 J. Biomedical Science and Engineering, 2013, 6, 1099-1108 JBiSE http://dx.doi.org/10.4236/jbise.2013.611138 Published Online November 2013 (http://www.scirp.org/journal/jbise/) 3D segmentation and visualization of lung and its structures using CT images of the thorax Pedro P. Rebouças Filho1, Paulo Cesar Cortez2, Victor Hugo C. de Albuquerque3* 1Industry Area, Federal Institute of Education Science and Technology of Ceará, Maracanaú, Brazil 2Teleinformatic Engineering Department, Federal University of Ceará, Fortaleza, Brazil 3Graduate Program in Applied Informatics, University of Fortaleza, Fortaleza, Brazil Email: pedrosarf@ifce.edu.br, cortez@deti.ufc.br, *victor.albuquerque@unifor.br Received 25 September 2013; revised 26 October 2013; accepted 7 November 2013 Copyright © 2013 Pedro P. Rebouças Filho et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. ABSTRACT Computing systems have been playing an important role in various medical fields, notably in image diag- nosis. Studies in the field of Computational Vision aim at developing techniques and systems capable of detecting various illnesses automatically. What has been highlighted among the existing exams that allow diagnosis aid and the application of computing sys- tems in parallel is Computed Tomography (CT). CT enables the visualization of internal organs, such as the lung and its structures. Computational Vision systems extract information from the CT images by segmenting the regions of interest, and then recognize and identify details in those images. This work fo- cuses on the segmentation phase of CT lung images with singularity-based techniques. Among these me- thods are the region growing (RG) technique and its 3D RG variations and the thresholding technique with multi-thresholding. The 3D RG method is appli- ed to lung segmentation and from the 3D RG seg- ments of the lung hilum, the multi-thresholding can segment the blood vessels, lung emphysema and the bones. The results of lung segmentation in this work were evaluated by two pulmonologists. The results ob- tained showed that these methods can integrate aid systems for medical diagnosis in the pulmonology field. Keywords: 3D Region Growing; Lungs segmentation; COPD; Pulmonary Structure Visualization; Computed Tomography 1. INTRODUCTION A large number of diseases that affect the world’s po- pulation are lung-related. Therefore, research in the field of pulmonology has great importance in public health studies and focuses mainly on asthma, bronchiectasis and Chronic Obstructive Pulmonary Disease (COPD) [1,2]. The World Health Organization (WHO) estimates that there are 300 million people who suffer from asthma, and this disease causes around 250 thousand deaths per year worldwide [3]. Also, WHO estimates that 210 million people have COPD. This disease caused the death of over 300 thousand people in 2005 [4]. Recent studies reveal that COPD is present in the 20- to 45-year- old age bracket, although it is characterized as an over- 50-year-old disease. Based on this, WHO estimates that the number of deaths due to COPD will increase 30% by 2015, and by 2030 COPD will be the third cause of mor- talities worldwide [5]. During the period from 1992 to 2006, 15% of all hospitalizations financed by the Brazilian Federal Health System (SUS) were due to pulmonary diseases, of which asthma and COPD together summed up 562,016 