated with BM predicted true BTB (P = 0.0542), while alleles 99 and 81 in the same marker were slightly significantly associated with increasing true BTB incidence; however, the LR test was not significant (P = 0.236). Alleles 102, 106 and 118 in INRA035 had significant effects on lesion occurrence, but allele 108 was marginal (P = 0.0917), with a somewhat more significant LR test (P = 0.0555). Alleles 133 and 141 in BM2113 were significantly associated with decreasing effect on both lesion occurrence and positive culture test, although the LR test was not significant. Alleles 185 and 189 in CSSM66 were significantly associated with increasing BTB positive culture test.
Table 2. Number of alleles and number of animals genotyped per marker across BTB traits considered.
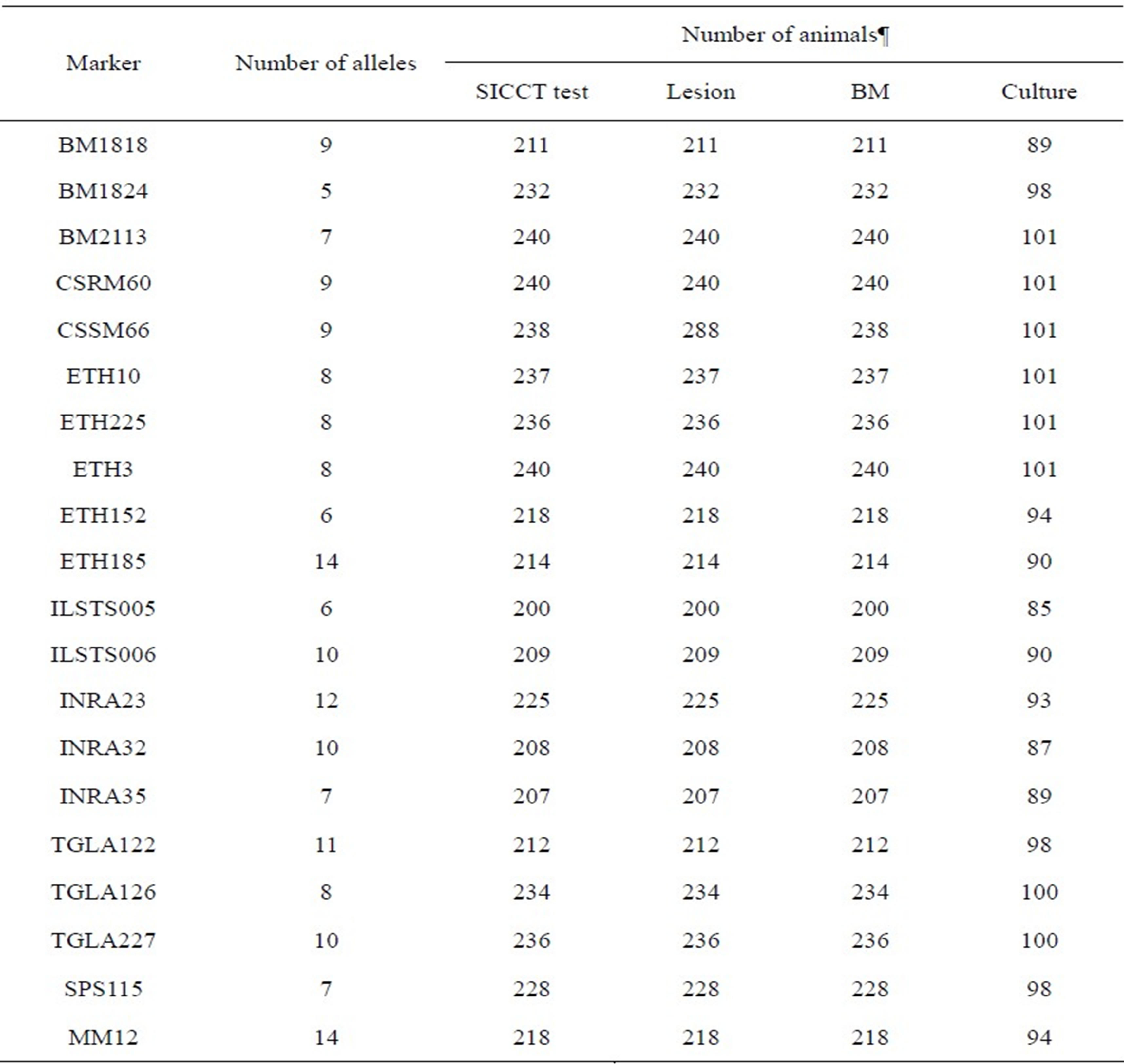
¶SICCT test = single intra-dermal comparative cervical tuberculin test; Lesion = post-slaughter lesion detected on various organs and lymph nodes; BM = Bayesian model estimate for the true BTB disease status of all sampled animals, and culture = bacteriological tissue culture for acid fast bacilli presence in culture confirmed by Ziehl-Neelsen staining metho.
