Open Journal of Ecology
Vol.3 No.4(2013), Article ID:35039,5 pages DOI:10.4236/oje.2013.34034
Enrichment of nitrous oxide reducing bacteria from coastal marsh sediments
![]()
School of Arts and Sciences, University of Houston-Victoria, Victoria, USA; *Corresponding Author: sobolevd@uhv.edu
Copyright © 2013 Khuiong B. T. Nguyen, Dmitri Sobolev. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 19 April 2013; revised 21 May 2013; accepted 28 June 2013
Keywords: Nitrous Oxide Reduction; Salt Marsh; Denitrification; DGGE; Gammaproteobacteria
ABSTRACT
We attempted to recover organisms capable of respiratory nitrous oxide reduction with acetate as an electron donor from a variety of coastal marine sediments from Lavaca Bay area, Texas by use of liquid enrichment cultures. Putative positive cultures were analyzed by amplifying eubacterial and archaeal 16S rRNA gene fragments and analyzing their diversity by separating them by a denaturing gradient gel electrophoresis (DGGE). No Archaea was detected in our enrichments; however, positive enrichments from coastal salt marsh indicated the presence of putative nitrous oxide reducing bacteria. DGGE patterns of the amplified DNA were similar between enrichments, with ca. 7 obvious bands. The dominant bands were tentatively identified as members of the Gammaproteobacteria class, closely related to various denitrifying pseudomonads. Our results indicate that coastal marine environments may sustain a nitrous oxide reducing community, although nitrous oxide reduction is probably an opportunistic form of metabolism in that environment.
1. INTRODUCTION
Nitrous oxide (N2O) is a trace gas primarily of biological origin [1,2]. Nitrous oxide present in the atmosphere has been implicated in global warming [3,4] and ozone layer destruction [5]. The concentration of nitrous oxide in the atmosphere has been increasing over the last few decades [3,6].
In biological systems, nitrous oxide is produced as an obligate intermediate in denitrification [7-9], being reduced to N2 by the action of Nos nitrous oxide reductase [10] in the final step of the same pathway [11].
Several processes can result in nitrous oxide release. Nos reductase is readily inhibited by hydrogen sulfide nearly omnipresent in suboxic marine environments [12]. A degree of transcriptional regulation of nitrous oxide reduction, similar to that described by Korner and Zumft [13] can be expected. Studies in culture have discovered a microbial genus (Ottowia) producing N2O as the sole end product of denitrification [14].
Suppression of nitrous oxide reductase activity by any mechanism results in release of nitrous oxide into the environment [15]. Once released, extracellular N2O could be used in energy metabolism; e.g., appreciable amount of energy could be extracted from organic matter by Pseudomonas [16] with N2O as an electron acceptor.
Consumption of nitrous oxide by microbes has been described in a variety of natural systems under artificial conditions, with N2O gas phase concentration far in excess of atmospheric values [17-20]. Data of Naqvi et al. [21] indicate that N2O consumption may also occur in ocean water where high concentrations (up to several hundreds nmol per L) of this compound are found. None of those studies attempted to identify the organisms responsible for that phenomenon, enzymes involved or the specific pathway through which consumption occurs.
Several assumptions about nitrous oxide utilization could be made based on the nature of the compound. Under certain conditions, environmental N2O could provide energy as an electron acceptor [16] in a process identical to the final step of denitrification. Conrad [22] as well as Bliecher-Mathiesen and Hoffman [23] suggest that denitrifying bacteria may be the only biological sink for nitrous oxide. As redox potential of N2O is lower than that of either oxygen or nitrate, this process will be expected to occur in the absence of those compounds according to the general scheme of electron acceptor utilization [24]. Lower redox potential of N2O compared to oxygen also allows a possibility of nitrous oxide utilized as an electron donor, producing nitric oxide, nitrite or nitrate via dissimilatory oxidation, a possibility not considered in the proposed study.
