Open Journal of Ecology
Vol.07 No.02(2017), Article ID:74008,15 pages
10.4236/oje.2017.72010
Are Invasive Genotypes Superior? An Experimental Approach Using Native and Invasive Genotypes of the Invasive Grass Phalaris Arundinacea
Jane Molofsky1*, Alexandra R. Collins1,2, Eric Imbert3, Tadas Bitinas4, Sébastien Lavergne5
1Department of Plant Biology, University of Vermont, Burlington, VT, USA
2Division of Information Technology & Sciences, Champlain College, Burlington, VT, USA
3Institut des Sciences de l’Evolution, Université de Montpellier, Montpellier, France
4Departamento de Ecología, Universidad de Alicante, Alicante, Spain
5Laboratoire d’Ecologie Alpine (LECA), CNRS-Université Grenoble Alpes, Grenoble, France

Copyright © 2017 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: December 1, 2016; Accepted: February 6, 2017; Published: February 9, 2017
ABSTRACT
The admixture and recombination of individuals from the native range into a new range may lead to the production of invasive genotypes that have higher fitness and wider climatic tolerances than the native genotypes. In this paper, we compare the survival and growth of native EU and invasive NA genotypes when planted back into the native EU range near where the EU genotypes were collected. We test this hypothesis using the invasive wetland grass Phalaris arundinacea. If invasive genotypes have evolved to have higher survival and growth, then they should outperform the native EU genotypes under field conditions that are better suited to the EU genotypes. Individual plants of the wetland grass, Phalaris arundinacea collected from native Europe (Czech Republic (CZ) and France (FR)) and North America (Vermont (VT) and North Carolina (NC)) were planted into common gardens in Trebon, Czech Republic (49.0042˚N, 14.7721˚E) and Moussac, France (43.9808˚N, 4.2241˚E). Invasive genotypes from North Carolina (NC) survived as well or better than native genotypes in both the Trebon and Moussac garden. Additionally, invasive NC genotypes suffered higher herbivore damage than native genotypes but their growth and survival were not significantly different than genotypes from the other regions. A companion field experiment that simulated biomass removal through grazing indicated that invasive NC genotypes recovered faster following grazing than genotypes from other regions. Our results suggest that not all invasive genotypes are superior and regional differences in aggressiveness between invasive genotypes are as great as differences between individuals from native and invasive populations. Introduction of genotypes leading to invasion depends upon the environmental conditions and the suitability of the climate for the introduced individuals.
Keywords:
Invasive Plants, Admixture, Invasive Genotypes, Enemy Release Hypothesis, Evolution of Increased Competitive Ability, Common Garden Experiment, Phalaris arundinacea

1. Introduction
An introduced species may behave invasively in the new range because it already possesses traits that confer invasiveness [1] [2] , or alternatively, it may evolve invasiveness in situ in the new range [3] [4] [5] [6] . Moreover, the introduction history of a species can affect the likelihood that it will become invasive [7] [8] . While many plant species suffer a genetic bottleneck when introduced in the new range [9] , multiple introductions of a plant species may make it more likely that a species becomes invasive in part because multiple introductions can inflate genetic diversity [8] [10] [11] . In addition, multiple introductions may result in the admixture of genomes that have never come into contact with each other creating novel, invasive genotypes that may express different traits and enhanced fitness [11] [12] [13] [14] .
A successful introduction can also depend upon the relationship between the introduced individuals and the new environment. Introduced individuals may have different climatic tolerances than their native counterparts [15] and/or wider climatic tolerances [16] [17] and this may contribute to their success. Testing whether invasive genotypes have a wider climatic tolerance than native genotypes requires planting clones of both known native and invasive genotypes in their home climate and in a different climate to test whether invasive genotypes have greater ability to survive and grow under a new climatic condition.
The increased growth of invasive genotypes found under controlled conditions may not be observed under field conditions [18] . Disentangling whether the reason is due to the traits of the introduced individuals or environmental factors in the field is difficult because traits favored in the new range may incur a cost in the native range. For example, reduced herbivore loads in the new range may have selected for genotypes that have reallocated resources from defense to growth resulting in invasive genotypes having faster growth than native ones [2] [19] [20] . But, if these genotypes are transported back to the native region in which herbivores are more abundant, they may experience higher levels of herbivore damage than their native counterparts and this can negate any increases in growth. Yet, if invasive genotypes are superior, then invasive genotypes may still experience greater growth even if they are preferentially preyed upon. Documenting higher growth following herbivore damage is difficult in a common garden because of variability in herbivore prevalence and local conditions. To document such a pattern requires simulating herbivore damage experimentally [21] [22] .
