American Journal of Molecular Biology
Vol.06 No.01(2016), Article ID:62575,12 pages
10.4236/ajmb.2016.61004
Phosphorylation on TRPV4 Serine Residue 824 Enhances Its Association with PGM1
Sung Hwa Shin1*, Eun Jeoung Lee2*, Sunghee Hyun3, Sang Sun Kang1#
1Department of Biology Education, Chungbuk National University, Cheongju, Republic of Korea
2Department of Internal Medicine, Chungbuk National University, Cheongju, Republic of Korea
3Department of Biomedical Laboratory Science, Eulji University, Daejeon, Republic of Korea

Copyright © 2016 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 15 September 2015; accepted 4 January 2016; published 7 January 2016
ABSTRACT
The TRPV4 cation channel is expressed in a broad range of tissues and participates in the generation of a Ca2+ signal and/or depolarization of membrane potential. Here, human phosphoglucomutase-1 (PGM1), an enzyme that converts glucose-6 phosphate to glucose-1 phosphate in the gly- colysis pathway, as the first auxiliary protein of TRPV4 Ca2+ channels, is identified with yeast two hybrid system, coimmunoprecipitation, confocal microscopy, and GST pull-down assays. TRPV4 forms a complex with PGM1 through its C-terminal cytoplasmic domain. Because it is demonstrated that TRPV4 serine residue 824 (S824) is phosphorylated by serum/glucocorticoid regulated kinase 1, we elucidate the effect of TRPV4 S824 phosphorylation on TRPV association with PGM1. Even an inactivated mutant version of TRPV4, S824A, exhibited a decreased ability to bind PGM1, an activated phosphomimetic mutant version of TRPV4, S824D, exhibited enhanced binding to PGM1. Thus, formation of the TRPV4/PGM1 complex and localization of this complex to the plasma membrane appear to be regulated by the phosphorylation status of residue S824 in TRPV4. The newly identified interactor of TRPV4 may help the molecular pathways modulating transport activity or glucose metabolism, respectively.
Keywords:
Membrane Localization, Phosphoglucomutase1, Phosphorylation, Protein-Protein Interaction, TRPV4

1. Introduction
The ubiquitously expressed TRPV4 cation channel, a member of the TRP vanilloid subfamily, generates a Ca2+ signal and/or depolarizes the membrane potential (see reviews [1] - [4] ). Through its contributions to osmo-and mechanotransduction, TRPV4 plays a role in cellular and systemic volume homeostasis, arterial dilation, nociception, epithelial hydroelectrolyte transport, bladder voiding, and regulation of ciliary beat frequency [5] -[10] . TRPV4 also responds to temperature, endogenous arachidonic acid metabolites, and phorbol esters, including the inactive 4α-phorbol 12, 13-didecanoate [11] [12] . In addition, TRPV4 also participates in receptor-operated Ca2+ entry; thus, showing multiple modes of activation [1] - [4] . In this sense, several proteins have been proposed to modulate TRPV4 subcellular localization and/or function, including microtubule-associated protein 7, calmodulin (CaM), PACSIN3, and no lysine protein kinase [13] - [16] . Furthermore, a close functional and physical interaction has been described between the inositol triphosphate receptor 3 (IP3R3) and TRPV4; this interaction was shown recently to sensitize the latter to the mechano- and osmotransducing messenger, 5′-6′-epox- ieicosatrienoic acid [17] [18] .
Glucose 1-phosphate is not a useful metabolic intermediate; however, phosphoglucomutase1 (PGM1) catalyzes conversion of glucose 1-phosphate to glucose 6-phosphate [19] . After glycogen phosphorylase catalyzes the phosphorolytic cleavage of a glucosyl residue from the glycogen polymer, the liberated glucose molecule retains a phosphate group at the 1-carbon position [20] . Because glucose 1-phosphate molecule is not a useful metabolic intermediate, PGM1 catalyzes further the conversion of glucose 1-phosphate to glucose 6-phosphate [21] . The metabolic fate of newly generated glucose 6-phosphate depends on the energy requirements of the cell at the time it is generated. If the cell is in a low-energy state, glucose 6-phosphate will be metabolized along the glycolytic pathway, eventually yielding two molecules of adenosine triphosphate [22] [23] . On the other hand, the cell is in need of biosynthetic intermediates, glucose 6-phosphate enters the pentose phosphate pathway, where it undergoes a series of reactions to yield ribose and/or NADPH, depending on cellular conditions [24] .
In the liver, glucose 6-phosphatase can also catalyze conversion of glucose 6-phosphate to glucose, which exits the liver and is transported to other cells. However, muscle cells lack glucose 6-phosphatase, so they cannot share their glycogen stores with the rest of the body [23] . PGM1 acts in the opposite fashion when blood glucose levels are high. In this case, PGM1 catalyzes the conversion of glucose 6-phosphate, a product generated from glucose by hexokinase, to glucose 1-phosphate. Glucose-1-phosphate then reacts with UTP to yield UDP-glu- cose in a reaction catalyzed by UDP-glucose-pyrophosphorylase [23] [24] . If activated by insulin, glycogen synthase will attach the glucose molecule from the UDP-glucose complex onto a glycogen polymer [23] . In yeast, PGM1 activity results in elevated Ca2+ in a PGM2 mutant grown in medium containing Gal as the carbon source. In that study, either Mg2+ or Ca2+ ions were required for PGM1 activity [25] .
The aim of the present study was to identify auxiliary proteins that specifically interact with TRPV4 [26] . To this end, we performed a putative motif searching screen to identify proteins associated with epithelial Ca2+ channels, and identified PGM1 as a candidate protein. Through validation studies, we found that the interaction between TRPV4 and PGM1 was enhanced by phosphorylation of TRPV4 serine residue 824 (S824). Functional interaction between TRPV4 and PGM1 was further substantiated by pull-down assays, immunohistological studies, and a Ca2+ ion imaging analysis, as TRPV4 subcellular localization and activity were shown to depend on PGM1. We propose that PGM1 preferentially associates with phosphorylated TRPV4 (the activated channel form); however, binding of PGM1 with TRPV4 negatively impacts its Ca2+ channel activity.
