American Journal of Molecular Biology
Vol.4 No.1(2014), Article ID:41628,6 pages DOI:10.4236/ajmb.2014.41004
Polysaccharides as initiators of nucleic acid polymerization
1Research and Production Association “Plazan”, Moscow, Russia
2Bashkir State University, Ufa, Russia
3Biochemistry and Genetics Institute, Russian Academy of Sciences, Ufa, Russia
4Institute of Organic Chemistry, Russian Academy of Sciences, Ufa, Russia
Email: *zanol@yandex.ru
Received 18 November 2013; revised 15 December 2013; accepted 2 January 2014
ABSTRACT
Quantum-chemical calculations and in vitro experiments have demonstrated that polysaccharides are able to adsorb mononucleotides due to the formation of hydrogen bonds. The subsequent dehydration of polysaccharide-nucleotide complexes in the physiologically acceptable temperature range results in the creation of “correct” DNA polymers that are “recognized” by the specific DNA-polymerases. DNA fragments abiogenically formed on polysaccharides vary in size and are characterized by a relatively “simple”, most probably, tandem structure. This research developed our previous concept of template-based polysaccharide synthesis with the participation of DNA tandem repeats (glycotranscription concept) making it possible to assume the existence of reverse glycotranscription in biological systems. Future research in the direction that we outlined experimentally may lead to a revolutionary approach in modern molecular biology—glycogenomics.
Keywords:Polysaccharides; Nucleic Acids; Polymerization; Complementarity; PCR
1. INTRODUCTION
Our previous papers presented the selectivity in nucleotide base interaction with acidic and neutral monoand oligosaccharides in silico and in vitro [1]. It was found that nucleic acids (NA) purine bases form complexes more effectively with acidic monoand oligosaccharides whereas NA pyrimidine bases—with neutral saccharides.
A single-stranded DNA was found to form helical structure with a polysaccharide fragment. The characteristic of such structure is identical to a double-stranded DNA. The analysis of the interaction between nucleic acids and oligosaccharides carried out alongside the study of the biosynthesis processes of the latter in vivo [2] has enabled us to suggest a concept of template-based synthesis of oligo and polysaccharide molecules with the participation of DNA tandem repeats (glycotranscription process) [3]. However, a question still remains undetermined whether DNA formation is possible on one-dimensional oligosaccharide molecule according to the reverse glycotranscription principle in the way that it was described for RNA-DNA tandem in modern classical molecular biology (the reverse transcription process).
Today, there are two generally acknowledged ways of a DNA polymer formation: the first one is the template synthesis method with the participation of enzymes, both in vivo and in vitro [4], and the second one is abiogenic synthesis carried out under the influence of thermal energy, described by L. Orgel [5]. Since the enzyme systems that would promote the assembly of oligonucleotide with polysaccharides acting as the template had never been described in the scientific papers, we followed the second approach to the formation of DNA chain in this paper.
It was of utmost importance for us to establish that polymer NA fragments acting as the template can be assembled out of mononucleotides on a polysaccharide, and in the case of objective registration of this process, to analyze the PCR-Amplified DNA fragments.
2. MATERIALS AND METHODS
As a rule, acidic polysaccharides and polyuronides in plants are represented by the monomers of two hexuronic acids—D-glucuronic acid and D-galacturonic acid. Dglucuronic acid is common in hemicellulose polysaccharides, whereas D-galacturonic acid in the form of galacturonans is the main component of pectic substances. It is worth noting that these polysaccharides can be easily obtained from oat seeds and apple pulp. Thus, acidic oligosaccharides were extracted from apples according to a standard method described earlier for oat seeds [3]. The extracted PU was additionally treated with deoxyribonuclease and proteases to prevent contamination with endogenous DNA. The enzymes from PU samples were then eliminated by three-fold treatment with phenolchloroform mixture followed by three-fold precipitation by triple volume cooled ethanol [6].