In this paper, we examine the performance of invasive genotypes when compared to that of native genotypes in the native’s own range in the invasive wetland plant, Phalaris arundinacea. The invasive grass Phalaris arundinacea is a good model system to address the issues of the emergence of novel and superior invasive genotypes [23] . Invasive genotypes of Phalaris arundinacea have been shown to be the product of multiple introductions and subsequent admixture [8] and in common greenhouse conditions, invasive genotypes were shown to have faster growth rate, to be taller, have more tillers and greater final biomass [8] . Additionally, invasive genotypes were also shown to have a smaller genome size [24] and had no consistent differences in genetic architecture [25] . However, in order to test whether these invasive genotypes are superior requires that native and invasive genotypes be compared in the native range to determine if the invasive genotypes created in the new range are superior (in survival, growth and/or reproduction) to native genotypes even under conditions in which native genotypes have evolved [26] [27] [28] . In this way, we can determine if the superior performance of invasive genotypes results in overall greater performance under conditions in which the native genotypes should be favored. In addition, by planting genotypes in a field common garden, we can assess in situ herbivore damage to determine if invasive genotypes suffer more herbivore damage than native ones [29] [30] . However, greater herbivore damage may not necessarily result in reduced growth and/or biomass as greater herbivore damage may be compensated for by a faster growth rate [21] . We test this idea in an experimental common garden in which the same genotypes are subject to biweekly biomass removal to simulate grazing by large herbivores. Finally, by planting genotypes collected from northern and southern populations into both northern and southern gardens, we can determine if differences in performance are due to genotypes having adapted to a similar climate rather than native and invasive differences.
2. Methods
Phalaris arundinacea is a C-3 perennial grass, native to wetlands and wet meadow habitats in Europe and Asia [23] . Considered highly invasive in the midwest and eastern United States, it was originally introduced repeatedly as a wet forage grass and has also been included as seed in conservation mixtures used for soil stabilization [23] . In Vermont, Phalaris has been used as a forage crop in wet pastures for over 50 years but in North Carolina it has been primarily introduced in conservation mix for ditch stabilization (Molofsky personal communication).
We selected three populations from each of four locations in both northern and southern regions of Phalaris’ range: two in the native European range (Czech Republic, 49.0042˚N, 14.7721˚E (CZ)) and southern France, 43.6108˚N, 3.8767˚E (FR) and two in the non-native United States range (Vermont 44.4759˚ N, 73.2121˚W (VT) and North Carolina, 35.4887˚N, 82.9887˚W (NC)) (for details of the population sampling see [8] (Table 1). Northern and southern populations were chosen to sample different climatic regions of Phalaris arundinacea’s range [8] . Thus, our sampling protocol allowed for a comparison of how genotypes from the native (CZ, FR) and invasive (VT, NC) range respond when planted outside their home climate (i.e. NC, FR in Trebon, Czech Republic and VT, CZ in Moussac, France).
Genotypes were identified through allozyme analysis [8] and a subset were selected for experimentation. We selected 18 invasive genotypes (9 VT and 9 NC) and 18 native genotypes (12 CZ and 6 FR). Fewer French genotypes were chosen because one French population contained only hexaploids, while all other populations contained tetraploids; we thus, eliminated hexaploid individuals from our study. Chosen genotypes were transplanted into pots in the University of Vermont greenhouse, where they were maintained, and then sequentially propagated prior to experimentation to remove any maternal environmental effects. Selected tillers of the chosen genotypes were placed into greenhouse flats, placed on their side and allowed to produce replicate tillers. In this way, we created identical copies of all genotypes used in the experiments.
We chose garden sites in the Czech Republic (near Trebon, 49.0042˚N, 14.7721˚E) and France (near Moussac, 43.9808˚N, 4.2241˚E) (Table 1). Because we were interested in whether introduced genotypes could outperform native genotypes under conditions where natives should be favored, we planted the native and invasive genotypes back into gardens near their original sampling location ensuring that climatic conditions in the garden were most similar and therefore favorable for the native genotypes. Our garden sites were within 50 km of the original collection of genotypes, ensuring that climatic conditions (if not soil conditions) would be similar to the genotypes collection location. Each garden site was a wet meadow and characterized by a mix of herbaceaous perennial plants and also had large stands of Phalaris arundinacea. Specific site measurements (soil moisture, nutrient status, plant cover) were not collected at either site. However, at each site we planted nine replicate blocks designed to span the range of conditions found in the wet meadow. Our only criteria for eliminating a potential location for a plot was that it could not contain native Phalaris since
Table 1. The geographical position of the two European gardens and the four regions from which the plants were collected is given in latitude/longitude (Lat/Long) and a set of descriptive climatic variables. Note the Czech Republic and French genotypes were collected from within 50 kms of the respective gardens.
the presence of established Phalaris might have made it difficult to locate our transplants. In our statistical models, we treated the nine blocks as a random factor to incorporate local unexplained environmental variation. After planting, the plots were left to regrow and we did not remove any of the native vegetation.