2. Materials and Methods
2.1. Site-Directed Mutagenesis
To generate phosphomimetic and other mutants, amino acid changes were introduced using the appropriate oligonucleotides for S/A (forward, 5′-agg gat cgttggGccGcggtggtgccccgc gta-3′; reverse, 5′-gcggggcaccaccgCggC ccaacgatccct acg-3′) or S/D (forward, 5′-agg gat cgttggGAc GAC gtggtgccccgc gta-3′; reverse, 5′-gcggggcaccac GTC gTCccaacgatccct acg-3′) and wild-type TRPV4 cDNA as a template. TRPV4 mutant constructs were prepared using a QuickChange® Multi Site-Directed Mutagenesis Kit (Stratagene). To generate the construct expressing a truncated version of TRPV4 (Δ718-871), the following primers were used: forward, 5′-ataggatcc- atgggt gag accgtgggc cag-3′; reverse, 5′-atactc gag cta cag tggggcatcgtc cgt-3′. All TRPV4 mutants were confirmed by DNA sequencing. Human embryonic kidney (HEK293) cells were transfected with TRPV4 and with the appropriate mutant constructs, as described previously.
2.2. Glutathione S-Transferase (GST)-TRPV4 Fusion Proteins and Pull-Down Assays
TRPV4 sequences were PCR-amplified, subcloned into pGEX-5X-1, sequenced, and expressed in Escherichia coli BL21 cells. S824A-agarose or GST-TRPV4 fusion proteins bound to glutathione-Sepharose resin were equilibrated in PBS buffer containing 0.1% Triton X-100 and either 1 mM CaCl2 or 2 mM EGTA. Incubation of immobilized fusion proteins with total cell lysates or recombinantly purified S824A fusion protein was followed by three washes with the appropriate buffers. Bound proteins were eluted with sample buffer, resolved by SDS-PAGE, and detected by chemiluminescence through exposure to X-ray film (Fuji Las 3000 mini).
2.3. Fluorescence Measurements of [Ca2+]i
Measurements of [Ca2+]i were obtained using the fluorescent Ca2+ indicator, Fluo4-acetoxymethyl ester (Fluo4- AM), as previously described. In brief, cells grown on cover slips were incubated in the dark for 40 min in DMSO solution containing 1 μM Fluo4-AM at 24˚C, and then washed and incubated for 15 min to allow hydrolysis of internalized Fluo4-AM. Measurements of [Ca2+]i in single cells were then obtained by examining the emitted fluorescence on a confocal microscope (LSM710 Zeiss, Germany) at wavelengths 495 nm (excitation) and 519 nm (emission). Absorption values (in arbitrary units) obtained with the argon-ion laser at 488 nm are expressed as relative intracellular Ca2+ ion concentrations [Ca2+]i. All experiments were carried out at 24˚C. After stimulation with mild heat (from 24˚C to 42˚C within 45 s for 2 min), [Ca2+]i was measured in single cells at 24˚C.
2.4. Confocal Microscopy
MDCK or HEK293 cells were seeded overnight at 60% confluence onto culture slides coated with human fibronectin (Becton Dickinson). Cells were washed several times with ice-cold PBS and then fixed in 3% paraformaldehyde for 10 min. Fixed cells were permeabilized with 0.1% Triton X-100 for 10 minutes and then blocked for 1 h in PBS containing 5% BSA (Sigma, USA) and 0.1% Tween. After an additional 20 minutes of incubation at 37˚C, cells were fixed, permeabilized, and immunostained with either anti-PGM1 or anti-TRPV4 antibodies. Cells were then incubated with secondary Alexa Fluor 568-conjugated donkey anti-rabbit or Alexa Fluor 488-conjugated goat anti-mouse antibodies (Molecular Probes, Inc., Eugene, OR, USA). Confocal microscopy analysis was performed with an LSM710 microscope (Zeiss, Germany) at the Center for Experimental Research Facilities at Chungbuk National University.
2.5. Plasmids and Protocol for Yeast Two-Hybrid Screening
The sequence encoding TRPV4 was subcloned into the bait plasmid (PCR primers: forward, 5’-CCCCAT ATGATGGGTGAGACCGTGGGC-3’; reverse, 5’-GGGGATCCGCAGTGGGGCATCGTCCGT-3’). The resultant plasmid was designated TRPV4-pGBKT7. A human liver cDNA library (Clontech) was screened for proteins that interacted with TRPV4 using the Matchmaker Gold Yeast Two-Hybrid System (Clontech protocol PT4084-1). The bait plasmid, TRPV4-pGBKT7, was transformed into the AH109 yeast strain; around 2 × 103 transformants were screened. Transformants were assayed for MEL1 activation by selection on high-stringency medium plates: SD/-Ade/-His/-Leu/-Trp/X-α-gal. Prey plasmids were recovered from positive yeast clones and retransformed into DH5α cells. Transformants harboring AD plasmids were selected on LB plates with 100 μg/ml ampicillin, and plasmid inserts were identified by DNA sequencing.
2.6. Solutions and Drugs
Cells were normally superfused with a solution containing (in mM): 88 NaCl, 5 KCl, 5.5 glucose, 1 CaCl2, 10 HEPES and 100 mannitol, adjusted to pH 7.4 with NaOH (300 mosm kg−1 H2O). The HTS was adjusted to 200 mosm kg−1 H2O by omitting mannitol. 4-αPDD was purchased from Sigma (St. Louis, MO, USA). Fluo-4AM and Fura-2AM were acquired from Molecular Probes, Inc. Stock solutions of phorbol esters were initially prepared in dimethyl sulphoxide (DMSO) at a concentration of 1 mM, and then stored at −20˚C. The final DMSO concentration in the experimental bath solution containing phorbol esters never exceeded 0.5%. Insulin and GSK650394 (an SGK1 inhibitor) were acquired from Tocris Bioscience (Ellisville, MO, USA), and used according to the manufacturer’s recommendations.
