Open Journal of Genetics
Vol.06 No.04(2016), Article ID:72971,9 pages
10.4236/ojgen.2016.64010
The Study of Apolipoprotein E4 Allele Distribution in Parents of Down’s Syndrome Children as a Risk Factor in Khorasan Razavi Province, Iran
Elmira Iranifar, Tayebeh Hamzehloie*
Department of Human Genetics, Mashhad University of Medical Science, Mashhad, Iran
Copyright © 2016 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: November 20, 2016; Accepted: December 20, 2016; Published: December 23, 2016
ABSTRACT
Backgrounds: Down syndrome (DS) is the most common chromosomal abnormality. The most important factor in DS is increased maternal age so after the age of 35, the risk of Down syndrome in pregnancy increases. Down syndrome can be diagnosed during pregnancy by prenatal screening. Nondisjunction in cell divisions is the main cause of the DS. Apo lipoprotein E is a 317 amino acid glycoprotein that plays an essential role in metabolism and cholesterol transport. Alzheimer’s disease (AD) is one of the symptoms of adults with DS. The apoE allele e4 has been identified as a risk factor for AD and also, played a main role in nondisjunction. An increased risk of AD in mothers of adults with DS has been reported. We hypothesized that young mothers of DS children (<35 age) could have an increased frequency of apoE allele E4 so we studied apoE allele distribution in cases of trisomy 21 and their parents. In present study, we investigated association of 112 codon of APOE gene C/T and 158 codon of APOE gene C/T with DS children and their parents in Northeast of Iran (Khorasan Razavi Provence). Methods: In this case-control study, 33 DS children and their parents were compared in case of age with 90 families without any history of DS. Genotyping was performed by ARMS-PCR technique. Statistical analysis was performed by SPSS v.21 software. Results: It indicated that there is a significant difference in allele distribution between case and control groups. The C allele for 112 codon of APOE gene and the C allele for 158 codon of APOE gene may associate with nondisjunction. In 112 codon of APOE gene, it seems having T allele reduces the risk of nondisjunction and in contrast C allele may be a risk factor in happening of nondisjunction. (p-value = 0.000006, OR = 2.66, 95% CI = 1.74 - 4.06). In 158 codon of APOE gene, it seems having T allele reduces the risk of nondisjunction and in contrast C allele may be a risk factor in happening of nondisjunction. (p-value = 0.0000, OR = 3.89, 95% CI = 2.38 - 6.34). E4 allele frequency in mothers of DS is about 14% more than those in control group. According to results of this study the C allele in 158 codon of APOE gene and the C allele in 112 codon of APOE gene could be considered as susceptibility genetic factors for nondisjunction in Northeast of Iran.
Keywords:
Down Syndrome, Nondisjunction, ApoE Allele e4, Polymorphism

1. Introduction
In 1959, Lejeune, Gautier, and Turpin discovered the association between Down’s syndrome and a third chromosome 21 [1] . Down syndrome (DS or DNS), also known as trisomy 21, is a genetic disorder caused by the presence of all, or part of a third copy of chromosome 21 [2] . It is typically associated with physical growth delays, characteristic facial features, and mild to moderate intellectual disability [3] . For example, the average IQ of a young adult with Down syndrome is 50 which is the same as mental age of an 8- or 9-year-old child, but this can vary widely. There is no known behavior or environmental factor that changes the risk. We can diagnose Down syndrome by prenatal tests and by direct observation and genetic testing after birth. The diagnosis of Down’s syndrome is made by chromosome analysis, which can be initiated prenatally due to identified risk factors, or postnatally due to the characteristic appearance of the infant [4] . Prenatal diagnosis for chromosomal anomalies was first introduced in the 1970s, and was initially restricted to amniocentesis in the second trimester [5] . Down syndrome can be identified during pregnancy by prenatal screening followed by diagnostic testing, or after birth by direct observation and genetic testing [2] . Research efforts now focus on improvement of the sensitivity and specificity of screening, to reduce or eliminate the number of women needing an invasive diagnostic test, such as chorionic-villus sampling or amniocentesis [1] . Use of fetal cells in the maternal circulation for prenatal diagnosis could eliminate the need for amniocentesis in diagnostic testing, however, isolation of fetal cells from maternal blood is still associated with several technical and biological difficulties [6] . Down syndrome is caused by having three copies of the genes on chromosome 21 [7] . Approximately 90% - 95% have free trisomy 21’, and in about 95% of these the extra chromosome is of maternal origin, as determined by DNA-poly- morphism analysis [8] . Among maternal errors, about 75% are a result of non-dis- junction in the first meiotic division and 25% of non-disjunction in the second meiotic division of the oocyte [9] . We still know little about the causes of non-disjunction that lead to Down’s syndrome. Calculation of the frequency of Down’s syndrome depends on whether maternal age, gestational timing of diagnosis, and case loss due