Plant quarantine rules prevented us from bringing tillers into Europe. Therefore, all plants were transported in moist paper towels as rhizome pieces without any soil present. In both Trebon and Moussac, rhizome pieces were transplanted into the common gardens approximately one week after they were prepared. Some early mortality may have resulted because of the lag between plant preparation and planting into the gardens. However, all individuals planted into each garden underwent the same preparation. Thus, transplants within a garden received the same treatment which allowed us to compare within a garden but not compare plants between gardens. In Moussac, we transplanted plants in October 2006 and harvested them in October 2008. However, because of the Mediterranean climate, which resulted in severe drought conditions in August, 85% of the plants died in August 2007. We therefore used the final growth measurements taken in May 2007 as our final experimental measurements. In Trebon, we transplanted the plants in May 2007 and harvested them in September 2009. However, in Trebon, high rainfall in August 2007 that flooded plots and disturbance by wild pigs resulted in high mortality. Wild pig damage was by trampling and not by eating. At the end of August 2007, only 33% of the original 324 plants remained. In Trebon, survival did differ among regions (see Table 2) but the differences were not related to the native/invasive dichotomy; rather the invasive region had both the highest (NC) and the lowest (VT) survivorship (see Results section).
In each garden, we transplanted nine replicates of each of 36 genotypes (i.e. a total of 324 plants per field site). The nine replicate 5 m × 5 m blocks were specifically chosen to encompass the natural variation present in our field sites. Individual rhizomes were planted into the existing vegetation with minimal disturbance to the surrounding soil and vegetation following the methods outlined
Table 2. Results of ordinal logistical model on survival for the Trebon and Moussac gar- dens. Values in Bold denote statistical significance. Overall model significance: Trebon: Log-Likelihood Ratio 27.325 Chi-Square 4.64, p = 0.1305; Moussac: Log-Likelihood Ratio 56.39 Chi-Square 112.78, p > 0.0001.
in [8] . Each of the experimental plants was measured monthly for survivorship, stem height, number of leaves and number of tillers. Plants did not flower in either garden during the duration of the experiment. Thus, our assessment of performance was survival and the three growth measures (tiller number, plant height, and leaf number). Tiller number, which measures a plant’s ability to spread and take over an area within the plant community following colonization, is a reasonable proxy for vegetative plant fitness. Because of low survival at the end of the experiment, we could not assess final biomass. Previous experimental work (Molofsky perscomm) indicated that tiller number is correlated with final biomass.
In the Moussac garden, we recorded the number of plants with evidence of herbivore damage. We did not record herbivore damage in the Trebon garden because of low overall herbivory. In the Moussac garden, because we visited the gardens monthly during the growing season, we could not accurately assess the identity of the herbivores that caused damage to our plants. We used a crude measure of herbivore damage. Evidence of herbivory was assessed as either damaged (1) or not damaged (0). We considered a plant damaged if it showed signs of tearing, holes in leaves or any other evidence of damage. We calculated the cumulative number of plants that experienced herbivory throughout the first year. We made the stipulation that once a plant had been counted as eaten it could not be counted again at a later survey. Dead plants that were not obviously eaten were removed from the analyses. Overall, we found that herbivory damage was low (45 plants out of 324). Therefore, we combined the regional genotypes data into two categories (native and invasive) and performed a contingency analysis (Chi Square test) on the number of plants eaten versus not eaten for native versus invasive genotypes.
We conducted an experiment to determine genotypic and regional differences in response to grazing by removing biomass of transplanted individuals in a garden in Burlington, Vermont (44.4759˚N, 73.2121˚W).
We transplanted tillers of each of the 36 genotypes into five experimental blocks on June 1-3, 2008. Tiller preparation followed methods previously described. Transplants were allowed to establish for four weeks. Following the initial establishment phase, we simulated grazing by removing all leaves from the main tiller and leaving only 1/2 of the youngest of the fully expanded shoot for each transplant ( [31] ); this treatment simulates grazing by mammalian herbivores. Simulated grazing treatments were implemented every two weeks from July 1st through August 15th, 2008. Removed biomass was dried at 60˚C and wei- ghed to assess the recovery of individuals from grazing.