3. Results
3.1. Identification of PGM1 as a TRPV4-Associated Protein-Protein Interaction
To identify proteins that interact with TRPV4, the C-terminal tail of TRPV4 (Figure 1(a)) was used to screen a mouse kidney cDNA library using a yeast two hybrid assay. One of the positive clones encoded PGM1, which catalyzes to convert glucose 6-phosphate (which is easily generated from glucose by the action of hexokinase) to glucose 1-phosphate. PGM1 strongly interacted with TRPV4, whereas no binding was observed with ENaC, indicating specificity of the PGM1-TRPV4 interaction. The subunit of the epithelial Na+ channel, ENaC, was used as a negative control. In addition β-galactosidase activity was not detectable in the absence of prey, or after co-trans- formation of the bait with the empty pACT2 (prey) vector (data not shown). The serine residue 824 in the C-ter- minal domain is indicated. Mutants of S824 (in 811shtvgrlrRdRwsSVvprvvel832), which was constructed for this study, are also detailed in Figure 1(b). EGFP (or GST)-tagged mouse TRPV4 WT, S824A (S824 nonphosphorylatable mutant), or S824D (S824 phospho-mimicking mutant) fusion proteins were expressed in the HEK293 cell line, purified with EGFP monoclonal antibody (or Glutathione beads), and utilized as antigen proteins.
Figure 1. Predicted topology of the TRPV4 C-terminal domain and yeast two-hybrid system. (a) Transmem- brane topology of mouse TRPV4 (871 residue). The three ankyrin binding repeats (ANK, gray circles), the six transmembrane regions (TM1 - TM6), the PGM1 binding site (PGM1), and the putative SGK1 phos- phorylation site (S824) are indicated. The C-terminal cytoplasmic region of TRPV4 (Δ718-871) is also indicated. The putative SGK1 phosphorylation site (S824) is present in the PGM1 domain (residues 812 - 832) of the TRPV4 channel. The line indicates the GST C-terminal cytoplasmic region of TRPV4 (Δ718- 871) or the mutant (S824A) fusion protein; the TRPV4 mutant site (S824A or S824D) is compared with the wild-type (WT, Gene Bank no. BC127052). (b) Alignment of TRPV4 WT, S824A, and S824D mutant ver- sions with the consensus SGK1 substrate motif. The putative SGK1 phosphorylation site (S824) of TRPV4 is located in the specific conserved SGK1 substrate region [R-X-R-X-X-(S/T) Φ]. The S824A and S824D mutant versions were constructed using site-directed mutagenesis. (c) Screening of TRPV4-interacting pro- teins by the yeast two-hybrid system. Yeast were transformed with a plasmid driving expression of TRPV4, and positive colonies were selected with leucine, tryptophan, adenine, histidine, and aureobasidin A. One, six, eight and nine are positive clones from the first round of selection from a human liver library using TRPV4- pGBKT7 as bait. All four clones contained the same gene (human PGM1), as revealed by DNA sequencing.
Homo sapiens phosphoglucomutase 1 (PGM1) was obtained from a human liver library (Clontech, Palo Alto, CA, USA) by polymerase chain reaction with the primers 5′-TAATACGACTCACTATAGGG-3′ and cloned into the pEGFP-C2 vector (Clontech) to generate the PGM1-pEGFP-C2plasmid. The entire coding region of the PGM1 was amplified and subcloned into the pGEX-5X-1 vector to construct the GST fusion protein.
3.2. Interaction between PGM1 and the C-Terminal Domain of TRPV4
GST pull-down binding assays were performed to further establish the interaction between TRPV4 and PGM1 (Figure 2(a)). The endogenous PGM1-containing MDCK homogenate was incubated with GST-TRPV4 or GST-TRPV4 (Δ718-871) fusion proteins immobilized on glutathione-Sepharose 4B beads. PGM1 bound specifically to the C-terminal tail of TRPV4, as no interaction was observed with GST-TRPV4 (Δ718-871) (Figure 2(b) left lane). Thus, the C-terminal tail of TRPV4 is required for the interaction with STM1. Taken together, our observations suggest that TRPV4 interacts directly with PGM1 through the C-terminal domain of TRPV4 in MDCK cells.
3.3. Phosphorylation of TRPV4 S824 Enhances the Interaction between TRPV4 and PGM1
We demonstrated previously that the interaction between TRPV4 and actin (STIM1 or tubulin) is differentially regulated by phosphorylation of TRPV4 S824 (Figure 1(b); [27] ). Similarly, we hypothesized that phosphorylation of TRPV4 on S824 regulates the interaction between TRPV4 and PGM1. Co-immunoprecipitation and GST pull-down assays of these proteins in their native forms were performed to investigate the consequences of TRPV4 S824 phosphorylation on the interaction between TRPV4 and PGM1 (Figure 3). HEK 293 cells, which do not express endogenous TRPV4, were transiently transfected with plasmids driving expression of either EGFP-TRPV4 WT or S824A. After 48 h, cell lysates were generated, and immunoprecipitation was conducted
Figure 2. Protein-protein interaction between PGM1 and the C-terminal domain of TRPV4. (a) Interaction between TRPV4 and PGM1 in MDCK cells. An immunoblot (IB) analysis was per- formed using antibodies against PGM1 (left) following immunoprecipitation (IP) using anti-TRPV4 antibodies. Conversely, anti-PGM1-immunoprecipitated complexes were subjected to immunoblot analysis using anti-TRPV4 antibodies (right). Co-immunoprecipitation of PGM1 with TRPV4 con- firmed the presence of the TRPV4-PGM1 complex. An unrelated antibody was used as a negative control for immunoprecipitation. Antibodies against actin were used as a control for immunoblot analysis (bottom). (b) The C-terminal domain of TRPV4 is required for its interaction with PGM1. After incubating GST-TRPV4 WT or a deletion mutant fusion protein (Δ718-871) with a HEK293 cell lysate, proteins bound to purified GST beads were detected by immunoblotting with antibodies against PGM1 (upper lane). The deletion mutant lacking the C-terminal region (Δ718-871) did not pull down PGM1 (right lane), whereas GST TRPV4 WT did (left lane).