hospita- lizations [3]. Thus, it is of fundamental importance for the public health system to obtain an early and correct diagnosis of any pulmonary disease. Diagnosis aid is important from a clinical point of view as it increases the amount of in- formation the specialist has concerning the patient’s state of health. Therefore, certain illnesses can be detected precociously, even saving lives in some cases. Besides, some techniques allow the clinical image of the disease to be tracked appropriately [6,7]. The segmentation stage pulmonology systems are es- sential for the correct and accurate medical diagnosis aid, as this stage delimits the lung area in CT images of the thorax, which must be analyzed by the system or by a specialist. The segmentation of objects and structures in medical images is a process that, in most cases, is more complex if compared to the segmentation of other image types, *Corresponding author. OPEN ACCESS  P. P. Rebouças Filho et al. / J. Biomedical Science and Engineering 6 (2013) 1099-1108 1100 such as metallographic and synthetic aperture radar re- mote sensing. This occurs due to the variability of struc- tures and/or internal organs, the visualization plane of these images and furthermore, the possibility of diseases affecting these organs. All of which contribute to in- creasing the difficulty to develop efficient techniques for medical image segmentation [8]. CT images of the lungs represent a slice of the ribcage, where a large number of structures are located, such as blood vessels, arteries, respiratory vessels, pulmonary pleura and parenchyma, each with its own specific infor- mation. Thus for analysis and for the pulmonary disease diagnosis aid, it is necessary to segment the lung structure. Lung segmentation techniques have been developed seeking optimization at this stage. Sluimer, Prokop and Ginneken in [9], and Felix et al. in [6], achieved lung segmentation of CT images using the region growing method. Felix et al. in [10], Hu, Hoffman and Reinhart in [11], and Silva et al. in [12], put together mathematical morphology, region growing, thresholding and edge detection methods to obtain even more accurate results. The Pulmonary Emphysema Detection system (PEDS) performs automatic lung segmentation through region growing associated with binary morphological operations of dilation and erosion [8]. Another methodology for lung segmentation is the 3D region growing approach, and this is applied to segment the lung and its internal structures, such as the vessels and airways [13-16]. This method works similarly to the 2D region growing technique that is initialized by a seed point and then expanded through its neighbors, obeying an aggregation rule. In addition, other computational tools have also been used to evaluate lung structures, for example, Chen et al. in [17], compared the diagnostic performances of an artificial neural network (ANNs) and multivariable logis- tic regression analyses for differentiating between malig- nant and benign lung nodules on computed tomography scans. Er, Yumusak and Temurtas in [18], also used ANNs to analyze chest disease diagnoses by using mul- tilayer, probabilistic, learning vector quantization, and generalized regression neural networks. These authors also used an artificial immune system classification [19], and obtained high accuracy in chest disease diagnoses. Fernández et al. in [20], evaluated serum biomarkers le- vels in lung cancer patients and non-cancer controls us- ing principal component analysis and ANNs modeling. Active contour methods have also been widely re- ported in the literature to segment lung structures from CT images. Annangi et al. in [21], proposed a region- based active contour method for x-ray lung segmentation using prior shape and low level features. Wu, Wang and Jia in [22], presented a novel external force, called adap- tive diffusion flow (ADF), with adaptive diffusion stra- tegies according to the characteristics of an image region in the parametric active contour model framework. Ke- shani et al. in [23], used a support vector machine com- bined with an active contour model to segment lung no- dules. Tan, Schwartz and Zhao in [24], combined image processing techniques of marker-controlled watershed, geometric active contours as well as the Markov random field to evaluate lung lesions considering their size, den- sity, and shape. Finally, Liu and Bovik in [25], proposed a novel external force for active contours, which we call neighborhood-extending and noise-smoothing gradient vector flow to extract the parenchyma and cancer re- gions. This work contemplates two singularity methods, the 3D region growing and the multi-thresholding algorithms, for the segmentation of the lung and its internal struc- tures. The methods aim to assist medical diagnosis aid in pulmonology diseases though 3D visualization and re- construction from bi-dimensional CT images. After ima- ge segmentation, it is possible to visualize and quantify the lung structures, as well as its internal structures tridimensionally. This facilitates the diagnosis by the medical experts and reduces/eliminates the subjectivity of interpretation of the test. The results obtained from the proposed method that performs the bi-dimensional seg- mentation and volumetric quantifying of all lung struc- tures were evaluated and validated by two medical spe- cialties (pulmonologists). One of the main contributions of this work is to use the proposed method on personal computers, maintaining, or in some cases, improving the performance. Also the use of a personal computer reduces the processing costs, the use of the CT scanner workstation and reduces the time to evaluate a test. 2. MATERIALS AND METHODS This section describes the digital image acquisition through Computed Tomography (CT) and, afterwards, the lung segmentation, reconstruction and visualization method using the 3D region growing method. Prior to the image acquisition, the tomograph is set to an air density of −1000 HU (Hounsfield Units). The cali- ation is carried out within three months of the exams, as specified by the manufacturer [26]. Also before the acquisition, a tomographic cut with a water phantom with a known density is performed for analysis and para- meter control of the system [2]. The images are quan- tified in 16 bits and stored in a DICOM format (Digital Imaging and Communications in Medicine). To read these image files, which are in the DICOM format, the free DICOM toolkit offered by OFFIS was used. This library is compatible with the C++ program- ming language, as used in this work. Copyright © 2013 SciRes. OPEN ACCESS  P. P. Rebouças Filho et al. / J. Biomedical Science and Engineering 6 (2013) 1099-1108 1101 Figure 1(a) shows an example of an image obtained