Table 3. Associations between microsatellite markers and single intra-dermal comparative cervical tuberculin (SICCT) test and Bayesian model (BM) estimate of true BTB disease status for bovine tuberculosis disease in Chadian zebu cattle.
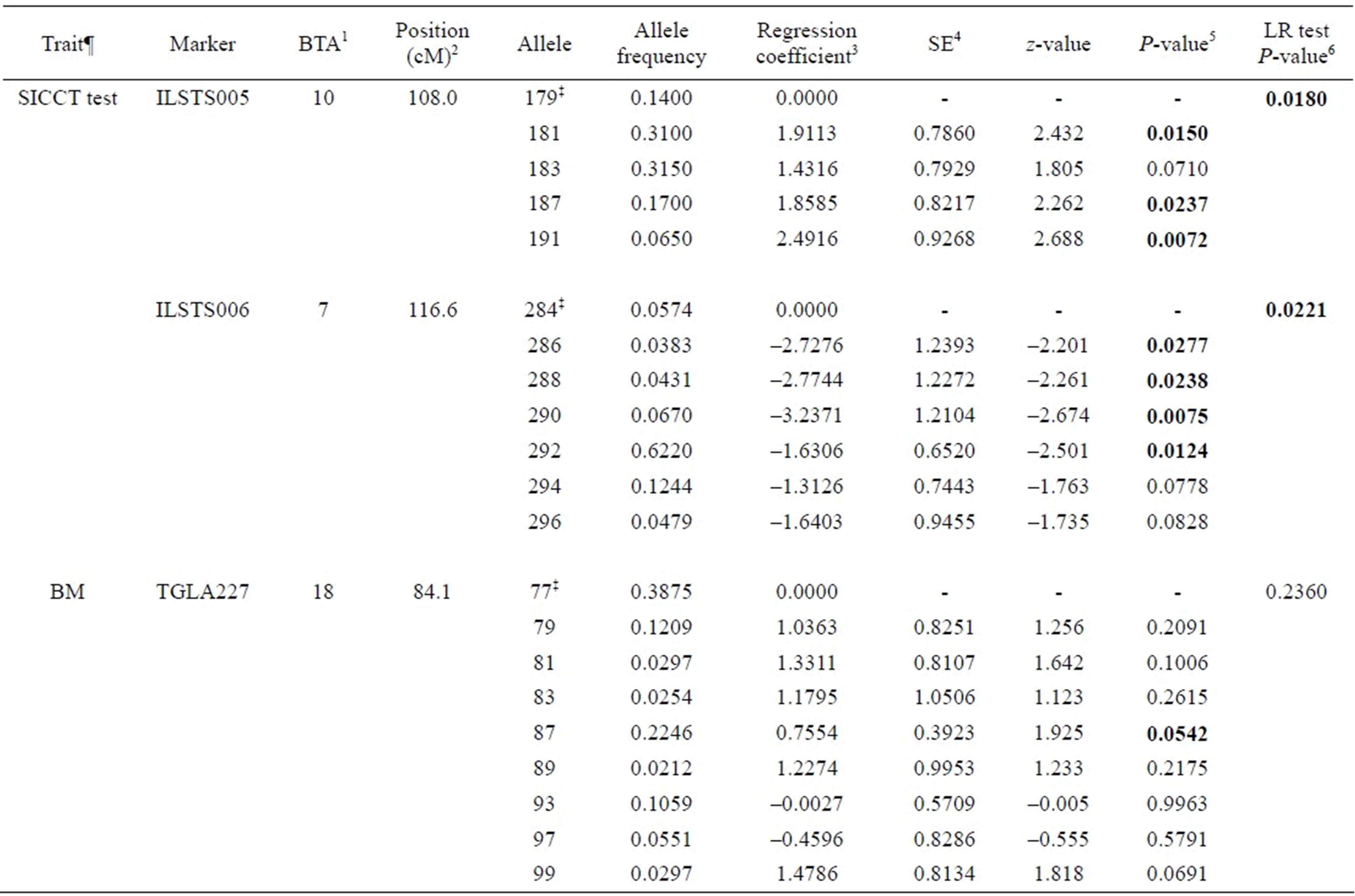
¶SICCT test = single intra-dermal comparative cervical tuberculin test; BM = Bayesian model estimate for the true BTB disease status; ‡Reference allele comparing to regression coefficients; 1Bovine autosomal chromosome; 2Genome position in centimorgan (cM), USDA MARC bovine linkage map; 3Regression coefficients due to the regression of the trait on number of copies of an allele in the marker, these regression coefficients are expressed in log odds ratios; 4Standard error of the regression coefficient; 5P-values for the association between allele in a marker and a trait; 6Likelihood ratio test to compare a full model and reduced model (a model without the marker being tested).
Table 4. Associations between microsatellite markers and lesion for bovine tuberculosis disease in Chadian zebu cattle.
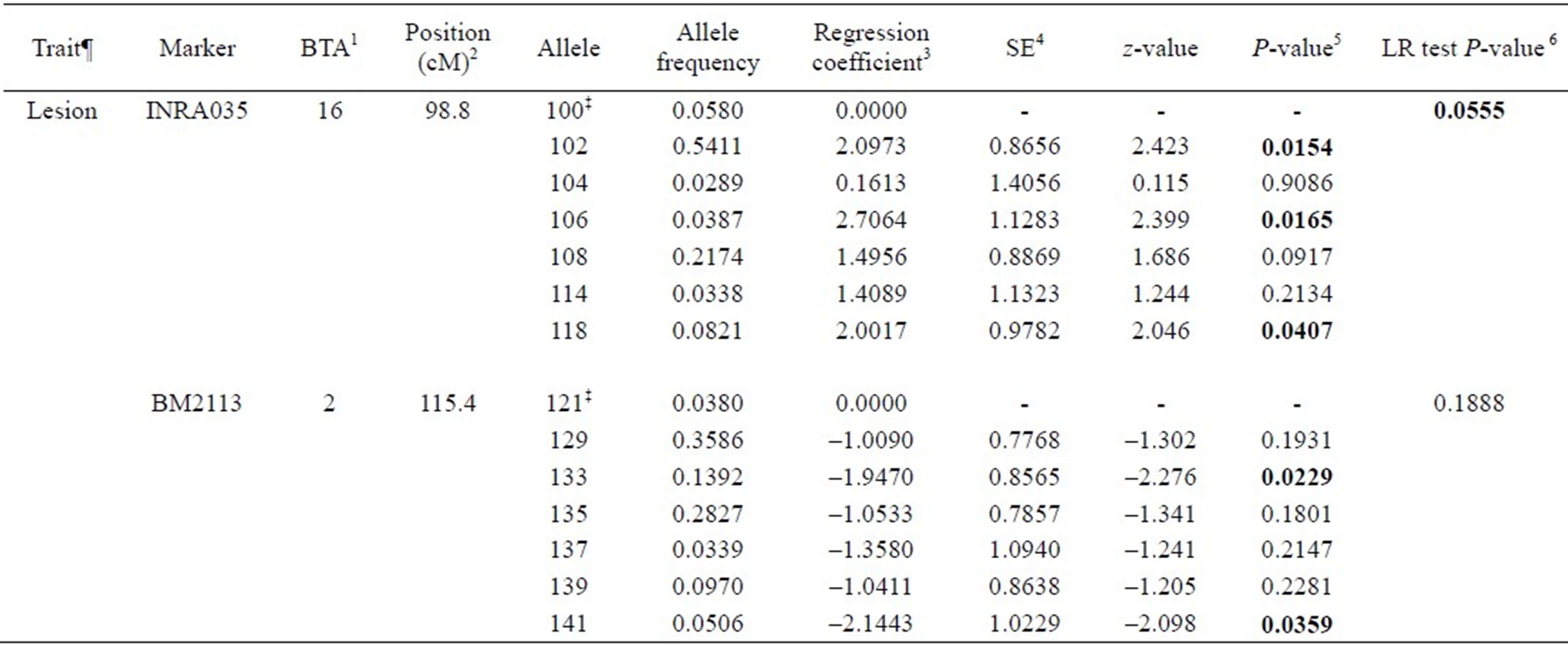
¶Lesion = post slaughter lesion detected on various organs and lymph nodes; ‡Reference allele comparing to regression coefficients; 1Bovine autosomal chromosome; 2Genome position in centimorgan (cM), USDA MARC bovine linkage map; 3Regression