Figure 3. Effects of phosphorylation of TRPV4 S824 on the interaction between TRPV4 and PGM1. (a) Co-immunoprecipitation of TRPV4 WT, S824A, or S824D mutant versions with PGM1. HEK 293 cells were transiently transfected with plasmids driving expression of EGFP-TRPV4 WT or S824A. Cell lysates were generated after 48 h, and pull-down assays from total protein extracts were performed with protein A-agarose beads. Western blot assays were performed with rabbit anti-TRPV4 or anti-PGM1 antibodies. (b) Pull-down analysis of PGM1 with GST-fusion C-terminal domains of TRPV4 WT, S824A, or S824D. GST-fusion proteins encompassing C-terminal TRPV4 domains were constructed and expressed in E. coli. Approximately 2 µg of WT, S824A, or S824D fusion protein bound to glutathione-Sepharose beads was incubated with HEK 293 cell lysates.
with mouse anti-TRPV4 or anti-PGM1 antibodies. Immunoblotting was then performed with rabbit anti-TRPV4 or anti-PGM1 antibodies (Figure 3(a)). Interestingly, TRPV4 S824A (a mutant mimicking unphosphorylated TRPV4) did not co-precipitate PGM1 (Figure 3(a), middle lane), whereas TRPV4 S824D (a phosphomimetic mutant form of TRPV4) and TRPV WT did. These results indicate that the phosphomimetic version of TRPV4 (S824D) exhibited a higher affinity for PGM1 than a version mimicking the unphosphorylated protein (S824A).
We performed pull-down assays of PGM1 using GST-fusion proteins of the C-terminal domain of TRPV4 WT, S824A, or S824D to further test the hypothesis that the interaction between TRPV4 and PGM1 is regulated by the phosphorylation status of TRPV4 S824 (Figure 3(b)). GST-fusion proteins encompassing C-terminal TRPV4 domains were constructed and expressed in E. coli. Approximately 2 µg of WT, S824A, or S824D fusion protein immobilized on glutathione-Sepharose resin was incubated with HEK 293 cell lysates. Similar to the results shown in Figure 3(a), PGM1 was captured from HEK 293 cell lysates by the GST-tagged C-terminal portion of TRPV4 when it was immobilized on glutathione-Sepharose 4B beads; thus, demonstrating a physical interaction between TRPV4 and PGM1 (Figure 3(b)). A greater amount of PGM1 was captured by TRPV4 S824A compared with TRPV4 WT, a result consistent with the data shown in Figure 2(a). Taken together, these data suggest that PGM1 preferentially binds TRPV4 when S824 is phosphorylated, and led us to hypothesize that the function of this interaction is to inhibit its channel activity (Figure 6).
3.4. Effects of PGM1 on Phosphorylated TRPV4 S824 and TRPV Binding with PGM1
As a complementary approach to examine whether phosphorylation of TRPV4 S824 enhances its binding with PGM1, HEK 293 cells were transiently cotransfected with EGFP-TRPV4 WT, S824A, or S824D, and pmCherry-PGM1 (pmCherry-N1, Clontech) expression plasmids. After 48 h, the cells were fixed, immunostained, and examined by confocal microscopy. Ectopically-expressed TRPV4 (green) and PGM1 (red) were colocalized in the cytosol (yellow) (Figure 4). As shown in Figure 4 (upper lane), we observed that ectopically-expressed TRPV4 WT was in close proximity to PGM1 in HEK293 cells. However, TRPV4 S824A did not colocalize with PGM1 (middle lane), whereas TRPV4 S824D colocalized with PGM1 (bottom lane), consistent with the results
Figure 4. Confocal microscopic images of cells transfected with EGFP-TRPV4 WT, S824A, or S824D. Cells were examined by direct immunofluorescence microscopy (n = 5). Confocal micros- copic images show EGFP-TRPV4 WT or mutant (S824A or S824D) (green), pmCherry-N1 PGM1 (red), and merged channels (yellow). EGFP-TRPV4 WT and S824D showed the greatest extent of colocalization with pmCherry-N1 PGM1 in the cytosol (upper and bottom panels). However, EGFP-TRPV4 S824A was primarily detected in the cytosol, and colocalized to a much lesser extent with PGM1(middle panel).
shown in Figure 3. Thus, these observations further support the hypothesis that phosphorylation on TRPV4 S824 regulates not only the interaction between TRPV4 and PGM1, but also subcellular localization of TRPV4.
3.5. Regulation of TRPV4 Subcellular Localization and Its Interaction with PGM1 by Phosphorylation of TRPV4 S824
We found previously that the C-terminal domain of TRPV4 interacts with either actin or tubulin, depending on the phosphorylation status of TRPV4 S824 [28] . As we demonstrated that PGM1 also interacts with the C-ter- minal domain of TRPV4, we hypothesized that a TRPV4 interaction with PGM1 is regulated by the phosphorylation status of the TRPV4serine residue 824. To determine whether TRPV4 localization and/or TRPV4 interaction with PGM1 are influenced by the phosphorylation status of TRPV4 S824, we examined localization of TRPV4 upon pharmacological treatment with GSK650394 (an SGK1 kinase inhibitor) or insulin (an SGK1 activator). As controls, TRPV4 S824A and S824D mutant versions were also used (Figure 5(a)). The subcellular locations of the TRPV4 mutants were not altered upon treatment with either GSK650394 or insulin, whereas the subcellular localization of the WT version was altered dramatically, according to the specific treatment (Figure 5(a)). The effect of PGM1 on TRPV4 activity was also determined by whole-cell Ca2+ image analysis in transiently-transfected HEK293 cells. As shown in Figure 5(b), the S824D mutant version of TRPV4 enhanced Ca2+ current significantly, whereas the channel activity of TRPV4 WT or S824A was unaffected. This result suggests that PGM1 prefers to bind the activated (phosphorylated) TRPV4, resulting in plasma membrane localization.