using CT with a multi-detector. This image is based on the principle that every internal structure of the human body possesses a determined radiologic density value, expressed in Hounsfield Units. The pulmonary densities adopted in this work are: hyper aerated (−1000 to −950 HU), normally aerated (−950 to −50 HU), poorly aerated (−500 to −100 HU), not aerated (−100 to 100 HU) and bone region (600 to 2000 HU) [27,28]. The distribution of these density bands is shown in Figure 1(b), in which the red color represents the hyper aerated region, the dark blue the normally aerated areas, the light blue represents the poorly aerated areas, and the black the non aerate areas, while the green represents the bone region and the white the non-classified area. 2.1. 3D Pulmonary Visualization and Reconstruction The graphic visualization system used in this work was the Open Graphics Library (OpenGL) Application Pro- gramming Interface (API). This API is an open and mul- tiplatform specification of a graphical and modeling rou- tine library, for the development of graphical computing applications, such as games and visualization systems. Two libraries that make up the OpenGL, GLU and GLUT, both open sources were used in this work. The OpenGL Utility Library contains various routines with low level OpenGL commands to execute tasks such as defining the matrix for the projection and orientation of the visualization, and render a surface. The OpenGL Uti- lity Toolkit (GLUT), which is a non-platform dependent toolkit and includes some graphic interface elements was also used here. Thus the routines for the 3D development system of this work were developed in C/C++ language, and called for the OpenGL library routines. The GLU library is applied to render objects, and configure their shape and illumination. The GLUT library, was also used to create windows and receive the user commands. This system is capable of being visualized in various platforms. In the case of specific CT images, the image spacing, (a) (b) Figure 1. CT lung image in the axial position: a) multi-detector CT scanner, and b) representation of the density ranges. or between planes, is one of the DICOM pattern proper- ties, defined by the slice thickness property. The pixel size is application specific when these images are 3D processed and in the DICOM pattern, and this is deter- mined by pixel spacing. Using these two parameters it is possible to reconstruct the CT on the same scale it was generated on. 2.2. Lung Segmentation Using 3D Region Growing Region growing is an image segmentation technique used to unite the regions of interest. This technique clusters the subgroups or groups of pixels in a deter- mined region. This is possible through the expansion of a given region initialized by a pixel, named seed. This expansion occurs by the aggregation of pixels to the re- gion through successive analysis iterations of the neigh- borhood of a given region [29]. In this technique, for a pixel to be grouped to the region, it is necessary for it to be in the region’s neigh- borhood and to obey pre-established criteria. This gene- rally bases itself on predefined parameters such as in- tensity of gray tone, and gray tone mean in the region, among others. An illustration of the technique’s application is pre- sented in Figure 2, in which the seed is shown in red, Figure 2(a), and the clustering ruling is