coefficients due to the regression of the trait on number of copies of an allele in the marker; 4Standard error of the regression coefficient; 5P-values for the association between allele in a marker and a trait; 6Likelihood ratio test to compare a full model and reduced model (a model without the marker being tested).
Table 5. Associations between microsatellite markers and bacteriological culture test for bovine tuberculosis disease in Chadian zebu cattle.
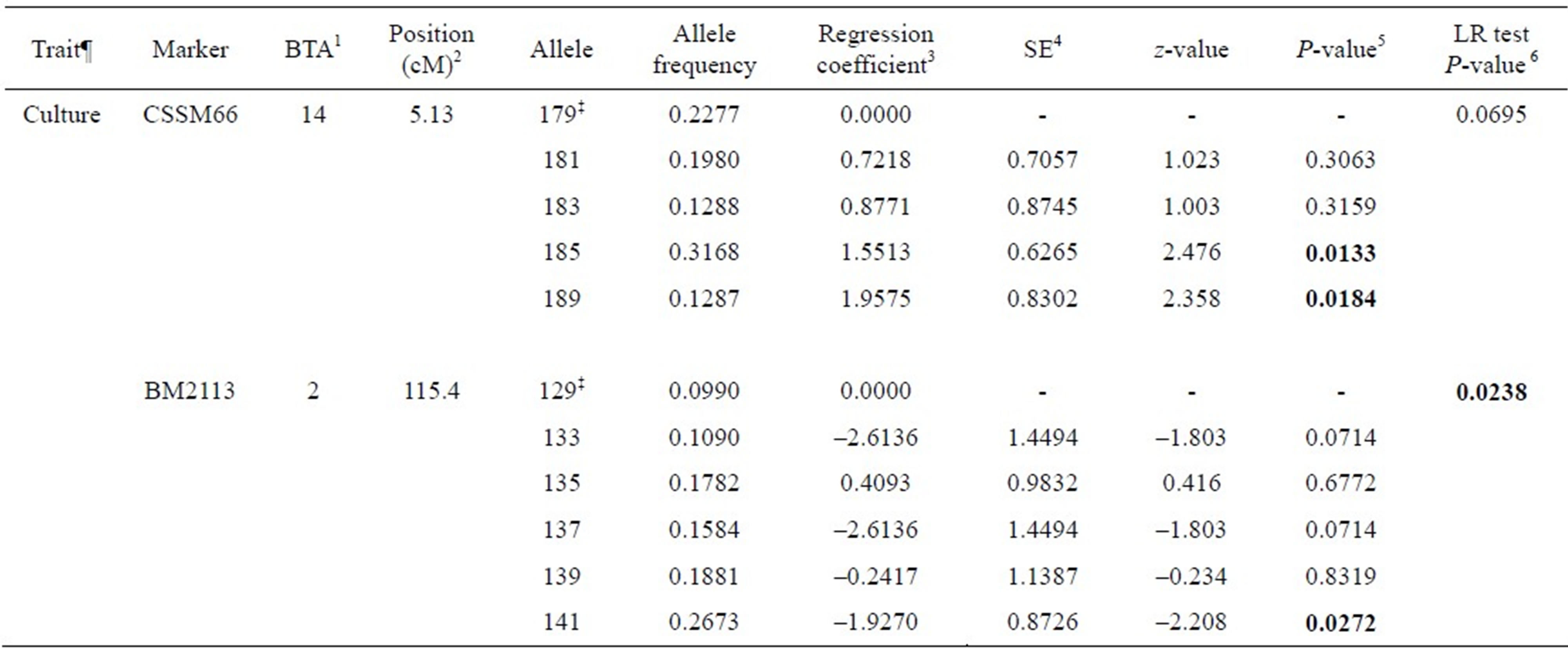
¶Culture = bacteriological tissue culture test; ‡Reference allele comparing to regression coefficients; 1Bovine autosomal chromosome; 2Genome position in centimorgan (cM), USDA MARC bovine linkage map; 3Regression coefficients due to the regression of the trait on number of copies of an allele in the marker; 4Standard error of the regression coefficient; 5P-values for the association between allele in a marker and a trait; 6Likelihood ratio test to compare a full model and reduced model (a model without the marker being tested).
4. DISCUSSION
The aim of the present study was to investigate whether genetic polymorphisms of microsatellite markers influence susceptibility/resistance to bovine tuberculosis in African zebu cattle. We have tested for possible associations between microsatellite markers and a number of clinical diagnostic tests for bovine