These findings suggest that the phosphorylation status of TRPV4 S824 regulates both the binding of the C-terminal domain of TRPV4 with PGM1, and also subcellular localization of TRPV4 (plasma membrane or focal adhesions).
3.6. Putative Role of PGM1 in TRPV4 Ca2+ Regulation
We demonstrated the presence of PGM1 in the TRPV4 channel complex, indicating a possible function for
Figure 5. Plasma membrane localization of TRPV4 WT, S824A, or S824D in cells after treatment with GSK650394 or insulin. (a) Confocal microscopic analysis of cells transfected with EGFP-TRPV4 S824A, WT, or S824D (green) after treatment with an SGK1 inhibitor (GSK 650397, left), an SGK1 activator (insulin, right) for 12 h, or a serum-free control (middle). Subcellular localization of TRPV4 WT (which can be phosphorylated on S824 by SGK1) was altered by treatment with either GSK 650394 or insulin. The image shown is representative of five independent experiments (n = 5). (b) Effects of PGM1 on TRPV4 WT, S824A, or S824D, resulting from expression of TRPV4 WT, S824A, or S824D, expressed as absorption values at 488 nm from an argon-ion laser in HEK 293 cells (in arbitrary % units). After transiently co-transfecting the EGFP-TRPV4 WT or mutant (S824A or S824D) (green), pmCherry-N1 PGM1 (red), [Ca2+]i was measured in single cells at 24˚C. Results shown are from one of five independent experiments.
PGM1 in the regulation of channel localization and/or activity. Because we demonstrated previously that S824 in TRPV4 is phosphorylated by SGK1, we propose a model in which the phosphorylation status of TRPV4 S824 enhances its interaction with PGM1, thereby resulting in inhibition of TRPV4 channel activity in the plasma membrane (Figure 6).
A bivalent metal ion, usually magnesium or cadmium (both of which complex directly with the phosphoryl group esterified to the PGM1 active site serine), is required for PGM1 enzymatic activity. Thus, it seems logical that association of PGM1 with the active Ca2+ channel (for example, via the S824-phosphorylated form of
Figure 6. Putative model for the regulation of the interaction between TRPV4 and PGM1 by phosphorylation on TRPV4 S824. PGM1 can be activated by Ca2+/Mg2+, which is transported by activated TRPV4. PGM1 preferentially associates with the phosphorylated form of the TRPV4 C-terminal cytoplasmic domain; thus, blocking the Ca2+ channel. PGM1 may play a role in TRPV4 channel regulation, similar to that played by calmodulin (CaM), thereby generating a feedback inhibition loop.
TRPV4) would be necessary to enhance PGM1 activity. However, association of PGM1 with the activated Ca2+ channel actually appears to inhibit channel activation.
In conclusion, we provide the first evidence that PGM1 interacts with the C-terminal domain of TRPV4. We also present data to indicate a regulatory role for the PGM1-TRPV4 complex in Ca2+ reabsorption, in particular, via TRPV4 subcellular localization. The interaction between PGM1 and TRPV4 was regulated by the phosphorylation status of TRPV4 S824, resulting in enhanced channel activity. The molecular mechanism that we propose involves tethering of the PGM1-TRPV4 complex to the Ca2+ channel, thereby inactivating its channel activity. This mechanism is likely applicable to other ion transporters given the broad tissue distribution of TRPV4 and PGM1. Thus, phosphorylation of TRPV4 S824 appears to be a major regulatory mechanism for its interaction with many proteins, including PGM1, IP3R3, CaM, actin, tubulin, and STIM1 [29] .
4. Discussion
We identified PGM1 as an auxiliary protein for the epithelial Ca2+ channel protein, TRPV4 (Figure 1). PGM1 catalyzes conversion of glucose 1-phosphate to glucose 6-phosphate, which is a useful metabolic intermediate [15] - [24] . Although the glucose 1-phosphate molecule is not a useful metabolic intermediate, PGM1 catalyzes conversion of glucose 1-phosphate to glucose 6-phosphate [23] . After glycogen phosphorylase catalyzes the phosphorolytic cleavage of a glucosyl residue from the glycogen polymer, the liberated glucose molecule retains a phosphate group on its 1-carbon atom [19] . The metabolic fate of glucose 6-phosphate depends on the needs of the cell at the time it is generated. If the cell is in a low-energy state, glucose 6-phosphate will be metabolized down the glycolytic pathway, eventually yielding two molecules of adenosine triphosphate [23] . If the cell is in need of biosynthetic intermediates, glucose 6-phosphate will enter the pentose phosphate pathway, where it will undergo a series of reactions that yield ribose and/or NADPH, depending on cellular conditions [29] . Thus, as PGM1 requires Ca2+/Mg2+ for activation, it tends to localize near Ca2+ channels, such as TRPV4 or the vacuolar Ca2+-ATPase, Pmc1p [28] [30] . In yeast, the role of Pmc1p appears to be similar to that of TRPV4 in human cells regarding PGM1 activation (Figure 6). Binding of PGM1 to the channel seems to block Ca2+ transport through the channel. However, it remains to be characterized whether the association with the activated or phosphorylated TRPV4 affects activity of PGM1 and the glycolytic pathway.
TRPV4 contains a similar aspartate-to-glutamate substitution at a different location in the selectivity filter, which may similarly account for somewhat lower 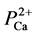 /PNa+~6 selectivity compared with
/PNa+~6 selectivity compared with  /PNa+~10 for TRPV1 and TRPV3. Permeability values relative to Na+ are 6 - 10 for Ca2+ and 2 - 3 for Mg2+ [31] . Their observations suggest that TRPV4 is a better channel for Mg2+ (which is required for PGM1 activation) than other TRP family members [32] . Furthermore, it was well characterized that TRPV4, which transports both Ca2+/Mg2+ ions, is the only active channel within body temperature [1] [2] [7] . Based on these reasons, we assume that this channel is selected for the activator for PGM1. Therefore, the interaction between these two proteins (TRPV4 and PGM1) is a major clue to answer why the mutant TRPV4 causes many human genetic diseases, including metatropic dysplasia [1] - [4] .