5’ nucleotide monophosphates (sodium salt) in ribo-, deoxyribo-form and oligo (dT)-cellulose were obtained from “Sigma-Aldrich”, amylose (neutral polysaccharide, NS) was obtained from “ServaFeinbiochemica GMBH”.
The procedure of AMP polymerization (sodium salt) was as follows. 10 mg of AMP and 10 mg of PU were dissolved in 50 ml of H2O. Then the solution was evaporated to dryness in vacuum at a temperature of 40˚C and the obtained nucleotide PS crystals were exposed to different temperatures (20˚C, 40˚C, 60˚C, 120˚C, 130˚C) for 6 hours. Poly-A fragments of NA were extracted on oligo (dT)-cellulose by a standard method [7].
Deoxynucleotide polymerization in the presence of polyuronides (PU) was carried out by dissolving 10 mg of PU in 40 ml of water and adding 2.5 mg of each of the four nucleotides. The mixture was made up to 50 ml with H2O. The solution was incubated at 40˚C or 60˚C for 1 hour, then it was evaporated to dryness in vacuum and dissolved in 5 ml of water.
The complexes were precipitated by adding triple volume of ethanol at +4˚C for 12 hours. The precipitate was collected by centrifugation and dried out. The quantity of nucleotides was estimated spectrophotometrically by measuring the absorbance at 260 nm. The correspondence of the absorbance to the quantity of nucleotides was determined by a calibration curve individually for each nucleotide. PS alcohol precipitation in the absence of nucleotides served as the control.
Gel-filtration on Sephadex G-25 was carried out on a column 30 cm long, 1.5 cm in diameter. The volume of the applied sample was 5 ml, fraction size was 3 ml. PU was detected spectrophtometrically by measuring the absorbance at 220 nm, whereas NA was detected by measuring the absorbance at 260 nm.
Polymerase chain reaction (PCR) was conducted with short oligonucleotides in non-specific conditions of hybridization (the DNA-RAPD method). The amplification process was preceded by a preliminary denaturation at 94˚C for 2 min and consisted of 40 cycles, each one including the following steps: denaturation at 94˚C for 20 sec, annealing of the primers at 30˚C for 20 sec, and elongation at 72˚C for 20 sec. The following oligonucleotides were used for DNA-amplification: (AFK-3— acggtggacg, AFK-1—gcgtccaattc, ADE-1—tcgaatagc).
Theoretical computations for the formation of nucleotide-polysaccharides (PS) complexes in this work were carried out by the PM3 semiempirical method [8] with the complete optimization of geometrical parameters employing the GAMESS program [9]. A global minimum of the total electron energy was sought by the Newton-Raphson method with an energy gradient of 0.1 kcal/mol ΔЕ, starting from various initial approximations of the complex structure.
3. RESULTS
It was found that both acidic and neutral polysaccharides are capable of forming hydrogen bonds with nucleic acid bases. Table 1 demonstrates energy estimates of hydrogen bonds between carboxyl group of polyuronides and hydroxymethyl group of neutral saccharides with nucleotides obtained by the PM3 method. The computations show that the energy of hydrogen bonds between nucleotides and acidic polysaccharides is more than 3 times higher than the corresponding estimates for neutral polysaccharides, thus, theoretically predetermining their higher capacity for nucleotide adsorption. The calculations were confirmed experimentally (Table 2).
Table 1. Energy of hydrogen bonds between carboxyl (PU) and hydroxymethyl groups (NS) of saccharides in a complex with nucleotides (PM3 method, kcal/mol).
Table 2. The quantity of deoxynucleotides detectable in the ethanol precipitation of oligosaccharides (mcg NA/mg PS).
Indeed, homopolymers of acidic saccharides (PU) adsorb 3.5 times more nucleotides as compared to homoglycans NS (amylose).