Although various hypotheses could be proposed, metabolic pathways for, and organisms involved in nitrous oxide consumption remain unknown. Contrary to suggestions of Conrad [22] and Bliecher-Mathiesen and Hoffman [23], oxidation of hydrogen sulfide with simultaneous reduction of nitrous oxide by thiobacilli [25] or hydrogen oxidation with concomitant nitrous oxide reduction described by Bock et al. [26] show that nitrous oxide fate in the environment is more complex than can be inferred from our current state of knowledge of nitrogen cycle.
2. MATERIALS AND METHODS
Sediment and water samples were collected from a freshwater bayou, a salt marsh and a bay in the general vicinity of Lavaca Bay on Texas Gulf Coast. The silty sediment was sieved to remove large particles and mixed with site water to produce slurry. Ten milliliters of the slurry were placed into a 100 mL serum bottles (Wheaton) and bottle headspace was flushed for 5 minutes with high-purity nitrogen gas. The bottle was then crimp-sealed with a gas-tight stopper. Negative controls received no additional treatment; experimental bottles were additionally flushed with nitrous oxide for 1 minute by means of a needle through the stopper. Experimental bottles were re-flushed with nitrous oxide weekly to replace gas consumed.
Enrichment cultures were produced by transferring of 0.1 mL on the slurry from bottles exposed to nitrous oxide for 3 weeks into 20 mL of mineral medium containing 10 mM HEPES buffer (pH 7.5), 0.05 g/L NaHPO4, 0.05 g/L NH4Cl, 1× vitamins for basal medium (MP Biomedicals, LLC) and 1× trace elements B (Mediatech, Inc.) in 100 mL serum bottle, with sodium sulfate to match expected in situ salinity (approx. 5, 16 and 22 ppt). Potassium acetate was added as an electron donor to the final concentration of 10 mM. Medium was placed into a serum bottle, bubbled with high purity nitrogen for 5 minutes, sealed, and flushed with nitrous oxide. The culture was re-flushed with nitrous oxide after one week and after 2 weeks of incubation 1 mL of culture was transferred to a fresh medium. Such transfer was repeated twice before the analysis. Negative controls were set up identically, with exception of nitrous oxide flush.
PCR mixture contained 10 μL 10× PerfectTaq buffer, 20 μL 5P solution, 0.2 μL each of 200 μM forward and reverse primer solutions (Operon Technologies), 2 μL of 20 mM dNTP solution (Fisher Scientific), 2.5 units PerfectTaq polymerase, and PCR-grade water (Fisher Scientific) to a total of 100 μL. All chemicals, unless otherwise noted, were supplied as a part of the PerfectTaq kit from 5 Prime, Inc. Ca. 400 bp 16S rRNA gene fragment was amplified with primers EUB f933 and EUB r1387 [27]. Primers A348Kf (identical to A348If, except that inositol was replaced with an equimolar G and T base mixture) and Arch915R [28] were used for archaeal 16S rRNA gene amplification attempt. A GC-tail [29] was added to the 5' end of forward primers for DGGE analysis.
No DNA extraction was performed; culture was used directly as a template by adding 1 μL of culture to 100 μL of the PCR mix. Reaction mixes were cycled in a heatedlid Eppendorf Mastercycler as follows: initial melting at 94˚C for 5 minutes, followed by 35 cycles of melting at 96˚C for 30 seconds, annealing at 60˚C for 30 seconds and extension for 2 minutes at 72˚C. The cycling was followed by a final extension step of 5 minutes at 72˚C. The PerfectTaq system allowed the use of the same annealing temperature for all of the primer sets.
DGGE gel was loaded with 12 uL of the PCR product and run was performed in a 20% - 60% gradient [29] at 120 volts for 4 hours. Gels were recovered, stained with 1 mg∙L−1 ethidium bromide and photographed under UV illumination.
Prominent bands were recovered from the DGGE gel by coring, reamplified with the same primers (minus GC tails) and sent out for sequencing to Sequetech, Inc, (Mountain View, CA) with the same primers. Resulting. ab1 files were reviewed by use of TraceEdit 1.1 software and trimmed to remove poorly resolved ends. Sequencing reaction was run multiple times forward and reverse and ambiguous bases called according to majority rule. GenBank database was searched for relatives of assembled sequences by use of BLAST [30]. Sequences were aligned using the CLUSTAL W algorithm implemented in MEGA5 [31] and used to generate a UPGMA tree [32]. The evolutionary distances were computed using the Maximum Composite Likelihood method according to the Tamura-Nei model [33]. Sequences from this study were deposited into the GenBank database with accession numbers KF031269-KF031277.