/PNa+~10 for TRPV1 and TRPV3. Permeability values relative to Na+ are 6 - 10 for Ca2+ and 2 - 3 for Mg2+ [31] . Their observations suggest that TRPV4 is a better channel for Mg2+ (which is required for PGM1 activation) than other TRP family members [32] . Furthermore, it was well characterized that TRPV4, which transports both Ca2+/Mg2+ ions, is the only active channel within body temperature [1] [2] [7] . Based on these reasons, we assume that this channel is selected for the activator for PGM1. Therefore, the interaction between these two proteins (TRPV4 and PGM1) is a major clue to answer why the mutant TRPV4 causes many human genetic diseases, including metatropic dysplasia [1] - [4] .
Our data provide the first evidence of a regulatory role for the PGM1-TRPV4 complex in Ca2+ reabsorption. Interaction between PGM1 and TRPV4 is also regulated by the phosphorylation status of TRPV4 S824 (Figure 1(b)). Thus, we suggest that phosphorylation of TRPV4 S824 is a major regulatory mechanism determining its interaction with many other proteins, including PGM1, STIM1, CaM, IP3R3, actin, and tubulin (Figure 6).
We demonstrated that the TRPV4 channel is an authentic substrate of SGK1, and that S824 of TRPV4 is phosphorylated by SGK1 (Figure 1(b)). Furthermore, we demonstrated that phosphorylation of S824 of TRPV4 is required for its interaction with F-actin, using TRPV4 mutants (S824D and S824A); this interaction also affected its subcellular localization. Here, we showed that phosphorylation of TRPV4 S824 promotes its association with PGM1; thus, inactivating TRPV4 (Figure 6). This signal convergence on the C-terminal domain of TRPV4 may constitute an important mechanism by which the timing and convergence of signal responses is integrated. Our results provide evidence for a functional role of plasma membrane-resident TRPV4 in the regulation of store-operated Ca2+ entry, mediated by protein-protein interactions of its C-terminal domain (Figure 1(a)).
We also reported that binding of Ca2+-CaM is prevented by SGK1-mediated phosphorylation on S824, a residue located within the CaM binding site [27] . Conversely, substitution of the target residue (S824) with aspartic acid (S824D) results in a more rapid and sustained activation of TRPV4-mediated current. These data show that Ca2+-CaM binding is inhibited by phosphorylation, indicating negative feedback regulation even at high Ca2+ concentrations. Even though phosphorylation of TRPV4 S824 was mediated by SGK1 (Figure 1(b)), this residue can also be phosphorylated by other protein kinases, including Akt kinase and protein kinase A. Further elucidation of the integration of extracellular signals with TRPV4 activation is required to better understand the mechanisms of TRPV4 regulation (Figure 6). TRPV4 plays an important role in pathological sensory perception and bone growth. As the potential effects of genetic mutations affecting TRPV4 function in human disease remain to be elucidated, characterization of TRPV4-interacting proteins may be useful for future efforts to cure or alleviate human diseases caused by TRPV4mutations.
CaM is a ubiquitously-expressed, dual-function protein that regulates the activity of multiple ion channels, Ca2+ pumps, and other proteins in a Ca2+-dependent manner [2] [14] . Thus, we hypothesized that the association between PGM1 and TRPV4 inhibits TRPV4 channel activity in the plasma membrane. The activated nature of TRPV4 S824D seems to be achieved by preventing the binding of inhibitory proteins, such as Ca2+-CaM or PGM1 (Figure 4(b)). As a bivalent metal ion, usually magnesium or cadmium (which both complex directly with the phosphoryl group esterified to the serine PGM1 active site) is required for PGM1 enzymatic activity, association of PGM1 with the active Ca2+ channel (via S824-phosphorylated TRPV4) is necessary for enhancing PGM1 activity (Figure 6). However, the association between PGM1 and the activated TRPV4 Ca2+ channel seems to inhibit channel function. TRPV4 contains a consensus sequence for SGK1-mediated phosphorylation within its CaM-binding domain (residues 811 - 850) (Figure 1(b)). The subcellular localization of TRPV4- S824A and S824D were independent of SGK1 activity (Figure 5(a)), whereas that of TRPV4-WT was affected. Thus, both PGM1 and CaM negatively affect TRPV4 channel function through protein-protein interactions (Figure 6). However, we do not know whether these two proteins affect TRPV4 function in a synergistic or competitive manner. As additional and as-yet unidentified proteins may also modulate TRPV4 function through protein-protein interactions with its C-terminal domain, further characterization of proteins interacting with this domain will be helpful to better understand its role (Figure 1). It also remains to be clarified whether the C-terminal domains of other TRP channel proteins (in addition to TRPV4) interact with PGM1. Because TRPV4 channels are often composed of heteromeric subunits in vivo, PGM1 may form a protein complex with other TRP channels.
5. Conclusion
The C-terminal domain of transient receptor potential vanilloid 4 (TRPV4) interacts with human phosphoglucomutase-1(PGM1). Phosphorylation on TRPV4 S824 enhances its association with PGM1. TRPV4-PGM1 interaction which is enhanced by the phosphorylation on TRPV4 serine 824 residue modulates its channel activity.
Acknowledgements
This work was supported by a National Research Foundation of Korea (NRF-2013R1A1A4A01005522 and BK21 plus) to S. S. Kang. Both Shin and Lee are Korean Research fellowship recipients (NRF-2014 R1A1A2009622, NRF-2013R1A1A2063994). We thank Dr. David M. Cohen (Oregon Health and Science University) for the TRPV4 gene and Dr. Joong-Kook Choi (Chungbuk National University) for the pmCherry-N1 PGM1 construct.