As can be seen from the above, the specific adsorption of nucleotides on PS is beyond any doubt, according to the presented results and our earlier publications describing the physicochemical principles of these processes [1,2]. However, the question of nucleotide polymerization on a linear PS molecule has been undetermined. Our in silico study of mononucleotide interaction with a polysaccharide molecule showed that nucleotide monomers on PS are ideally oriented to form covalent bonds between them [2]. This should significantly stimulate the process of polynucleotide formation from monomers bound with PS. This assumption was verified in the series of experiments for the dehydration of PU complexes with AMP as it was described in the experimental section. These experiments were aimed at obtaining poly (A) oligomers with their further isolation using the specific oligo (dT) cellulose sorbent.
The control of our experiment was the approach used by L. Orgel for the synthesis of polynucleotides under the influence of thermal energy [5]. Polyuronides were used in the empirical sample in parallel with AMP. The results of the experiment are shown in Figure 1.
The results shown in Figure 1 demonstrate that AMP polymerization is practically undetected in the control sample dehydrated at a temperature below 100˚C, which corresponds to the data provided by L. Orgel. Indeed, the free AMP polymerizes at a temperature above 100˚C, while the nucleotide in the presence of PU shows the maximum NA polymerization rate in the temperature
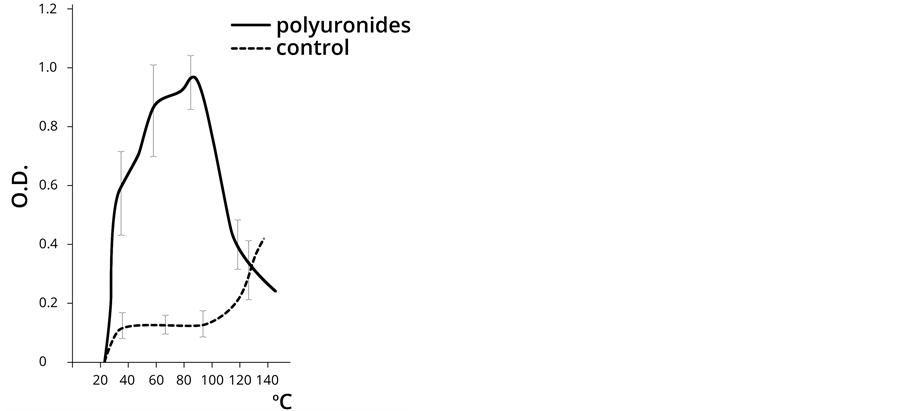
Figure 1. The dynamics of poly (A) NA formation (chromatography on the oligo (dT) cellulose column) at different temperatures in the absence (control) and presence of PU (OD at 260 nm). Note: PU dehydration was carried out only in the presence of AMP.
range 30˚C to 80˚C. Further temperature increase in the empirical sample resulted in the destruction of the polynucleotide.
As can be seen from the above, PU demonstrated a significant catalytic activity in the process of nucleotide polymerization within the physiological temperature range (20˚C - 60˚C), and apparently acted as the template for the arrangement of NA polymer molecules. It seems fair to assume that, that PU does not only adsorb nucleotides, but also facilitate the process of their polymerization when dehydrated. Abiogenic polymerization of nucleotides is about 10 times more intensive in the presence of PU. NA concentration is increased in the PU-NA complexes in the subsequent processes of solution and repeated dehydration of the complexes with the mixture of nucleotides, as it is demonstrated in Table3
Our next objective was to isolate NA fragments formed on the PU by chromatography for their further analysis using the PCR method. As it is shown in Figure 2(A), the results of PU-NA complex chromatography on Sephadex G-25 demonstrate that high-polymer PU components were eluted predominantly before fraction 15. Low-polymer components were eluted between fractions 16 and 22 in smaller quantities than the high-polymer ones. The monomers eluted after the 23th fraction predictably were not represented in the samples, since ethanol precipitation of PS-NA leaves nucleotide monomers in the solution.