3. RESULTS AND DISCUSSION
After approx. 2 weeks, several of the cultures inoculated with nitrous oxide exposed sediment samples from the salt marsh developed turbidity; no such turbidity was observed in uninoculated controls (not shown). Visible turbidity developed consistently at 12 - 14 days in the subsequent transfers. Both freshwater and bay enrichments failed to produce growth signs and were discarded from further analysis. Lack of sulfate reduction, detected by the lack of characteristic sulfide smell was taken as a sign that sulfate was not utilized as an electron acceptor.
All growth-positive enrichments (identified visually by their turbidity) yielded appropriately sized amplicons when amplified with bacteria-specific primers. Amplification of archaeal 16S rRNA gene produced no signal in any of the samples. DGGE separation of the amplicons produced fundamentally similar patterns, with small variations sample to sample (Figure 1). A total of 9 bands marked with numbers were successfully recovered from the DGGE gel (Figure 1). All of the organisms identified from the DNA sequencing were closely related to each other and to the known pseudomonads, falling within the Gammaproteobacteria class.
An UPGMA tree of the recovered sequences is presented in Figure 2. Sequences designated DGGE-Nitrous-01 (KF031269) and DGGE-Nitrous-03 (KF031271) were indistinguishable from each other and 99% identical to phthalate esther degrading Pseudomonas sp. p6-4 + 2 (EF066358). Curiously, Pseudomonas sp. p6-4 + 2 was also 99% identical to sequence DGGE-Nitrous-04 (KF031272) (Figure 2) and similar to clone DGGE-Nitrous-07 (KF031275). Those clones formed a sister clade to oilfield-associated pseudomonads GE7GXPU01- AOUFH (HM974652) and GE7GXPU01B8OHF (HM- 972903) and clone F5OHPNU07H3W8N (HQ091939)
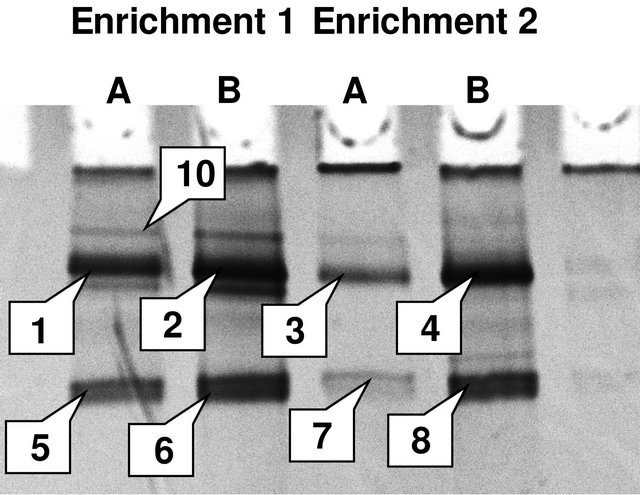
Figure 1. Community structure of nitrous oxide enrichments. Band numbers correspond to clone numbers in Figure 2.
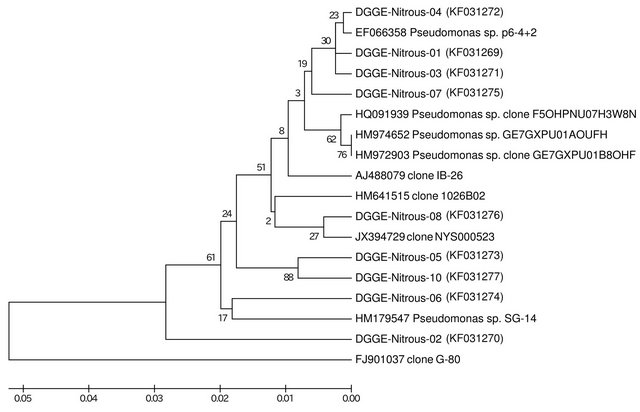
Figure 2. UPGMA phylogenetic tree of enrichment-recovered organisms.