tuberculosis in a Chadian cattle population. An association study between genetic markers and bovine tuberculosis (BTB) is an important first step toward the identification of resistance/susceptibility genes to BTB. The ultimate goal of such a study is to identify genetically resistant animals for breeding or cattle keeping purposes and reduce costs of cattle production.
The BTB incidence estimates in the present study were higher than previously reported in the original dataset (954 animals) by [15,16]. This may be due to the difference in the number of animals tested in the original dataset and our subsample dataset. The breed and age of animals were important risk factors for post-slaughter lesion presence in this population: the probability of lesion occurrence increases with age, and BTB lesions are more likely be detected in Mbororo breed compared to Arab breed; this finding is consistent with findings from the original dataset [16].
We identified six microsatellite markers significantly associated with at least one BTB trait (see Tables 3-5). ILSTS005 (alleles 181, 187 and 191), TGLA227 (allele 87), INRA035 (alleles 102, 106, and 118) and CSSM66 (alleles 185 and 189) were significantly associated with increasing effect on SICCT test, Bayesian estimate for true BTB disease status, lesion and tissue culture test outcome, respectively. Animals carrying these alleles are likely to be susceptible to BTB disease. ILSTS006 (alleles 286, 288, 290 and 292) and BM2113 (alleles 133 and 1410) were significantly reducing SICCT, lesion and culture positive outcomes.
A recent study of genome-wide association which used high density SNPs (single nucleotide polymorphisms) has identified three SNPs in a 65 kb genomic region on chromosome 22 associated with bovine tuberculosis susceptibility in Irish dairy cattle [14]. This genomic region has been found to contain gene SLC6A6 which is known to function in host immune systems. A study on footrot in sheep revealed animals homozygous at BMC5221 marker were susceptible to virulent footrot, compared with heterozygous individuals [22]. Also, a study in a natural population of wild pigs observed that individuals with relatively high homozygosity in a panel of neutral microsatellite markers were likely to be susceptible to bovine tuberculosis infection [23]. The effect of alleles 286, 288, 290 and 292 in ILSTS006 on SICCT were significantly associated with decreasing SICCT test positive, whereas alleles 133 and 141 in BM2113 were associated with decreasing lesion presence and tissue culture positive for BTB disease. Allele 141 in BM2113 showed consistent association for reducing both lesion presence and confirmed tissue culture positive in this cattle population.