3. Statistical Analyses
We performed separate analyses on each of our gardens to compare the performance of individuals within the garden but not between gardens. To assess survival differences among individuals, we used an ordinal logistic regression [32] . The effects in the model included block, range, region nested within range, population nested within region and range, and genotype nested within population, region and range.
In addition to survival, we assessed overall plant performance for three plant traits (tiller number (log transformed), leaf number and stem height (log-trans- formed). We analyzed leaf number and stem height separately because they were weakly correlated (less than 0.49) and each trait may represent a different gro- wth strategy. In Phalaris arundinacea, flowering only occurs once a plant reaches a threshold height (~30 cm Collins pers. observation). Thus, height represents an important feature of the plants that can influence competitive ability (for light) and reproductive ability.
We performed a mixed ANOVA with range and region within range as the main effects and block, population nested within range and genotypes nested within population region, range as random factors.
We also recorded the presence (1) or absence of herbivory on each plant in the Moussac garden. Data on the presence/absence of herbivory was analyzed using a Chi-Square test.
In the simulated grazing experiment, we analyzed how each plant recovered following biomass removal. Total accumulated removed biomass was analyzed using a general linear model with range and region within range as main effects. The biomass data was square root transformed to achieve normality.
4. Results
The overall performance of the plants in the gardens differed. First of all, survival varied greatly between gardens. In the Trebon garden, only 34% of the plants survived whereas in the Moussac garden 66% of the plants survived. In Trebon, the full logistic regression model including all main effects was not significant, most likely because of the overall low survival of all individuals (Table 2, Figure 1); however, despite the overall low survival, we still found significant differences in regional survival (Table 2) with the greatest difference between NC and VT genotypes (44% survival vs. 25% survival for NC and VT, respectively). Survival for the CZ and FR genotypes was intermediate and similar (35% vs. 30% for CZ and FR, respectively). In the Moussac garden, the overall model was significant (Table 2) but neither of our main explanatory factors (range and region within range) significantly affected survivorship; rather the random factors of block, population and genotype were all highly significant (Table 2). However, average survivorship did differ between the northern and southern genotypes and cut across invasive and native and regional differences (Figure 1). The CZ and VT genotypes had lower survivorship (58% and 55% respectively) than the FR and NC genotypes (78% and 80% respectively).
In both the Trebon and Moussac gardens, tiller number was low (approximately 3 and 2, in Trebon and Moussac, respectively). The small number of tillers per plant made it unlikely that our main effects of range or region within range were significant and in fact, neither of our main effects were significant in either garden (Table 3(a) and Table 3(b)). However, in Trebon, the random
Figure 1. Mean number of individuals aggregated by region (VT, NC, CZ, FR) surviving in the common gardens a. Trebon, Czech Republic, and b. Moussac, France. Black bars denote invasive range; grey bars denote native range.
Table 3.Results of the mixed-ANOVA for the three growth measurements in the A Trebon garden and B. Moussac garden. Significant values are indicated in bold.
variable of block explained 14% of the variation and population accounted for another 4%. In Moussac, block explained approximately 20% of the variation in tiller number and genotype explained approximately 4% of the variation.
For the trait of plant height, in the Trebon garden, differences were not explained by differences in either range or region within range (Table 3(a)). Nor did population or genotypes within population account for the variation. Rather, the random factor of block accounted for 44% of the variation with the rest of the variation being unexplained. In the Moussac garden, the variability in plant height was not explained by either of the main effects of range or region within range. However in contrast to the Trebon garden, population (approximately 10%) and genotypes within population explained the variation (approximately 11%); thus, approximately 21% of the total variation was explained by plant identity although not correlated with our expected explanatory factors. The random variable of block accounted for approximately 4% of the variation.
For leaf number, in the Trebon garden, we found a modest difference between native and invasive individuals; native genotypes had slightly more leaves than invasive individuals (native LSMean = 3.2, S.E. 0.029 vs. invasive LSMean = 2.6, S.E. = 0.29, p = 0.085). However, regional differences were not significant. Genotype within population accounted for approximately 7% of the variation and block accounted for approximately 13% of the variation. In the Moussac garden, we found no significant variation in leaf number for our main effects of range or region within range. Likewise, population differences were not significant but genotypic differences accounted for 6% of the variation and variation among blocks for another 15%.
Herbivory in the Moussac garden was overall very low; only 45 of our original transplants experienced herbivory. Invasive plants had a higher probability of being eaten than native plants (X2 = 12.26, p = 0.0005) (Figure 2). Overall, NC genotypes suffered the highest herbivory (17 plants) and FR genotypes the least (2 plants) (Figure 2). Plants that are eaten may respond by producing more tillers [21] . In the Moussac garden, we found that invasive NC genotypes produced slightly more tillers per plant than native FR genotypes (invasive NC 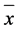 = 2.3, s.e.m. = 0.1211 vs. native FR
= 2.3, s.e.m. = 0.1211 vs. native FR 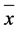 =1.9, s.e.m. = 0.1284, p = 0.026) but the differences were modest and likely of little import in the field.