from oil sands tailing ponds [34]. Clone DGGE-Nitrous- 08 (KF031276) was closely related to airborne clone NYS000523 (JX394729) from New York City subway, with which it formed a sister clade to clone 1026B02 (HM641515) associated with nitrate-dependent pyrite oxidation. Sequences DGGE-Nitrous-05 (KF031273) and DGGE-Nitrous-10 (KF031277) formed their own unique clade. Clone DGGE-Nitrous-06 (KF031274) was similar to sequence HM179547 of Pseudomonas sp. SG-14. Sequence DGGE-Nitrous-02 (KF031270) was 95% identical to clone G-80 from a petroleum reservoir [35].
Incomplete denitrification to nitrous oxide has been observed under a variety of circumstances. Generally, this process may occur when nitrous oxide reductase is inhibited by hydrogen sulfide [12] or when organic matter is present in stoichiometric excess to nitrate [16]; some organisms produce nitrous oxide constitutively. Based on our results, saltwater marsh has the right combination of factors for nitrous oxide production and concomitant consumption.
The organisms recovered in our enrichments fell squarely among Gammaproteobacteria, while being closely related to various pseudomonads. Members of this group of organisms are known to be capable of complete denitrification of nitrate to dinitrogen. As nitrous oxide is obligately produced and consumed in this process, it is not surprising that this gas can enter the metabolism of denitrifying bacteria at an intermediate point.
The fact that the organisms recovered are closely related to the ones capable of complete denitrification pathway raises an interesting question: are the organisms responsible for nitrous oxide consumption capable of consuming this compound constitutively, or are we dealing with opportunistic organisms taking advantage of electron acceptors as they become available? This question is beyond the scope of this paper and will be addressed in the future studies.
4. CONCLUSION
Our investigation of a denitrifying microbial community by a combination of culturing and molecular approaches demonstrated the presence of organisms capable of nitrous oxide reduction with acetate as an electron donor, either constitutively or opportunistically. Those organisms belong to the class Gammaproteobacteria, are closely related to pseudomonads from various environments potentially capable of denitrification and found under a specific set of environmental conditions. Overall, our results support conclusions by Koike and Hattori [16] that Pseudomonas-like organisms are capable of consuming nitrous oxide as a part of their energy metabolism and those by LaMontagne et al., Mengis et al., Mei et al., and Schuster and Conrad [17-20] that those organisms could be ubiquitous in the environment. Our results agree well with those by Conrad [22] as well as Bliecher-Mathiesen and Hoffman [23], who suggested that the organisms responsible for nitrous oxide consumption are likely to be the ones capable of denitrification.
5. ACKNOWLEDGEMENTS
This work was supported in part by the University of Houston-Victoria Office of the Provost Junior Faculty Research Grant to DS. Data processing, image manipulation and manuscript preparation were largely accomplished by use of free open source software; authors are indebted to Open Office, TraceEdit, MEGA and GIMP development communities.
REFERENCES
- Lindau, C.W., DeLaune, R.D., Jiraporncharoen, S. and Manajuti, D. (1991) Nitrous oxide and dinitrogen emissions from Panicum hemitomon S. fresh-water marsh soils following addition of N-15 labeled ammonium and nitrate. Journal of Freshwater Ecology, 6, 191-198. doi:10.1080/02705060.1991.9665293
- Mengis, M., Gächter, R. and Wehrli, B. (1997) Sources and sinks of nitrous oxide (N2O) in deep lakes. Biogeochemistry, 38, 281-301. doi:10.1023/A:1005814020322
- Dickinson, R.E. and Cicerone R. J. (1986) Future global warming from atmospheric trace gases. Nature, 319, 109- 115. doi:10.1038/319109a0
- Wang, W.C., Yung, Y.L., Lacis, L.A., Mo, T. and Hansen G.E. (1976) Greenhouse effects due to man-made perturbations of trace gases. Science, 194, 685-690. doi:10.1126/science.194.4266.685
- Crutzen, P.J. (1970) The influence of nitrogen oxides on the atmospheric ozone content. Quarterly Journal of the Royal Meteorological Society, 96, 320-325. doi:10.1002/qj.49709640815
- Rasmussen, R.A. and Khalil, M.A.K. (1986) Atmospheric trace gases: Trends and distributions over the last decade. Science, 232, 1623-1624. doi:10.1126/science.232.4758.1623
- Anderson, I.C. and Levine, J.S. (1986) Relative rates of nitric oxide and nitrous oxide production by nitrifiers, denitrifiers, and nitrate respirers. Applied and Environmental Microbiology, 51, 938-945.