that the gray tonality needs to be equal to the chosen seed’s tonality, resulting in the shown segmentation in Figure 2(b). The efficacy of this method depends directly on the choice of the seed and the grouping rule. If these choices are not adequate, the segmentation could present failures. The 3D expansion of this technique is possible assuming each element’s neighborhood is analyzed considering nearby planes. This technique is common for objects and medical image segmentations, principally in CT images. A set of images from such exams follows a predeter- mined sequence, obeying a standard pattern [13,16]. In these cases the smallest element is the voxel and the lo- cale begins being analyzed in the “x”, “y” and “z” axes. An example of this evolution is presented in Figure 3. (a) (b) Figure 2. Illustration of the region growing method: a) original image (seed represented by red color), and b) result of the segmentation (red color). Copyright © 2013 SciRes. OPEN ACCESS 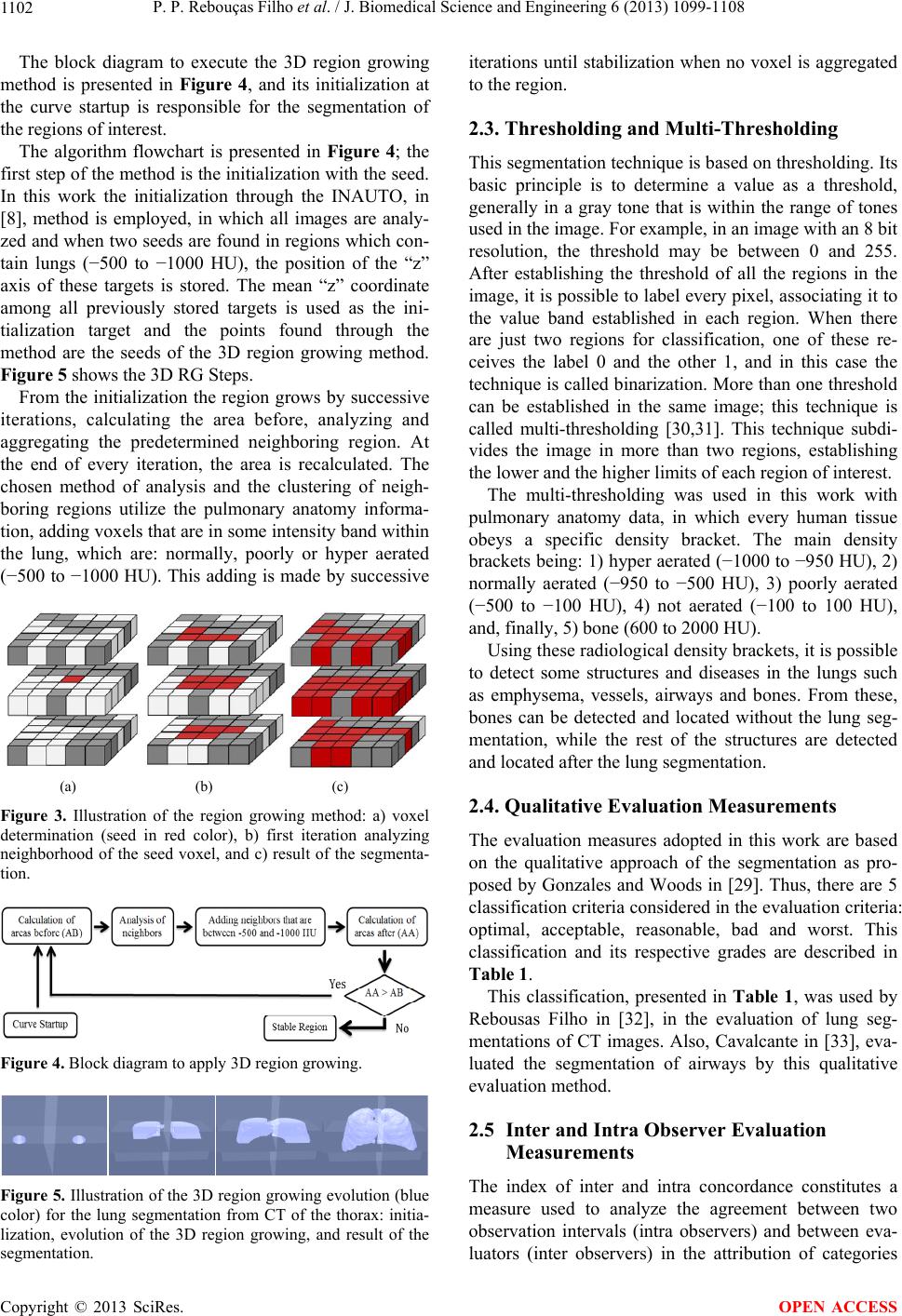 P. P. Rebouças Filho et al. / J. Biomedical Science and Engineering 6 (2013) 1099-1108 1102 The block diagram to execute the 3D region growing method is presented in Figure 4, and its initialization at the curve startup is responsible for the segmentation of the regions of interest. The algorithm flowchart is presented in Figure 4; the first step of the method is the initialization with the seed. In this work the initialization through the INAUTO, in [8], method is employed, in which all images are analy- zed and when two seeds are found in regions which con- tain lungs (−500 to −1000 HU), the position of the “z” axis of these targets is stored. The mean “z” coordinate among all previously stored targets is used as the ini- tialization target and the points found through the method are the seeds of the 3D region growing method. Figure 5 shows the 3D RG Steps. From the initialization the region grows by successive iterations, calculating the area before, analyzing and aggregating the predetermined neighboring region. At the end of every iteration, the area is recalculated. The chosen method of analysis and the clustering of neigh- boring regions utilize the pulmonary anatomy informa- tion, adding voxels that are in some intensity band within the lung, which are: normally, poorly or hyper aerated (−500 to −1000 HU). This adding is made by successive (a) (b) (c) Figure 3. Illustration of the region growing method: a) voxel determination (seed in red color), b) first iteration analyzing neighborhood of the seed voxel, and c) result of the segmenta- tion. Figure 4. Block diagram to apply 3D region growing. Figure 5. Illustration of the 3D region growing evolution (blue color) for the lung segmentation from CT of the thorax: initia- lization, evolution of the 3D region growing, and result of the segmentation. iterations until stabilization when no voxel is aggregated to the region. 