The BM2113 marker is located on BTA2 at 115.4 cM, and a number of genomic regions within BTA2 have been linked to resistance/tolerance to parasite and bacterial disease in cattle. [24] identified loci on chromosome 2 to be associated with tolerance to Johne’s disease in Holstein cows. NRAMP1 gene, which is also located on BTA2, was linked to bovine tuberculosis resistance [12]. Furthermore, four loci on BTA2 located from 110 to 126 cM have been found to be associated with bovine viral disease [25]. It is likely that BTA2 harbours genes or regulatory genes that are affecting the resistance to infectious disease in cattle. Fine mapping studies of the regions on BTA2 would assist to characterise the loci associated with disease resistance. Further immuno-genetic investigation in and around these genomic regions may help to understand the genes and downstream innate immune defence mechanism involved in resistance or susceptibility to BTB. Once these genes are identified, selection for these genes could produce significant benefits, particularly through the use of artificial insemination (AI) from sires with genomic variants associated with disease resistance.
Breeding for enhanced resistance to infectious disease such as BTB is also an effective means of improving the health and fitness of livestock as well as reducing incidence of human TB in both developing and underdeveloped countries. The main problem in host genetic studies for chronic infectious disease with multiple stages such as BTB is the accurate determination of cases. A dataset compiled from bacteriological culture results and subsequently determining the exact Mycobacterium species by PCR (polymerase chain reaction) technique would be considered the gold standard; in this case, genetic markers can be selected with a high degree of certainty. In practice, however, large-scale phenotypic data for BTB using the combined culture test and PCR-based genetic profiling of a pathogen may not be feasible. The phenotypic data used to characterise animals in this study are more detailed than currently available literature which reports on genetic resistance/susceptibility to BTB. However, the limitations of the data made it impossible to track pedigree information of the animals, as it is not common practice in many developing and underdeveloped countries; therefore, our assumption of no relationships between animals may not be true. In addition, the number of microsatellite markers used was neither sufficient to cover a large genomic regions nor targeted to a genomic region of interest.
5. CONCLUSION
The aim of this paper was to investigate a possible association between a panel of microsatellite markers and BTB phenotypes. These phenotypes were a single intradermal comparative cervical tuberculin test performed on live animals, post-slaughter lesion presence and postslaughter tissue culture outcomes, and a Bayesian model estimate of true BTB disease status in a Chadian cattle population. We have identified six markers: BM2113 (BTA2), ILSTS006 (BTA7), ILSTS005 (BTA10), CSSM66 (BTA14), INRA035 (BTA16) and TGLA227 (BTA18) which were significantly associated with at least one phenotype considered. The effect of alleles in ILSTS006 and BM2113 markers on BTB phenotypes considered were significantly associated with decreasing BTB disease incidence; hence they could be considered in a markerassisted selection of tolerant animals. However, further studies with large validation population would be required to confirm the effect of these alleles. This is the first study to report association of microsatellite markers with bovine tuberculosis traits in Chadian cattle.
6. ACKNOWLEDGEMENTS
The authors would like to acknowledge the Swiss National Science Foundation for financial support and Laboratoire de Recherche Veterinaires et Zootechniques de Farcha, N’Djamena, Chad for the collection and processing of samples. We are grateful to Professor Jakob Zinsstag, Dr. B. Müller and Dr. B. N. R. Ngandolo for help with collection of BTB phenotypic data in Chad for this study. A. A. Ali acknowledges the financial support from the Commonwealth Scientific and Industrial Research Organisation (CSIRO) Animal, Food and Health Sciences for his Ph.D. scholarship.