- Schmidt, H.L., Werner, R.A., Yoshida, N. and Well, R. (2004) Is the isotopic composition of nitrous oxide an indicator for its origin from nitrification or denitrification? A theoretical approach from referred data and microbiological and enzyme kinetic aspects. Rapid Communications in Mass Spectrometry, 18, 2036-2040. doi:10.1002/rcm.1586
- Firestone, M.K., Smith, M.S., Firestone, R.B. and Tiedje, J.M. (1979) The influence of nitrate, nitrite, and oxygen on the composition of the gaseous products of denitrification in soil. Soil Science Society of America Journal, 43, 1140-1144.
- Ferguson, S.J. (1998) Nitrogen cycle enzymology. Current Opinion in Chemical Biology, 2, 182-193. doi:10.1016/S1367-5931(98)80059-8
- Otte, S., Grobben, N.G., Robertson, L.A., Jetten, M.S. and Kuenen, J.G. (1996) Nitrous oxide production by Alcaligenes faecalis under transient and dynamic aerobic and anaerobic conditions. Applied and Environmental Microbiology, 62, 2421-2426.
- Sørensen, J., Tiedje, J.M. and Firestone, R.B. (1980) Inhibition by sulfide of nitric and nitrous oxide reduction by denitrifying Pseudomonas fluorescens. Applied and Environmental Microbiology, 39, 105-108.
- Korner, H. and Zumft, W.G. (1989) Expression of denitrification enzymes in response to the dissolved oxygen levels and respiratory substrate in continuous cultures of Pseudomonas stutzeri. Applied and Environmental Microbiology, 137, 74-78.
- Spring, S., Jäckel, U., Wagner, M. and Kämpfer, P. (2004) Ottowia thiooxydans gen. nov., sp. nov., a novel facultatively anaerobic, N2O-producing bacterium isolated from activated sludge, and transfer of Aquaspirillum gracile to Hylemonella gracilis gen. nov., comb. nov. International Journal of Systematic and Evolutionary Microbiology, 54, 99-106. doi:10.1099/ijs.0.02727-0
- Betlach, M.R. and Tiedje, J.M. (1981) Kinetic explanation for accumulation of nitrite, nitric oxide, and nitrous oxide during bacterial denitrification. Applied and Environmental Microbiology, 42, 1074-1084.
- Koike, I. and Hattori, A. (1975) Energy yield of denitrification: An estimate from growth yield in continuous cultures of Pseudomonas denitrificans under nitrate-, nitriteand oxide-limited conditions. Journal of General Microbiology, 88, 11-19. doi:10.1099/00221287-88-1-11
- LaMontagne, M.G., Duran, R. and Valiela, I. (2003) Nitrous oxide sources and sinks in coastal aquifers and coupled estuarine receiving waters. Science of the Total Environment, 309, 139-149. doi:10.1016/S0048-9697(02)00614-9
- Mengis, M., Gächter, R. and Wehrli, B. (1997) Sources and sinks of nitrous oxide (N2O) in deep lakes. Biogeochemistry, 38, 281-301. doi:10.1023/A:1005814020322
- Mei, L., Yang, L., Wang, D., Yin, B., Hu, J. and Yin, S. (2004) Nitrous oxide production and consumption in serially diluted soil suspensions as related to in situ N2O emission in submerged soils. Soil Biology & Biochemistry, 36, 1057-1066. doi:10.1016/j.soilbio.2004.03.001
- Schuster, M. and Conrad, R. (1992) Metabolism of nitric oxide and nitrous oxide during nitrification and denitrification in soil at different incubation conditions. FEMS Microbiology Letters, 101, 133-143. doi:10.1111/j.1574-6968.1992.tb05769.x
- Naqvi, S.W., Jayakumar, D.A., Narvekar, P.V., Naik, H., Sarma, V.V., D’Souza, W., Joseph, S. and George, M.D. (2000) Increased marine production of N2O due to intensifying anoxia on the Indian continental shelf. Nature, 408, 346-349. doi:10.1038/35042551
- Conrad, R. (1996) Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiology Letters, 60, 609-640.