2.3. Thresholding and Multi-Thresholding This segmentation technique is based on thresholding. Its basic principle is to determine a value as a threshold, generally in a gray tone that is within the range of tones used in the image. For example, in an image with an 8 bit resolution, the threshold may be between 0 and 255. After establishing the threshold of all the regions in the image, it is possible to label every pixel, associating it to the value band established in each region. When there are just two regions for classification, one of these re- ceives the label 0 and the other 1, and in this case the technique is called binarization. More than one threshold can be established in the same image; this technique is called multi-thresholding [30,31]. This technique subdi- vides the image in more than two regions, establishing the lower and the higher limits of each region of interest. The multi-thresholding was used in this work with pulmonary anatomy data, in which every human tissue obeys a specific density bracket. The main density brackets being: 1) hyper aerated (−1000 to −950 HU), 2) normally aerated (−950 to −500 HU), 3) poorly aerated (−500 to −100 HU), 4) not aerated (−100 to 100 HU), and, finally, 5) bone (600 to 2000 HU). Using these radiological density brackets, it is possible to detect some structures and diseases in the lungs such as emphysema, vessels, airways and bones. From these, bones can be detected and located without the lung seg- mentation, while the rest of the structures are detected and located after the lung segmentation. 2.4. Qualitative Evaluation Measurements The evaluation measures adopted in this work are based on the qualitative approach of the segmentation as pro- posed by Gonzales and Woods in [29]. Thus, there are 5 classification criteria considered in the evaluation criteria: optimal, acceptable, reasonable, bad and worst. This classification and its respective grades are described in Table 1. This classification, presented in Table 1, was used by Rebousas Filho in [32], in the evaluation of lung seg- mentations of CT images. Also, Cavalcante in [33], eva- luated the segmentation of airways by this qualitative evaluation method. 2.5 Inter and Intra Observer Evaluation Measurements The index of inter and intra concordance constitutes a measure used to analyze the agreement between two observation intervals (intra observers) and between eva- luators (inter observers) in the attribution of categories Copyright © 2013 SciRes. OPEN ACCESS  P. P. Rebouças Filho et al. / J. Biomedical Science and Engineering 6 (2013) 1099-1108 1103 for a determined classification, as shown in Table 1. The statistical concordance measurement used in this work is the Cohen’s Kappa index. This index is an intra and inter observer concordance measurement and meas- ures the degree of concordance between two observers and varies from 0 to 1, in what 1 means a greater con- cordance and 0 suggests the concordance is attributed randomly [34]. The Kappa index is calculated from a confusion matrix, an example of this is presented in Ta- ble 2. The Kappa analysis is a discrete multivariable used in a thematic precision evaluation and uses all the elements of the confusion matrix. The Kappa coefficient (K) is a measurement of the real concordance, indicated by the diagonal confusion matrix elements, minus the concor- dance by chance, indicated by the total line versus the column result, which does not include non-recognized inputs. This means it is a measure of how much the classification is in accordance with the reference data. Further details on the calculation of the kappa index can be seen in [35]. Although the Kappa coefficient is quite often used to evaluate the exactness of mapping, there are no minimal acceptable levels of this coefficient in a classification [35]. However, Table 3 presents