As it is shown in Figure 2(B), the spectrophotometric analysis of the eluted complexes demonstrates that the low-polymer fraction is rich in nucleotides, as the OD ratio A260/A220 fluctuates within the range of 0.8 - 1.0 in these samples, whereas for the high-polymer fraction it is 0.5. These results suggest the presence of abiogenically formed DNA in the fractions containing low polymer fragments.
To verify the presence of “correct” DNA chains recognized by specific enzymes in the test sample and its characteristic, we carried out an analysis using PCR. The NA polymers of fractions 16 - 22 after the chromatography oh Sephadex G-25 were used in the experiment. The results of the experiment are shown in Figure 3.
As Figure 3 demonstrates, the primers themselves (Lanes 1) can serve as the template for the synthesis of
Table 3 . Optical characteristics of PU-NA complexes subjected to dehydration once (I), twice (II), three times (III) (260/220 nm).
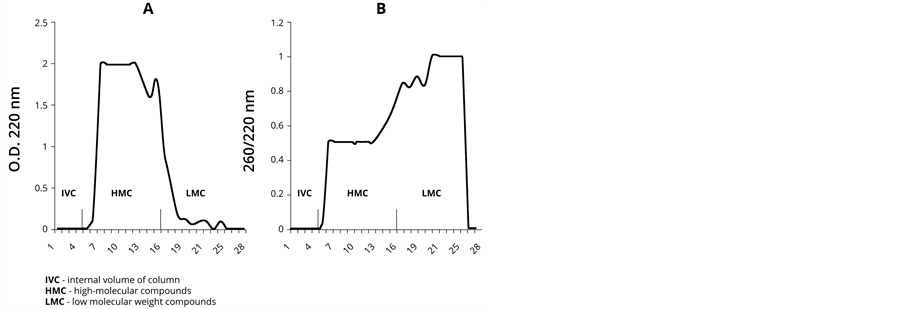
Figure 2. PU-NA complex chromatography on Sephadex G-25. (A) represents detection based on OD at 220 nm, and (B) represents detection based on OD 260/220 nm measurements. Note: in this experiment PU dehydration was conducted once in the presence of 4 deoxynucleotide monophosphates at a temperature of 60˚C.
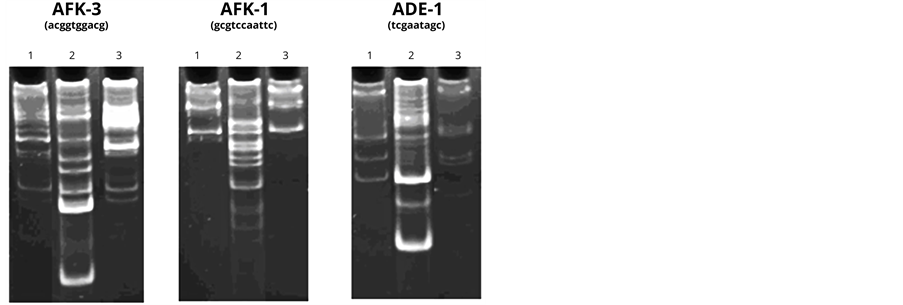
Figure 3. The results of DNA-RAPD with the DNA preparation synthesized on PU. 1. Control (primer), 2. Human DNA + primer, 3. DNA synthesized on PU, + primer.
high-polymer DNA. The reaction conditions facilitate the accumulation of the products at initially low template concentration due to the properties of the primers and to the presence of some DNA oligomers in the reaction mixture. Moreover, the introduction of an additional control—human DNA (Lane 2) to the experiment allows us to deny the presence of human DNA in the tested samples and to observe low ability of the primers to “self-prime” in the presence of the given DNA.
The acceleration of the polymerase activity is observed in the test sample (Lane 3) in the presence of AFK-3 primer. The new product, which is a few hundreds of nucleotide pairs in length, cannot be explained by human DNA contamination (most likely in the PCR process). Since the DNA product in the tested sample is formed in the presence of only one of the primers, it can be assumed that the amplicon, and therefore, the original DNA have low “complexity”, i.e. have limited number of primary structure variants. The latter, as is well-known, is typical for DNA tandem repeats.