to prenatal diagnosis and termination of pregnancy are taken into account [10] . Maternal age affects the chances of having a pregnancy with Down syndrome and advanced maternal age remains the only well documented risk factor in DS [11] . There is no clear evidence of paternal age effect. The number of terminated pregnancies with Down’s syndrome has increased, and the prevalence of Down’s syndrome births has decreased from one in 700 to about one in 1000 [12] . Many (15%) who live 40 years or longer develop Alzheimer disease [13] . Apolipoprotein E (apoE) is a plasma protein involved in cholesterol transport and metabolism. The apoE gene is located on chromosome 19, and the three most common alleles are e2, e3, and E4 [14] . The apoE allele e4 has been identified as a risk factor for early-onset and late-onset Alzheimer’s disease (AD) in both familial and sporadic cases [15] [16] . Increased frequency of Down syndrome (DS) births in the families of individuals with AD and increased frequency of AD in relatives of DS probands suggest a shared genetic susceptibility to DS and AD [17] . An increased risk of AD in mothers of adults with Down syndrome (DS) has been reported. A shared genetic susceptibility to DS and AD leading to an accelerated ageing process had been hypothesized. Subsequently, an increased e4 allele frequency has been found in mothers (£32 years of age) of DS children arising from a meiosis II (MII) error in the Danish population [18] . We hypothesized that young mothers of DS children could have an increased frequency of apoE allele E4, and studied apoE allele distribution in cases of trisomy 21 and their parents. Wehypothesized that young mother of DS children could have an increased frequency of Apolipoprotein E (apoE) allele E4. Apolipoprotein E4 allele distribution in parents of Down’s syndrome children as a risk factor in KhorasanRazaviprovince, Iran was studied in this paper. The present study is to investigate association of 112 codon of APOE gene C/T and 158 codon of APOE gene C/T with Down syndrome in population of Northeast Iran (Khorasan Razavi).
2. Material and Methods
NCBI SNP database (dbSNP) was explored to discover frequency of these polymorphisms (112 codon of APOE gene C/T and 158 codon of APOE gene C/T) (http://www.ncbi.nlm.nih.gov/projects/SNP). No frequency data was registered for these variations in dbSNP. To determine power of study, information about frequency is essentials. There for 99 individuals (33 families with Down syndrome “trio study”) with Persian origin from Northeast of Iran was selected and genotyped for mentioned SNPs in order to find out allele frequency in our population. 33 mothers, and their corresponding fathers and DS children (due to free trisomy 21) from the population of North-east of Iran have been included in this study. All samples were collected after informed consent during the period between 2013 and 2015.Basic demographic data of these participators are about age, sex, race. The average age of Down syndrome children was 6 years (range 0 - 35). Average maternal age was 33.2 years. Inclusion criteria is thatall patients with Down syndrome who are already using Karyotyping for detecting trisomy 21 has been confirmed in them. In addition, 270 subjects from the general population (paternity testing cases with an a Exclusion criteria is all patients who do not have Down syndrome or, there is no evidence of a definite diagnosis. verage age of 36 years; 90 males, 90 females and 90 children) chosen as control group. Control individuals have no clinical evidence for Down syndrome. The study received the approval of the hospital Ethics Research Committee and all the investigated participants provided informed consent before sampling.
2.1. DNA Extraction & Genotyping
Peripheral blood from 99 patients and 270 controls was collected in EDTA CBC tube and genomic DNA was extracted and purified from whole blood lymphocytes 5PRIME kit according to the manufacturer’s instructions. Specific primers for detection of SNP by Amplification Refractory Mutation System-Polymerase Chain Reaction (ARMS- PCR) were designed by using PRIMER 3 software (http://primer3.ut.ee). Specificity of designed primers was checked for human genome by the Primer-BLAST tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast). Finally, Oligo Analyzer software was usedto check the absence of Hairpin, homodimeric and heterodimeric in PCR conditions (http://eu.idtdna.com/calc/analyzer). The sequences of primers are shown in Table 1. In study genotyped in a single reaction tube with six primers consisting of two common primers and two specific primers for each of two single nucleotide polymorphism (SNP) sites by tetra-primer amplification refractory mutation system (multiplex T ARMS) polymerase chain reaction (Table 2). ARMS-PCR was performed initially on 99 individuals selected for case groups then control groups. Micro-tube components and thermal protocol of PCR reaction are available in Table 3. PCR products were loaded directly onto 2% agarose gels (containing green viewer), electrophoreses and visualized by photography under UV illumination. The product sizes for 112 codon were 115 bp, 444 bp and for 158 codon were 253 bp, 307bp for the C & T alleles, while the product size for the internal control was 514 bp.