Cite this paper
Sung HwaShin,Eun JeoungLee,SungheeHyun,Sang SunKang, (2016) Phosphorylation on TRPV4 Serine Residue 824 Enhances Its Association with PGM1. American Journal of Molecular Biology,06,33-44. doi: 10.4236/ajmb.2016.61004
References
- 1. Nilius, B. and Voets, T. (2013) The Puzzle of TRPV4 Channelopathies. EMBO Reports, 14, 152-163.
http://dx.doi.org/10.1038/embor.2012.219 - 2. Kang, S.S., Shin, S.H., Auh, C.K. and Chun, J. (2012) Human Skeletal Dysplasia Caused by a Constitutive Activated Transient Receptor Potential Vanilloid 4 (TRPV4) Cation Channel Mutation. Experimental & Molecular Medicine, 44, 707-722.
http://dx.doi.org/10.3858/emm.2012.44.12.080 - 3. Nishimura, G., Lausch, E., Savarirayan, R., Shiba, M., Spranger, J., Zabel, B., Ikegawa, S., Superti-Furga, A. and Unger, S. (2012) TRPV4-Associated Skeletal Dysplasias. American Journal of Medical Genetics Part C: Seminars in Medical Genetics, 160C, 190-204.
http://dx.doi.org/10.1002/ajmg.c.31335 - 4. Lang, F. and Shumilina, E. (2013) Regulation of Ion Channels by the Serum- and Glucocorticoid-Inducible Kinase SGK1. FASEB Journal, 27, 3-12.
http://dx.doi.org/10.1096/fj.12-218230 - 5. Willette, R.N., Bao, W., Nerurkar, S., Yue, T.L., Doe, C.P., Stankus, G., Turner, G.H., Ju, H., Thomas, H., Fishman, C.E., Sulpizio, A., Behm, D.J., Hoffman, S., Lin, Z., Lozinskaya, I., Casillas, L.N., Lin, M., Trout, R.E., Votta, B.J., Thorneloe, K., Lashinger, E.S., Figueroa, D.J., Marquis, R. and Xu, X. (2008) Systemic Activation of the Transient Receptor Potential Vanilloid Subtype 4 Channel Causes Endothelial Failure and Circulatory Collapse: Part 2. Journal of Pharmacology and Experimental Therapeutics, 326, 443-452.
http://dx.doi.org/10.1124/jpet.107.134551 - 6. Earley, S., Heppner, T.J., Nelson, M.T. and Brayden, J.E. (2005) TRPV4 Forms a Novel Ca2+ Signaling Complex with Ryanodine Receptors and BKCa Channels. Circulation Research, 97, 1270-1279.
http://dx.doi.org/10.1161/01.RES.0000194321.60300.d6 - 7. Suzuki, M., Mizuno, A., Kodaira, K. and Imai, M. (2003) Impaired Pressure Sensation in Mice Lacking TRPV4. Journal of Biological Chemistry, 278, 22664-22668.
http://dx.doi.org/10.1074/jbc.M302561200 - 8. Arniges, M., Vazquez, E., Fernandez-Fernandez, J.M. and Valverde, M.A. (2004) Swelling-Activated Ca2+ Entry via TRPV4 Channel Is Defective in Cystic Fibrosis Airway Epithelia. Journal of Biological Chemistry, 279, 54062-54068.
http://dx.doi.org/10.1074/jbc.M409708200 - 9. Lorenzo, I.M., Liedtke, W., Sanderson, M.J. and Valverde, M.A. (2008) TRPV4 Channel Participates in Receptor-Operated Calcium Entry and Ciliary Beat Frequency Regulation in Mouse Airway Epithelial Cells. Proceedings of the National Academy of Sciences USA, 105, 12611-12616.
http://dx.doi.org/10.1073/pnas.0803970105 - 10. Jia, Y., Wang, X., Varty, L., Rizzo, C.A., Yang, R., Correll, C.C., Phelps, P.T., Egan, R.W. and Hey, J.A. (2004) Functional TRPV4 Channels Are Expressed in Human Airway Smooth Muscle Cells. American Journal of Physiology-Lung Cellular and Molecular Physiology, 287, L272-L278.
http://dx.doi.org/10.1152/ajplung.00393.2003 - 11. Watanabe, H., Vriens, J., Prenen, J., Droogmans, G., Voets, T. and Nilius, B. (2003) Anandamide and Arachidonic Acid Use Epoxyeicosatrienoic Acids to Activate TRPV4 Channels. Nature, 424, 434-438.
http://dx.doi.org/10.1038/nature01807 - 12. Birder, L., Kullmann, F.A., Lee, H., Barrick, S., de Groat, W., Kanai, A. and Caterina, M. (2007) Activation of Urothelial Transient Receptor Potential Vanilloid 4 by 4α-Phorbol 12,13-Didecanoate Contributes to Altered Bladder Reflexes in the Rat. Journal of Pharmacology and Experimental Therapeutics, 323, 227-235.
http://dx.doi.org/10.1124/jpet.107.125435 - 13. Suzuki, M., Hirao, A. and Mizuno, A. (2003) Microtubule-Associated Protein 7 Increases the Membrane Expression of Transient Receptor Potential Vanilloid 4 (TRPV4). The Journal of Biological Chemistry, 278, 51448-51453.
http://dx.doi.org/10.1074/jbc.M308212200 - 14. Strotmann, R., Schultz, G. and Plant, T.D. (2003) Ca2+-Dependent Potentiation of the Nonselective Cation Channel TRPV4 Is Mediated by a C-Terminal Calmodulin Binding Site. The Journal of Biological Chemistry, 278, 26541-26549.
http://dx.doi.org/10.1074/jbc.M302590200 - 15. Fu, Y., Subramanya, A., Rozansky, D. and Cohen, D.M. (2006) WNK Kinases Influence TRPV4 Channel Function and Localization. American Journal of Physiology—Renal Physiology, 290, F1305-F1314.
http://dx.doi.org/10.1152/ajprenal.00391.2005 - 16. Cuajungco, M.P., Grimm, C., Oshima, K., d’Hoedt, D., Nilius, B., Mensenkamp, A.R., Bindels, R.J., Plomann, M. and Heller, S. (2006) PACSINs Bind to the TRPV4 Cation Channel. PACSIN 3 Modulates the Subcellular Localization of TRPV4. The Journal of Biological Chemistry, 281, 18753-18762.