REFERENCES
- Smith, N.H., Gordon, S.V., Rua-Domenech, R., CliftonHadley, R.S. and Hewinson, R.G. (2006) Bottlenecks and broomsticks: The molecular evolution of Mycobacterium bovis. Nature Reviews Microbiology, 4, 670-681. doi:10.1038/nrmicro1472
- Garnier, T., Eiglmeier, K., Camus, J.C., Medina, N., Mansoor, H., Pryor, M., Duthoy, S., Grondin, S., Lacroix, C., Monsempe, C., Simon, S., Harris, B., Atkin, R., Doggett, J., Mayes, R., Keating, L., Wheeler, P.R., Parkhill, J., Barrell, B.G., Cole, S.T., Gordon, S.V. and Hewinson, R.G. (2003) The complete genome sequence of Mycobacterium bovis. Proceedings of the National Academy of Sciences of the United States of America, 100, 7877-7882. doi:10.1073/pnas.1130426100
- Mathers, C.D., Boerma, T. and Fat, D.M. (2009) Global and regional causes of death. British Medical Bulletin, 92, 7-32. doi:10.1093/bmb/ldp028
- Houlihan, C.F., Mutevedzi, P.C., Lessells, R.J., Cooke, G.S., Tanser, F.C. and Newell, M.L. (2010) The tuberculosis challenge in a rural South African HIV programme. BMC Infectious Diseases, 10, 1-9. doi:10.1186/1471-2334-10-23
- Ayele, W.Y., Neill, S.D., Zinsstag, J., Weiss, M.G. and Pavlik, I. (2004) Bovine tuberculosis: An old disease but a new threat to Africa. International Journal of Tuberculosis and Lung Disease, 8, 924-937.
- Zinsstag, J. (2008) Towards effective control of bovine tuberculosis in Africa: A case for public engagement in science. Ethiopian Journal of Health Development, 22, 117-118.
- Ngandolo, B.N., Diguimbaye-Djaibe, C., Müller, B., Didi, L., Hilty, M., Schiller, I., Schelling, E., Mobeal, B., Toguebaye, B.S., Akakpo, A.J. and Zinsstag, J. (2009) Antemortem and postmortem diagnoses of bovine tuberculosis in southern Chad: Case of slaughter cattle. Diagnostics ante et post mortem de la tuberculose bovine au sud du Tchad: Cas des bovins destines a l’abattage. Revue d'Elevage et de Medecine Veterinaire des Pays Tropicaux, 62, 5-12.
- Mackintosh, C.G., Qureshi, T., Waldrup, K., Labes, R.E., Dodds, K.G. and Griffin, J.F.T. (2000) Genetic resistance to experimental infection with Mycobacterium bovis in red deer (Cervus elaphus). Infection and Immunity, 68, 1620-1625. doi:10.1128/IAI.68.3.1620-1625.2000
- Bermingham, M.L., More, S.J., Good, M., Cromie, A.R., Higgins, I.M., Brotherstone, S. and Berry, D.P. (2009) Genetics of tuberculosis in Irish Holstein-Friesian dairy herds. Journal of Dairy Science, 92, 3447-3456. doi:10.3168/jds.2008-1848
- Brotherstone, S., White, I.M.S., Coffey, M., Downs, S.H., Mitchell, A.P., Clifton-Hadley, R.S., More, S.J., Good, M. and Woolliams, J.A. (2010) Evidence of genetic resistance of cattle to infection with Mycobacterium bovis. Journal of Dairy Science, 93, 1234-1242. doi:10.3168/jds.2009-2609
- Vordermeier, M., Gobena, A., Berg, S., Bishop, R., Robertson, B.D., Abraham, A., Hewinson, R.G. and Young, D.B. (2012) The influence of cattle breed on susceptibility to bovine tuberculosis in Ethiopia. Comparative Immunology, Microbiology & Infectious Diseases, 35, 227-232. doi:10.1016/j.cimid.2012.01.003
- Kadarmideen, H.N., Ali, A.A., Thomson, P.C., Müller, B. and Zinsstag, J. (2011) Polymorphisms of the SLC11A1 gene and resistance to bovine tuberculosis in African Zebu cattle. Animal Genetics, 42, 656-658. doi:10.1111/j.1365-2052.2011.02203.x