Table 1. Primers sequence.
Table 2. Micro-tube components of ARMS-PCR reaction.
2.2. Statistical Analysis
The differences between the two groups were compared using the Student’s t test for continuous variables and the chi-square (χ2) test for categorical variables. Allele and genotype frequencies between case and controls were obtained using the chi-square (χ2) test. Odds ratio (OR) and 95% confidence intervals (CI) were calculated using logistic regression. p < 0.05 was considered statistically significant. Statistical analyses were performed with SPSS for Windows software package version 21.0 (SPSS Inc., Chicago, IL, USA).
3. Results
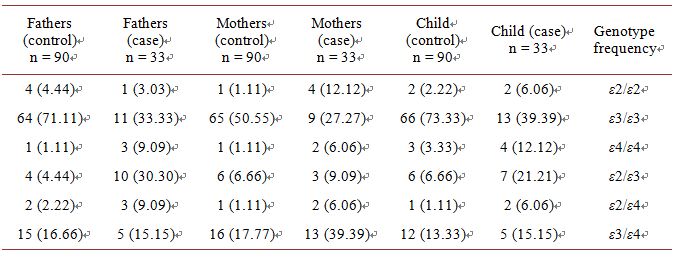
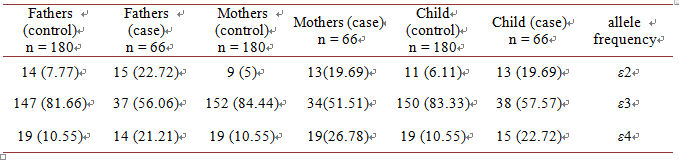
Allele frequencies for 112 and 158 codons obtained from 99 individuals in case group and 270 individuals in control group Persians of northeastern part of Iran. 99 cases and 270 controls were genotyped for SNPs loci 112 and 158 codon of APOE gene by ARMS- PCR assay. Genotype and allele frequencies in both groups of patient and controls were compared separatelyas shown in Table 4 and Table 5. In 112 codon of APOE gene it seems having T allele reduces the risk of nondisjunction and in contrast C allele may be a risk factor in happening of nondisjunction. (p-value = 0.000006, OR = 2.66, 95% CI = 1.74 - 4.06) (Table 6). In 158 codon of APOE gene it seems having T allele reduces the risk of nondisjunction and in contrast C allele may be a risk factor in happening of nondisjunction. (p-value = 0.00 OR = 3.89, 95% CI = 2.38 - 6.34) (Table 7). E4 allele frequency in mothers of Down syndrome is about 14 percent more than those in
Table 3. Thermal protocol of PCR reaction
Table 4. Genotype distributions of 112 codon APOE gene for case and the control group.
control group as shown in Table 8 and Table 9. According to results of the present study, 112 codon of APOE gene and 158 codon of APOE gene can be considered as susceptibility genetic factors for nondisjunction in Northeast of Iran. E4 allele has main role in nondisjunction.
Table 5. Genotype distributions of 158 codon APOE gene for case and the control group.
Table 6. Allele distributions of 112 codon APOE gene for the case and the control group.
Table 7. Allele distributions of 158 codon APOE gene for the case and the control group.
Table 8.Genotype frequency of APOE in case and control group.
Table 9.Allelefrequency of APOE in case and control group.