=1.9, s.e.m. = 0.1284, p = 0.026) but the differences were modest and likely of little import in the field.
Our experimental grazing experiment provides evidence for how plants recover following major herbivore damage such as grazing. We predicted that invasive genotypes would recover faster than native genotypes and, in fact, we did find that invasive genotypes regrew more biomass following biomass removal than native genotypes (SS1,102 = 96.34, F = 4.78, p = 0.0312). Although regional differences were not significant in the full model, multiple comparison tests indicate that NC genotype produced the most cumulative total biomass and FR genotypes the least (Figure 3).
5. Discussion
Introduced individuals can become invasive in a new range through a process of
Figure 2. The proportion of plants from the different regions with herbivore damage from the four different regions (VT, NC, CZ,FR) in the Moussac garden. Black bars denote the invasive range; grey bars denote the native range. Chi-Square test indicated that invasive plants had higher herbivore damage than native plants (X2 = 12.26, p = 0.0005).
Figure 3. Total accumulated biomass following bi-weekly harvesting in Burlington, Vermont garden (44o.46/’73o.15’). Error bars represent + SE and letters represent significant differences (p < 0.05). Black bars denote the invasive range; grey bars denote the native range.
selection for greater growth and higher fitness [1] . This selection process can occur through genotypic sorting post introduction or evolution in situ may select for traits that can enhance performance, promoting invasiveness in the new range [5] . The introduction of new genetic variants into a new range can also allow genomes that have never been in contact with each other to hybridize, potentially resulting in new combinations with different traits that have higher fitness than the original native counterparts [11] [14] . Introduced species that have been introduced multiple times for economic or conservation uses such as Phalaris arundinacea, may have already been selected prior to introduction for traits that confer advantages such as higher growth rate, greater biomass production, and greater ability to withstand herbivore damage [8] [23] .
To understand whether introduced individuals have acquired traits that make them more invasive, one needs to compare native and invasive individuals when grown together under environmental conditions for which the natives has been selected but the introduced individuals have not. For the invasive and native genotypes studied here, we find only minimal differences between native and invasive genotypes; the only significant native/invasive difference was leaf number in one garden. Thus, for the individuals in this study, invasive individuals were not superior to their native counterparts. In fact, population differences may predominate over any native/invasive difference. In a common garden study on invasive Solidago canadensis in Europe, van Kluenen and Schmidt 2003 [33] found no consistent native/invasive differences among the 9 invasive European and 10 native United States populations when planted into a common garden in the invasive range. In our study, genotypes from NC outperformed other genotypes but these differences may not necessarily be related to their invasive status. Invasive VT genotypes enjoyed no such advantages, even when grown in similar climatic conditions such as Trebon, Czech Republic.
Invasive phenotypes may only be expressed in the correct environmental context [34] . For the same genotypes used in this study, Molofsky and Collins 2015 [18] found that invasive genotypes only outperformed native genotypes under ideal growing conditions but not under more stressful growing conditions. In this study, differences in survival among genotypes occurred but were not linked to invasion status. Rather in the northern garden in Trebon, the invasive genotypes had both the highest (NC genotypes) and the lowest (VT) survivorship. In the southern garden in Moussac, the southern genotypes (NC and FR) had higher survival than the northern genotypes (VT and CZ) suggesting that Phalaris genotypes had adapted to higher temperatures. Heat tolerance in this cool season grass is likely to be a stronger selective force than cold tolerance since plants can become dormant during the winter months.
Several plant traits such as faster growth rate, taller and greater leaf production are all assumed to be higher in invasive individuals [35] . In the common gardens here, no such trend was found. Plant height was primarily environmentally determined in Trebon (44% of the variation) and only correlated with plant identity in Moussac (22% of the variation). Tiller number, a measure of vegetative spread in a clonal plant was also not correlated with native/invasive dichotomy and was primarily environmentally determined. For leaf number, we did find a native/invasive dichotomy, but the differences were only present in one garden and too slight to be of biological significance.