- Vázquez-Flores, F., Alonso, R., Villegas-Sepúlveda, N., Arriaga, C., Pereira-Suárez, A.L., Mancilla, R. and EstradaChávez, C. (2006) A microsatellite study of bovine solute carrier family 11 a1 (Slc11a1) gene diversity in Mexico in relation to bovine tuberculosis. Genetics and Molecular Biology, 29, 503-507. doi:10.1590/S1415-47572006000300019
- Finlay, E.K., Berry, D.P., Wickham, B., Gormley, E.P. and Bradley, D.G. (2012) A genome wide association scan of bovine tuberculosis susceptibility in Holstein-Friesian dairy cattle. Plos One, 7, e30545. doi:10.1371/journal.pone.0030545
- Müller, B., Vounatsou, P., Ngandolo, B.N.R., Diguimbaye-Djaibe, C., Schiller, I., Marg-Haufe, B., Oesch, B., Schelling, E. and Zinsstag, J. (2009) Bayesian receiver operating characteristic estimation of multiple tests for diagnosis of bovine tuberculosis in Chadian cattle. Plos One, 4, e8215. doi:10.1371/journal.pone.0008215
- Ngandolo, B.N.R., Müller, B., Diguimbaye-Djaibe, C., Schiller, I., Marg-Haufe, B., Cagiola, M., Jolley, M., Surujballi, O., Akakpo, A.J., Oesch, B. and Zinsstag, J. (2009) Comparative assessment of fluorescence polarization and tuberculin skin testing for the diagnosis of bovine tuberculosis in Chadian cattle. Preventive Veterinary Medicine, 89, 81-89. doi:10.1016/j.prevetmed.2009.02.003
- Interantion Office of Epizootic (OIE) (2006) Manual of diagnostic tests and vaccines for terrestrial animals, 2004.
- Herenda, D., Chambers, P.G., Ettriqui, A., Seneviratna, P. and da Silva, T.J.P. (1994) Manual on meat inspection for developing countries. Food and Agriculture Organisation (FAO) of the United Nations, Rome.
- FAO (2004) Secondary guidelines for development of national farm animal geneticresources management plans. Rome, 55.
- Kadarmideen, H.N., Janss, L.L.G. and Dekkers, J.C.M. (2000) Power of quantitative trait locus mapping for polygenic binary traits using generalized and regression interval mapping in multi-family half-sib designs. Genetical Research, 76, 305-317. doi:10.1017/S001667230000481X
- R Development Core Team (2011) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, 2011. http://www.R-project.org/ .
- Smith, E.M., Hoffman, J.I., Green, L.E. and Amos, W. (2012) Preliminary association of microsatellite heterozygosity with footrot in domestic sheep. Livestock Science, 143, 293-299. doi:10.1016/j.livsci.2011.10.009
- Acevedo-Whitehouse, K., Vicente, J., Gortazar, C., Hofle, U., Fernandez-de-Mera, I.G. and Amos, W. (2005) Genetic resistance to bovine tuberculosis in the Iberian wild boar. Molecular Ecology, 14, 3209-3217. doi:10.1111/j.1365-294X.2005.02656.x
- Zanella, R., Settles, M.L., McKay, S.D., Schnabel, R., Taylor, J., Whitlock, R.H., Schukken, Y., Kessel, J.S.V., Smith, J.M. and Neibergs, H.L. (2011) Identification of loci associated with tolerance to Johne’s disease in Holstein cattle. Animal Genetics, 42, 28-38. doi:10.1111/j.1365-2052.2010.02076.x
- Neibergs, H., Zanella, R., Casas, E., Snowder, G.D., Wenz, J., Neibergs, J.S. and Moore, D. (2011) Loci on Bos taurus chromosome 2 and Bos taurus chromosome 26 are linked with bovine respiratory disease and associated with persistent infection of bovine viral diarrhea virus. Journal of Animal Science, 89, 907-915. doi:10.2527/jas.2010-3330