- Bliecher-Mathiesen, G. and Hoffmann, C.C. (1999) Denitrification as a sink for dissolved nitrous oxide in a freshwater riparian fen. Journal of Environmental Quality, 28, 257-262. doi:10.2134/jeq1999.00472425002800010031x
- Cord-Ruwisch, R., Seitz, H.-J. and Conrad, R. (1988) The capacity of hydrogenotrophic anaerobic bacteria to compete for traces of hydrogen depends on the redox potential of the terminal electron acceptor. Archives of Microbiology, 149, 350-357. doi:10.1007/BF00411655
- Dalsgaard, T. and Bak, F. (1992) Effect of acetylene on nitrous oxide reduction and sulfide oxidation in batch and gradient cultures of Thiobacillus denitnificans. Applied and Environmental Microbiology, 58, 1601-1608.
- Bock, E., Schmidt, I, Stüven, R. and Zart, D. (1995) Nitrogen loss caused by denitrifying Nitrosomonas cells using ammonium or hydrogen as electron donors and nitrite as electron acceptor. Archives of Microbiology, 163, 16- 20. doi:10.1007/BF00262198
- Kawai, M., Matsutera, E., Kanda, H., Yamaguchi, N., Tani, K. and Nasu, M. (2002) 16S ribosomal DNA-based analysis of bacterial diversity in purified water used in pharmaceutical manufacturing processes by PCR and denaturing gradient gel electrophoresis. Applied and Environmental Microbiology, 68, 699-704. doi:10.1128/AEM.68.2.699-704.2002
- Hori, T., Haruta, S., Ueno, Y., Ishii, M. and Igarashi, Y. (2006) Direct comparison of single-strand conformation polymorphism (SSCP) and denaturing gradient gel electrophoresis (DGGE) to characterize a microbial community on the basis of 16S rRNA gene fragments. Journal of Microbiological Methods, 66, 165-169. doi:10.1016/j.mimet.2005.11.007
- Muyzer, G. (1999). DGGE/TGGE a method for identifying genes from natural ecosystems. Current Opinion in Microbiology, 2, 317-322. doi:10.1016/S1369-5274(99)80055-1
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z. Miller, W. and Lipman, D.J. (1997) Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research, 25, 3389-3402. doi:10.1093/nar/25.17.3389
- Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M. and Kumar, S. (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28, 2731-2739. doi:10.1093/molbev/msr121
- Sneath, P.H.A. and Sokal, R.R. (1973) Numerical taxonomy. Freeman, San Francisco, 573 p.
- Tamura, K. and Nei, M. (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution, 10, 512-526.
- Ramos-Padrón, E., Bordenave, S., Lin, S., Bhaskar, I.M., Dong, X., Sensen, C.W., Fournier, J., Voordouw, G. and Gieg, L.M. (2011) Carbon and sulfur cycling by microbial communities in a gypsum-treated oil sands tailings pond. Environmental Science & Technology, 45, 439-446. doi:10.1021/es1028487
- Ren, H.-Y., Zhang, X.-J., Song, Z.-Y., Rupert, W., Gao, G.-J., Guo, S.-X. and Zhao, L.-P. (2011) Comparison of microbial community compositions of injection and production well samples in a long-term water-flooded petroleum reservoir. PLoS ONE, 6, Article ID: 23258. doi:10.1371/journal.pone.0023258