4. Discussion
Chromosomal non-disjunction is one of the questions of human genetics. Because of high incidence of Down syndrome in newborns and its socioeconomic consequences, we focus on this syndrome. Increased maternal age has main role, but the biological mechanisms of this occurrence are imprecise. No risk factor has been detected in young mothers, but they give birth to majority of Down syndrome children. Our study shows an increase in the frequency of apoE allele e4 in young mothers. We hypothesize that apoE allele E4 is a risk factor for non-disjunction of chromosome 21 in young mothers. The frequency of allele e4 in young mothers with DS children supports their increased risk of Alzheimer disease as allele e4 is a genetic susceptibility factor in AD. ApoE has a main role in the oocyte and we know that pathological role of allele E4 in chromosomal nondisjunction remain to be investigated. ApoE is produced in most organs, like ovary, in which it involved in chromosomal nondisjunction. In our study, the frequency of apoE allele e4 in fathers of Down syndrome children was 21.21% significantly higher than 10.55% in control group. But in other study, the frequency of apoE allele e4 in Danish fathers of DS children lower than control group. The biological importance of this finding is not clear. In other studies, there aren’t any differences in the frequency of allele e4 between case and control groups. So it is difficult to imagine that the apoE genotype of father has a main role in non-disjunction of chromosome 21. Our finding show that the frequency of apoE allele E4 in Down syndrome children is about 22.72%. The frequency of this allele in control group is 10.55%. In other studies there were no differences in apoE allele distribution in people with Down syndrome compare with controls. We can see that there is agreement between small and large number of case- control groups. In other studies, mothers in case and controls divided to two subgroups: young mothers (<35) and older mothers (>35). But in our study, all mothers are young and findings show that the e4 frequency in mothers of DS children (26.78%) was higher as compared with controls (11%). In other studies findings are different in subgroups. Avramopoulos and et al found that in young mothers with a meiosis II error; e4 frequency was (30.0%) higher than in older mothers with a meiosis II error (13.0%). There is no clear difference in e4 frequency in both mothers (case and controls) that participate in M. ezquerra and et al., study. Genotyping methods that detect sequences in apoE alleles are RFLP, real-time PCR, mass spectrometry and T-ARMS PCR. All techniques are costly and time-comsuming, exect T-ARMS PCR. With this method, two different single nucleotide polymorphisms are detectable. So far no association study has been investigated for this polymorphism and no frequency data is reported for it. Genotype and allele frequencies in both groups of patient and controls were compared separately. In 112 codon of APOE gene it seems having T allele reduces the risk of nondisjunction and in contrast C allele may be a risk factor in happening of nondisjunction. (p-value = 0.000006, OR = 2.66, 95% CI = 1.74 - 4.06). In 158 codon of APOE gene it seems having T allele reduces the risk of nondisjunction and in contrast C allele may be a risk factor in happening of nondisjunction (p-value = 0.0000 OR = 3.89, 95% CI = 2.38 - 6.34).
5. Conclusion
This study indicates that there is a significant difference in allele distribution between case and control groups. The C allele for 112 codon of APOE gene and the C allele for 158 codon of APOE gene shows association with nondisjunction. E4 allele frequency in mothers of Down syndrome is about 14% more than those in control group. According to results of the present study, 112 codon of APOE gene and 158 codon of APOE gene can be considered as susceptibility genetic factors for nondisjunction in Northeast of Iran. E4 allele has main role in nondisjunction.
Acknowledgements
The authors would like to declare their gratitude to all contributors who have made the completion of this study. This study was part of a student thesis, supported financially by Grant No.: 139400 of International Medical Sciences University, Mashhad, Iran.
Cite this paper
Iranifar, E. and Hamzehloie, T. (2016) The Study of Apolipoprotein E4 Allele Distribution in Parents of Down’s Syndrome Children as a Risk Factor in Khorasan Razavi Province, Iran. Open Journal of Genetics, 6, 87-95. http://dx.doi.org/10.4236/ojgen.2016.64010
References
- 1. Roizen, N.J. and Patterson, D. (2003) Down’s Syndrome. The Lancet, 361, 1281-1289.
https://doi.org/10.1016/S0140-6736(03)12987-X - 2. Patient, U. (1991) Down Syndrome. Congenital Heart Disease, 40, 21.