http://dx.doi.org/10.1074/jbc.M602452200 - 17. Vriens, J., Owsianik, G., Fisslthaler, B., Suzuki, M., Janssens, A., Voets, T., Morisseau, C., Hammock, B.D., Fleming, I., Busse, R. and Nilius, B. (2005) Modulation of the Ca2 Permeable Cation Channel TRPV4 by Cytochrome P450 Epoxygenases in Vascular Endothelium. Circulation Research, 97, 908-915.
http://dx.doi.org/10.1161/01.RES.0000187474.47805.30 - 18. Fernandes, J., Lorenzo, I.M., Andrade, Y.N., Garcia-Elias, A., Serra, S.A., Fernandez-Fernandez, J.M. and Valverde, M.A. (2008) IP3 Sensitizes TRPV4 Channel to the Mechano- and Osmotransducing Messenger 5’-6’-Epoxyeicosa-trienoic Acid. The Journal of General Physiology, 131, i2.
http://dx.doi.org/10.1085/JGP1315OIA2 - 19. Putt, W., Ives, J.H., Hollyoake, M., Hopkinson, D.A., Whitehouse, D.B. and Edwards, Y.H. (1993) Phosphoglucomutase 1: A Gene with Two Promoters and a Duplicated First Exon. Biochemical Journal, 296, 417-422.
http://dx.doi.org/10.1042/bj2960417 - 20. Foster, J.M., Davis, P.J., Raverdy, S., Sibley, M.H., Raleigh, E.A., Kumar, S. and Carlow, C.K. (2010) Evolution of Bacterial Phosphoglycerate Mutases: Non-Homologous Isofunctional Enzymes Undergoing Gene Losses, Gains and Lateral Transfers. PLoS ONE, 5, e13576.
http://dx.doi.org/10.1371/journal.pone.0013576 - 21. Mitra, S., Cui, J., Robbins, P.W. and Samuelson, J. (2010) A Deeply Divergent Phosphoglucomutase (PGM) of Giardia Lamblia Has both PGM and Phosphomannomutase Activities. Glycobiology, 20, 1233-1240.
http://dx.doi.org/10.1093/glycob/cwq081 - 22. Hindle, A.G., Karimpour-Fard, A., Epperson, L.E., Hunter, L.E. and Martin, S.L. (2011) Skeletal Muscle Proteomics: Carbohydrate Metabolism Oscillates with Seasonal and Torpor-Arousal Physiology of Hibernation. American Journal of Physiology—Regulatory, Integrative and Comparative Physiology, 301, R1440-R1452.
http://dx.doi.org/10.1152/ajpregu.00298.2011 - 23. Kletzien, R.F., Harris, P.K. and Foellmi, L.A. (1994) Glucose-6-Phosphate Dehydrogenase: A “Housekeeping” Enzyme Subject to Tissue-Specific Regulation by Hormones, Nutrients, and Oxidant Stress. The FASEB Journal, 8, 174- 181.
- 24. Onteru, S.K., Gorbach, D.M., Young, J.M., Garrick, D.J., Dekkers, J.C. and Rothschild, M.F. (2013) Whole Genome Association Studies of Residual Feed Intake and Related Traits in the Pig. PLoS ONE, 8, e61756.
http://dx.doi.org/10.1371/journal.pone.0061756 - 25. Chen, Y. and Tye, B.K. (1995) The Yeast Mcm1 Protein Is Regulated Posttranscriptionally by the Flux of Glycolysis. Molecular and Cellular Biology, 15, 4631-4639.
http://dx.doi.org/10.1128/MCB.15.8.4631 - 26. Takahashi, K., Inuzuka, M. and Ingi, T. (2004) Cellular Signaling Mediated by Calphoglin-Induced Activation of IPP and PGM. Biochemical and Biophysical Research Communications, 325, 203-214.
http://dx.doi.org/10.1016/j.bbrc.2004.10.021 - 27. Shin, S.H., Lee, E.J., Hyun, S., Chun, J., Kim, Y. and Kang, S.S. (2012) Phosphorylation on the Ser 824 Residue of TRPV4 Prefers to Bind with F-Actin than with Microtubules to Expand the Cell Surface Area. Cellular Signalling, 24, 641-651. http://dx.doi.org/10.1016/j.cellsig.2011.11.002
- 28. Li, X., Qian, J., Wang, C., Zheng, K., Ye, L., Fu, Y., Han, N., Bian, H., Pan, J., Wang, J. and Zhu, M. (2011) Regulating Cytoplasmic Calcium Homeostasis Can Reduce Aluminum Toxicity in Yeast. PLoS ONE, 6, e21148.
http://dx.doi.org/10.1371/journal.pone.0021148 - 29. Chun, J., Shin, S.H. and Kang, S.S. (2012) The Negative Feedback Regulation of TRPV4 Ca2+ Ion Channel Function by Its C-Terminal Cytoplasmic Domain. Cellular Signalling, 24, 1918-1922.
http://dx.doi.org/10.1016/j.cellsig.2012.06.008 - 30. Ninfali, P., Accorsi, A., Palma, F., Fazi, A., Piatti, E., Chiarantini, L. and Fornaini, G. (1984) Human Erythrocyte Phosphoglucomutase: Comparison of the Kinetic Properties of PGM1 and PGM2 Isoenzymes. Biochimie, 66, 617-623.
http://dx.doi.org/10.1016/0300-9084(84)90115-9 - 31. Nilius, B., Vriens, J., Prenen, J., Droogmans, G. and Voets, T. (2004) TRPV4 Calcium Entry Channel: A Paradigm for Gating Diversity. American Journal of Physiology—Cell Physiology, 286, C195-C205.
http://dx.doi.org/10.1152/ajpcell.00365.2003 - 32. Voets, T., Prenen, J., Vriens, J., Watanabe, H., Janssens, A., Wissenbach, U., Bodding, M., Droogmans, G. and Nilius, B. (2002) Molecular Determinants of Permeation through the Cation Channel TRPV4. The Journal of Biological Chemistry, 277, 33704-33710.
http://dx.doi.org/10.1074/jbc.M204828200
NOTES
*Both contribute equally.
#Corresponding author.