The results of PCR with AFR-3 primer on PU and on the isolated NA fragments synthesized on PU are shown in Figure 4.
The Figure 4 demonstrates that the primer itself (Lane 1) can serve as the template for high-polymer DNA synthesis as per the above procedure, promoting the product accumulation at initially low concentration of the template due to the properties of the primer itself and the presence of the admixture of DNA fragments of an unidentified origin.
The activity of the Taq-polymerase used in the PCR reaction was not detected in the samples containing PU and nucleotide (Lane 3) as well as in the pure PU (Lane 2). It may suggest the absence of NA fragments in the original PU samples and the ability of PU in high concentrations to suppress enzyme activity. Alternatively, the PU from fractions 16 - 22 (Figure 2(A)) stimulated enzyme activity. This fact attests the presence of DNA molecules in the given polysaccharide fractions (Lane 4). The synthesized product, which is a few hundred nucleotide pairs in length, does not result from human DNA contamination, as was mentioned above (Figure 3). No visible quantities of the product are detectable in Lane 5 demonstrating the PCR result in the conditions of self-priming of isolated NA synthesized on PU. It can be caused by significant variations throughout the length of

Figure 4. DNA-RAPD results with the preparations of NA synthesized on PU. Lanes: 1—control (AFK-3 primer), 2—AFK-3 primer + PU, 3—AFK-3 primer + PU-nucleotide mixture, 4—AFK-3 primer + oligo nucleotides from PU-NA complex after dehydration, 5—isolated oligonucleotides from PU-NA complex in the absence of the primer.
the analyzed NA sequences formed on PU.
As can be seen from the above, the PCR method confirmed the presence of DNA oligomers of varying sizes synthesized on PU. According to preliminary data they have a “simple”, most probably tandem structure. We found that that the given NA sequences do not relate to human DNA. The nature of the given NA calls for further research.
4. DISCUSSION
Our studies have demonstrated via computational method in silico that DNA repeats are complementary to certain homoand heteroglycans which has later been proved experimentally in vitro [1]. Our further research has shown that synthesis initiation of glycoside moiety of the proteoglycans is associated with DNA and takes place in eukaryote nucleus in vivo system [3]. With oligoand polysaccharides being of utmost importance for tissue specificity, organ specificity, immunity and other vitally important functions of living matter set on the genetic level and no clear explanations of DNA repeats role, a concept of template-based polysaccharide synthesis has been proposed, in which DNA tandem repeats play an integral information role [2]. The postulated process of genetic information transfer from DNA repeats to polysaccharides (glycotranscription) allows to assume in principle the possibility of the reverse process of template-based assembly of nucleotides on polysaccharide molecules (reverse glycotranscription). This question is being studied in the given materials.
As have been experimentally proven polysaccharides are not only able to adsorb nucleotides (see Tables 1 and 2), but also help to form specific bonds between monomers creating polynucleotide fragments within physiologically acceptable temperature range (see Figure 1). In fact, polysaccharides catalyze the process of polymer DNA chain formation by increasing the intensiveness of polymerization process and shifting the temperature range to physiologically acceptable values (30˚C - 60˚C). The mentioned quality of PS makes it possible to reconsider their potential role in molecular evolution of biopolymers of living matter. In view of the above and taking into account the ability of PS to ensure proper nucleotide assembly to form functionally active DNA fragments recognizable by specific DNA polymerases (Figures 3 and 4), we have every reason to assume the existence of DNA assembly process on PS in vivo.
The present research only verifies the assembly of NApolymeric chains on PS in vitro. However, the existence of such a process in vivo, its role and characteristics of the obtained DNA fragments require additional studies which could lead to a new discipline in modern molecular biology—glycogenomics—with its own tasks, objectives and subjects of research [10].