Invasive populations have been postulated to have greater growth at the cost of reduction in defense [19] [36] [37] . If invasive individuals have reallocated resources away from defense and into growth, then we would expect that invasive individuals would be preferentially attacked in the native range. In common garden studies of Silene latifolia in the native European range, Wolfe et al. 2004 [37] showed that although invasive individuals had greater herbivore and fungal damage than natives in the native European garden, the invasive populations still outperformed native populations. Similarly, in a study on the invasive liana kudzu (Peuraria montana var. lobata) where invasive and native populations were planted into the native range, invasive individuals suffered higher herbivore damage than the native individuals, but produced greater biomass [30] . Here we find a similar result; although NC genotypes are the most heavily damaged, they survive as well as the native locally adapted FR genotypes. A companion experiment documented the response of native and invasive genotypes to experimental grazing and found that NC genotypes produced the greatest biomass and FR genotypes produced the least. Brodersen et al. 2008 [38] showed that FR genotypes have higher photosynthetic rates but slower growth rates in a common greenhouse study suggesting that increased carbon gained through photosynthesis is being allocated to defense.
Introduction history can have a large influence on the subsequent success of an introduced species [11] . Phalaris has been used as a forage crop in wet pastures for over 50 years in Vermont but in North Carolina it has been primarily introduced in conservation mix for ditch stabilization (Molofsky pers com). We therefore expected that the genotypes used for feed to have greater growth rates and greater ability to recover following grazing. Surprisingly, we find that NC genotypes have higher growth rates following grazing than genotypes from VT or the native range. However, the superior performance of the NC genotypes may be somewhat arbitrary and not related to their invasive status.
Our results have implications for the continued spread of invasive plant species. Firstly, if invasive plant species have wider climatic tolerances then native species than invasive species may spread at the expense of native ones. In our study, only the NC genotypes had greater climatic tolerances as evidence by their relatively high survival in both gardens as compared to the VT genotypes. Secondly, although biological control can be an important component to controlling invasive plant species, our results show that some invasive genotypes may be able to compensate with higher growth rates in the face of even extreme grazing pressure and may be able to maintain their fitness in the field even when preferentially preyed upon. Thirdly, an important conclusion of this research is the wide range of responses from presumably invasive genotypes when planted into field settings. Thus, studies that look for traits associated with invasiveness may find them in “ideal” settings but these same traits may not be expressed under more stressful field conditions. Finally, our study emphasizes that not all introduced populations are equally invasive and that it is important to examine multiple populations of introduced genotypes before drawing conclusions about whether invasive individuals outperform native ones [39] . Therefore, any conclusions about invasive behavior must be experimentally verified and tested in a range of habitats and for multiple populations.
Acknowledgements
We thank Jitka Klimesova, Jordi Cortina, Neil Buckley, Thomas Buckley, Carey Suehs, David Carbonell, Manisha Patel, William Teague, Mike Fitzgerald, Christine O’Brien, Lenka Malikova and Monika Sosnova for help arranging and setting up the common garden experiments.
Funding
This research was funded by USDA NRI-2006-03645 and USDA HATCH to J.M.
Cite this paper
Molofsky, J., Collins, A.R., Imbert, E., Bitinas, T. and Lavergne, S. (2017) Are Invasive Genotypes Superior? An Experimental Approach Us- ing Native and Invasive Genotypes of the Invasive Grass Phalaris arundinacea. Open Journal of Ecology, 7, 125-139. https://doi.org/10.4236/oje.2017.72010
References
- 1. Bossdorf, O., Auge, H., Lafuma, L., Rogers, W.E., Siemann, E. and Prati, D. (2005) Phenotypic and Genetic Differentiation between Native and Introduced Plant Populations. Oecologia, 144, 1-11.
https://doi.org/10.1007/s00442-005-0070-z - 2. Bossdorf, O., Lipowsky, A. and Prati, D. (2008) Selection of Preadapted Populations Allowed Senecio inaequidens to Invade Central Europe. Diversity and Distributions, 14, 676-685.
https://doi.org/10.1111/j.1472-4642.2008.00471.x - 3. Sakai, A.K., et al. (2001) The Population Biology of Invasive Species. Annual Review of Ecology and Systematics, 32, 305-332.
https://doi.org/10.1146/annurev.ecolsys.32.081501.114037 - 4. Muller-Scharer, H., Schaffner, U. and Steinger, T. (2004) Evolution in Invasive Plants: Implications for Biological Control. Trends in Ecology & Evolution, 19, 417-422.
https://doi.org/10.1016/j.tree.2004.05.010 - 5. Prentis, P.J., Wilson, J.R.U., Dormontt, E.E., Richardson, D.M. and Lowe, A.J. (2008) Adaptive Evolution in Invasive Species. Trends in Plant Science, 13, 288-294.
https://doi.org/10.1016/j.tplants.2008.03.004 - 6. Drenovsky, R.E., Grewell, B.J., D’Antonio, C.M., et al. (2012) A Functional Trait Perspective on Plant Invasion. Annals of Botany, 110, 141-153.