- 3. Weijerman, M.E. and De Winter, J.P. (2010) Clinical Practice. European Journal of Pediatrics, 169, 1445-1452.
https://doi.org/10.1007/s00431-010-1253-0 - 4. Dzurova, D. and Pikhart, H. (2005) Down Syndrome, Paternal Age and Education: Comparison of California and the Czech Republic. BMC Public Health, 5, 1.
https://doi.org/10.1186/1471-2458-5-69 - 5. Yaron, Y. and Mashiach, R. (2001) First-Trimester Biochemical Screening for Down Syndrome. Clinics in Perinatology, 28, 321-331.
https://doi.org/10.1016/S0095-5108(05)70086-5 - 6. Bianchi, D.W. (1999) Fetal Cells in the Maternal Circulation: Feasibility for Prenatal Diagnosis. British Journal of Haematology, 105, 574-583.
https://doi.org/10.1046/j.1365-2141.1999.01383.x - 7. Patterson, D. and Cabelof, D.C. (2012) Down Syndrome as a Model of DNA Polymerase Beta Haploinsufficiency and Accelerated Aging. Mechanisms of Ageing and Development, 133, 133-137.
https://doi.org/10.1016/j.mad.2011.10.001 - 8. Antonarakis, S.E. (1991) Parental Origin of the Extra Chromosome in Trisomy 21 as Indicated by Analysis of DNA Polymorphisms. New England Journal of Medicine, 324, 872-876.
https://doi.org/10.1056/NEJM199103283241302 - 9. Antonarakis, S.E., Petersen, M.B., McInnis, M.G., Adelsberger, P.A., Schinzel, A.A., Binkert, F., et al. (1992) The Meiotic Stage of Nondisjunction in Trisomy 21: Determination by Using DNA Polymorphisms. American Journal of Human Genetics, 50, 544.
- 10. Morris, J., Mutton, D. and Alberman, E. (2002) Revised Estimates of the Maternal Age Specific Live Birth Prevalence of Down’s Syndrome. Journal of Medical Screening, 9, 2-6.
https://doi.org/10.1136/jms.9.1.2 - 11. Penrose, L.S. (1933) The Relative Effects of Paternal and Maternal Age in Mongolism. Journal of Genetics, 27, 219-224.
https://doi.org/10.1007/BF02984413 - 12. Natoli, J.L., Ackerman, D.L., McDermott, S. and Edwards, J.G. (2012) Prenatal Diagnosis of Down Syndrome: A Systematic Review of Termination Rates (1995-2011). Prenatal Diagnosis, 32, 142-153.
https://doi.org/10.1002/pd.2910 - 13. Mansfield, C., Hopfer, S. and Marteau, T.M. (1999) Termination Rates after Prenatal Diagnosis of Down Syndrome, Spina Bifida, Anencephaly, and Turner and Klinefelter Syndromes: A Systematic Literature Review. Prenatal Diagnosis, 19, 808-812.
https://doi.org/10.1002/(SICI)1097-0223(199909)19:9<808::AID-PD637>3.0.CO;2-B - 14. Davignon, J., Gregg, R.E. and Sing, C.F. (1988) Apolipoprotein E Polymorphism and Atherosclerosis. Arteriosclerosis, Thrombosis, and Vascular Biology, 8, 1-21.
https://doi.org/10.1161/01.ATV.8.1.1 - 15. Chartier-Hariln, M.-C., Parfitt, M., Legrain, S., Pérez-Tur, J., Brousseau, T., Evans, A., et al. (1994) Apolipoprotein E, ε4 Allele as a Major Risk Factor for Sporadic Early and Late-Onset Forms of Alzheimer’s Disease: Analysis of the 19q13.2 Chromosomal Region. Human Molecular Genetics, 3, 569-574.
https://doi.org/10.1093/hmg/3.4.569 - 16. Strittmatter, W.J., Saunders, A.M., Schmechel, D., Pericak-Vance, M., Enghild, J., Salvesen, G.S., et al. (1993) Apolipoprotein E: High-Avidity Binding to Beta-Amyloid and Increased Frequency of Type 4 Allele in Late-Onset Familial Alz-heimer Disease. Proceedings of the National Academy of Sciences of the United States of America, 90, 1977-1981.
https://doi.org/10.1073/pnas.90.5.1977 - 17. Van Duijn, C., Clayton, D., Chandra, V., Fratiglioni, L., Graves, A., Heyman, A., et al. (1991) Familial Aggregation of Alz-heimer’s Disease and Related Disorders: A Collaborative re-Analysis of Case-Control Studies. International journal of epidemiology, 20, S13-S20.
https://doi.org/10.1093/ije/20.supplement_2.s13 - 18. Richards, S., Holland, A., Palmer, C., Schupf, N., Kapell, D., Lee, J., et al. (1994) Increased Risk of Alzheimer’s Disease in Mothers of Adults with Down’s Syndrome. The Lancet, 344, 1092-1095.
https://doi.org/10.1016/S0140-6736(94)91753-1