5. CONCLUSION
In addition to the conventional way of genetic information transfer from DNA through RNA to polypeptide, we postulated the glycotranscription process in which genetic information is transferred according to the following scheme: DNA-RNA-polysaccharide. The information about the initial polysaccharide structure is coded by DNA tandem repeats [2]. The experimental data obtained in the present research confirm the possibility of a reverse glycotranscription process in vitro. “Correct” DNA fragments formed on polysaccharides and recognized by specific DNA polymerases are characterized by a “low level” of complexity typical of DNA tandem repeats. Further in vivo investigation of this phenomenon, which we observed experimentally, may lead to a revolutionary discipline in modern molecular biology—glycogenomics.
ACKNOWLEDGEMENTS
This work was supported by the “Plazan” Closed Joint-Stock Company, Moscow.
REFERENCES
- Zimnitskii, A.N., Bashkatov, S.A. and Urazbaev, V.N. (2007) Interaction between nucleic acids and glycans: In silico and in vitro. Biophysics, 52, 282-287. http://nanoderm.ru/img/userimg/statyi/magazine_biofizika_eng.pdf http://dx.doi.org/10.1134/S0006350907030049
- Zimnitskii, A.N. (2012) Concept of template synthesis of proteoglycans. In: Desiree, N.K., Ed., The Complex World of Polysaccharides, InTech. http://www.intechopen.com/books/the-complex-world-of-polysaccharides/concept-of-template-synthesis-of-proteoglycans http://dx.doi.org/10.5772/48085
- Zimnitsky, A.N. and Bashkatov, S.A. (2006) Association of glycosaminoglycan biosynthesis with the nuclear and microsomal cell apparatuses. Molecular Biology, 40, 289- 299. http://nanoderm.ru/technology/nauchnye-stati-a_n_zimnicko-go/association_of_glucosaminoglycan_biosynthesis_with_the_nuclear_and_microsomal_cell_apparatuses http://dx.doi.org/10.1134/S0026893306020117
- Singer, M. and Berg P. (1998) Genes and genomes. Mir, Moscow, 764 p.
- Orgel, L.E. (1998) The origin of life—A review of facts and speculations. Trends in Biochemical Sciences, 23, 491-495. http://dx.doi.org/10.1016/S0968-0004(98)01300-0
- Osterman, L.A. (1981) Methods of investigating proteins and nucleic acids: Electrophoresis and ultracentrifugation. Nauka, Мoscow, 286 p.
- Gilyazetdinov, S.Y. and Zimnitskii, A.N. (1986) Composition of poly(А+)-RNA population in the roots of corn heterotic hybrids and their original strains. Physiology of Plant, 33, 769-777.
- Sterwart, J.J.P. (1989) Optimization of parameters for semiempirical methods. II. Applications. Journal of Computational Chemistry, 10, 221-261.
- Schmidt, M.W., Baldridge, K.K., Boatz, J.A., Elbert, S.T., Gordon, M.S., Jensen, J.H., Koseki, S., Matsunaga, N., Nguyen, K.A., Su, S.J., Windus, T.L., Dupuis, M., Montgomery, J.A. (1993) General atomic and molecular electronic structure system. Journal of Computational Chemistry, 14, 1347-1363. http://dx.doi.org/10.1002/jcc.540141112
- Chemeris, A.V., Magdanov, E.G., Garafutdinov, R.R., Matniyazov, R.T., Baimiev, A.K. Bikbulatova, S.M., Gimalov, F.R. and Vakhitov, V.A. (2013) Some technological past, present and future of modern biology by 2030. Biomics, 5, 10-43. http://biomics.ru/nomera/2013/54-nekotoroe-tehnologicheskoe-proshloe-nastoyaschee-a-takzhe-buduschee-sovremennoy-biologii-k-2030-godu-chast-pervaya.html
NOTES
*Corresponding author.