https://doi.org/10.1093/aob/mcs100 - 7. Keller, S.R., Sowell, D.R., Neiman, M., Wolfe, L.M. and Taylor, D.R. (2009) Adaptation and Colonization History Affect the Evolution of Clines in Two Introduced Species. New Phytologist, 183, 678-690.
https://doi.org/10.1111/j.1469-8137.2009.02892.x - 8. Lavergne, S. and Molofsky, J. (2007) Increased Genetic Variation and Evolutionary Potential Drive the Success of an Invasive Grass. Proceedings of the National Aca-demy of Sciences of the United States of America, 104, 3883-3888.
https://doi.org/10.1073/pnas.0607324104 - 9. Dlugosch, K.M. and Parker, I.M. (2008) Invading Populations of an Ornamental Shrub Show Rapid Life History Evolution despite Genetic Bottlenecks. Ecology Letters, 11, 701-709.
https://doi.org/10.1111/j.1461-0248.2008.01181.x - 10. Ellstrand, N.C. (2009) Evolution of Invasiveness in Plants Following Hybridization. Biological Invasions, 11, 1089-1091.
https://doi.org/10.1007/s10530-008-9389-9 - 11. Keller, S.R. and Taylor, D.R. (2010) Genomic Admixture Increases Fitness during a Biological Invasion. Journal of Evolutionary Biology, 23, 1720-1731.
https://doi.org/10.1111/j.1420-9101.2010.02037.x - 12. Facon, B., Genton, B.J., Shykoff, J., Jarne, P., Estoup, A. and David, P. (2006) A General Eco-Evolutionary Framework for Understanding Bioinvasions. Trends in Ecology and Evolution, 21, 130-135.
https://doi.org/10.1016/j.tree.2005.10.012 - 13. Facon, B., Pointier, J.P., Jarne, P., Sarda, V. and David, P. (2008) High Genetic Variance in Life-History Strategies within Invasive Populations by Way of Multiple Introductions. Current Biology, 18, 363-367.
https://doi.org/10.1016/j.cub.2008.01.063 - 14. Culley, T.M. and Hardiman, N.A. (2009) The Role of Intraspecific Hybridization in the Evolution of Invasiveness: A Case Study of the Ornamental Pear Tree Pyrus calleryana. Biological Invasions, 11, 1107-1119.
https://doi.org/10.1007/s10530-008-9386-z - 15. Broennimann, O. and Guisan, A. (2008) Predicting Current and Future Biological Invasions: Both Native and Invaded Ranges Matter. Biology Letters, 4, 585-589.
https://doi.org/10.1098/rsbl.2008.0254 - 16. Broennimann, O., Treier, U.A., Muller-Scharer, H., Thuiller, W., Peterson, A.T. and Guisan, A. (2007) Evidence of Climatic Niche Shift during Biological Invasion. Ecology Letters, 10, 701-709.
https://doi.org/10.1111/j.1461-0248.2007.01060.x - 17. Gallagher, R.V., Beaumont, L.J., Hughes, L. and Leishman, M.R. (2010) Evidence for Climatic Niche and Biome Shifts between Native and Novel Ranges in Plant Species Introduced to Australia. Journal of Ecology, 98, 790-799.
https://doi.org/10.1111/j.1365-2745.2010.01677.x - 18. Molofsky, J. and Collins, A.R. (2015) Using Native and Invasive Populations as Surrogate “Species” to Predict the Potential for Native and Invasive Populations to Shift Their Range. Evolutionary Ecology Research, 16, 505-516.
- 19. Blossey, B. and Notzold, R. (1995) Evolution of Increased Competitive Ability in Invasive Nonindigenouse Plants—A Hypothesis. Journal of Ecology, 83, 887-889.
https://doi.org/10.2307/2261425 - 20. Maron, J.L., Vila, M. and Arnason, J. (2004) Loss of Enemy Resistance among Introduced Populations of St. John’s Wort (Hypericum perforatum). Ecology, 85, 3243-3253.
https://doi.org/10.1890/04-0297 - 21. Alward, R.D. and Joern, A. (1993) Plasticity and Overcompensation in Grass Response to Herbivory. Oecologia, 95, 358-364.
https://doi.org/10.1007/BF00320989 - 22. Bossdorf, O., Prati, D., Auge, H. and Schmid, B. (2004). Reduced Competitive Ability in an Invasive Plant. Ecology Letters, 7, 346-353.
https://doi.org/10.1111/j.1461-0248.2004.00583.x - 23. Lavergne, S. and Molofsky, J. (2004) Reed Canary Grass (Phalaris arundinacea) as a Biological Model in the Study of Plant Invasions. Critical Reviews in Plant Sciences, 23, 415-429.
https://doi.org/10.1080/07352680490505934 - 24. Lavergne, S., Muenke, N.J. and Molofsky, J. (2010) Genome Size Reduction Can Trigger Rapid Phenotypic Evolution in Invasive Plants. Annals of Botany, 105, 109-116.
https://doi.org/10.1093/aob/mcp271 - 25. Calsbeek, B., Lavergne, S., Patel, M. and Molofsky, J. (2011) Comparing the Genetic Architecture and Potential Response to Selection of Invasive and Native Populations of Reed Canary Grass. Evolutionary Applications, 4, 726-735.
https://doi.org/10.1111/j.1752-4571.2011.00195.x - 26. Gilchrist, G.W. and Lee, C.E. (2007) All Stressed Out and Nowhere to Go: Does Evolvability Limit Adaptation in Invasive Species? Genetica, 129, 127-132.
https://doi.org/10.1007/s10709-006-9009-5 - 27. Moloney, K.A., Holzapfel, C., Tielborger, K., Jeltsch, F. and Schurr, F.M. (2009) Rethinking the Common Garden in Invasion Research. Perspectives in Plant Ecology Evolution and Systematics, 11, 311-320.
https://doi.org/10.1016/j.ppees.2009.05.002 - 28. Williams, J.L., Auge, H. and Maron, J.L. (2008) Different Gardens, Different Results: Native and Introduced Populations Exhibit Contrasting Phenotypes across Common Gardens. Oecologia, 157, 239-248.
https://doi.org/10.1007/s00442-008-1075-1 - 29. Blair, A.C. and Wolfe, L.M. (2004) The Evolution of an Invasive Plant: An Experimental Study with Silene latifolia. Ecology, 85, 3035-3042.
https://doi.org/10.1890/04-0341 - 30. Yang, X., Huang, W., Tian, B. and Ding, J. (2014) Differences in Growth and Herbivory Damage of Native and Invasive Kudzu (Peuraria Montana var lobata) Populations Grown in the Native Range. Plant Ecology, 215, 339-346.
https://doi.org/10.1007/s11258-014-0304-4 - 31. Zhao, W., Chen, S.P. and Lin, G.H. (2008) Compensatory Growth Responses to Clipping Defoliation in Leymus chinensis (Poaceae) under Nutrient Addition and Water Deficiency Conditions. Plant Ecology, 196, 85-99.
https://doi.org/10.1007/s11258-007-9336-3 - 32. Software SS (2008) SAS/STAT Software: Changes and Enhancements Release 8.2. Cary.
- 33. Van Kleunen, M. and Schmid, B. (2003) No Evidence for an Evolutionary Increased Competitive Ability in an Invasive Plant. Ecology, 84, 2816-2323.
https://doi.org/10.1890/02-0494 - 34. González-Moreno, P., Pino, J., Carreras, D., Basnou, C., Fernández-Rebollar, I. and Vila, M. (2013) Quantifying the Landscape Influence on Plant Invasions in Mediterranean Coastal Habitats. Landscape Ecology, 28, 891-903.
https://doi.org/10.1007/s10980-013-9857-1 - 35. Pysek, P. and Richardson, D.M. (2007) Traits Associated with Invasiveness in Alien Plants: Where Do We Stand? In: Nentwig, W., Ed., Biological Invasions, Springer, Berlin, 97-125.
https://doi.org/10.1007/978-3-540-36920-2_7 - 36. Wolfe, L.M. (2002) Why Alien Invaders Succeed: Support for the Escape-from-Enemy Hypothesis. American Naturalist, 160, 705-711.
- 37. Wolfe, L.M., Elzinga, J.A. and Biere, A. (2004) Increased Susceptibility to Enemies Following Introduction in the Invasive Plant Silene latifolia. Ecology Letters, 7, 813-820.
https://doi.org/10.1111/j.1461-0248.2004.00649.x - 38. Brodersen, C., Lavergne, S. and Molofsky, J. (2008) Genetic Variation in Photosynthetic Characteristics among Invasive and Native Populations of Reed Canarygrass (Phalaris arundinacea). Biological Invasions, 10, 1317-1325.
https://doi.org/10.1007/s10530-007-9206-x - 39. Colautti, R.I., Maron, J.L. and Barrett, S.C.H. (2009) Common Garden Comparisons of Native and Introduced Plant Populations: Latitudinal Clines Can Obscure Evolutionary Inferences. Evolutionary Applications, 2, 187-199.
https://doi.org/10.1111/j.1752-4571.2008.00053.x