classification perfor- mance levels, normally accepted by the scientific com- munity. Nowadays, the Kappa index is largely used to calcu- late the concordance level between observers in various applications such as, classification and image analysis of Synthetic Aperture Radar [36], classification of subcel- lular molecules [37], classification of breast lesions [38] and the evaluation of multimedia quality [39]. Table 1. Evaluative criteria for qualitative image segmentation. Note Classification Description 5 Optimal High quality: as good as might be desired 4 Acceptable Good quality with minor errors 3 Reasonable Intermediate quality with outliers 2 Bad Only a small portion of the object of interest 1 Worst Does not segment the object of interest Table 2. Illustration of a confusion matrix [35]. Classification 1 2 c Line total ni+ 1 X11 X 12 X 1c X 1+ 2 X21 X 22 X 2c X 2+ c Xc1 X c2 X cc X c+ Line total n+i X +1 X +2 X +c n 3. RESULTS AND DISCUSSION In this section, the method of medical image acquisition is presented first. Then, the segmentation using the 3D region growing is evaluated on CT lung images of the thorax of healthy patients and patients with COPD and fibrosis. Finally, some applications are presented to pro- ve the robustness, effectiveness and efficacy of the pro- posed method. 3.1. Acquisition of the Medical Images The images used to evaluate the algorithms were ac- quired from different CT scanners, some of the images were saved as samples of a complete CT exam, while other images were saved together with the complete exam. The models used to acquire the complete exams were the Toshiba Aquilion (TA), the GE Medical system LightSpeed 16 (GEMSL) and the Phillips Brilliance 10 (PB). All images were of 512 × 512 resolution and 16 bits. Table 4 gives the characteristics of these complete Table 3. Kappa’s index and classification performance [34]. Kappa index (K) Performance 0 No concordance 0 to 0.20 Slightly 0.21 to 0.40 Reasonable 0.41 to 0.60 Moderated 0.61 to 0.80 Substantial 0.81 to 1 Excellent (full concordance) Table 4. Description of full exam used to evaluate the 3D methods. N˚ of examN˚ of imagesSlice thickness Model of the tomography Disease 1 908 0.5 mm TA Normal 2 297 0.5 mm TA Normal 3 685 0.5 mm TA Normal 4 760 0.5 mm TA Normal 5 229 3.0 mm TA COPD 6 278 1.25 mm GEMSB Normal 7 267 1.25 mm GEMSB Fibrosis 8 239 1.25 mm GEMSB Normal 9 276 2.0 mm PB Fibrosis 10 296 2.0 mm PB Normal 11 597 1.0 mm PB COPD Copyright © 2013 SciRes. OPEN ACCESS 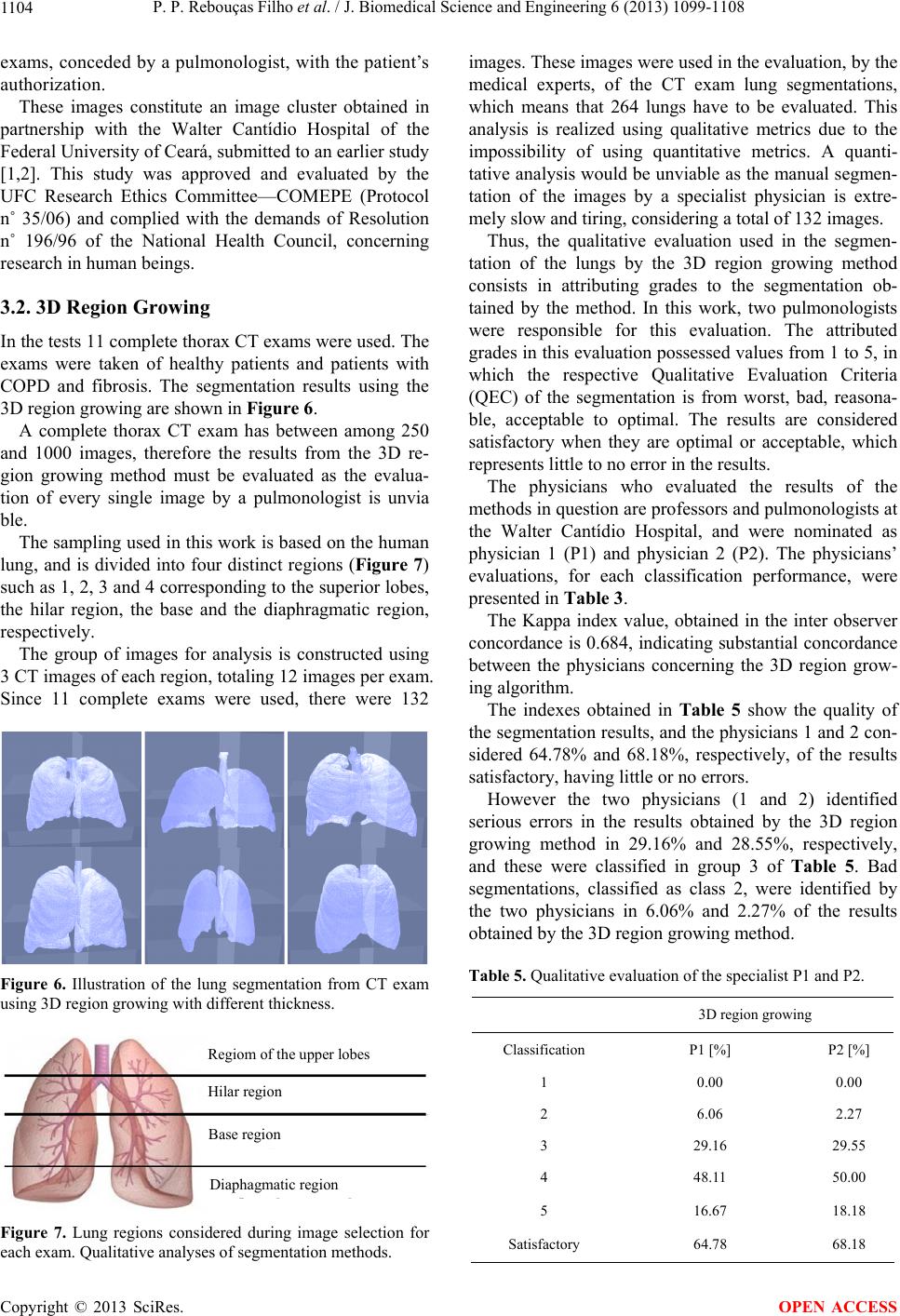 P. P. Rebouças Filho et al. / J. Biomedical Science and Engineering 6 (2013) 1099-1108 1104 exams, conceded by a pulmonologist, with the patient’s authorization. These images constitute an image cluster obtained in partnership with the Walter Cantídio Hospital of the Federal University of Ceará, submitted to an earlier study [1,2]. This study was approved and evaluated by the UFC Research Ethics Committee—COMEPE (Protocol n˚ 35/06) and complied with the demands of Resolution n˚ 196/96 of the National Health Council, concerning research in human beings. 3.2. 3D Region Growing In the tests 11 complete thorax CT exams were used. The exams were taken of healthy patients and patients with COPD and fibrosis. The segmentation results using the 3D region growing are shown in Figure 6. A complete thorax CT exam has between among 250 and 1000 images, therefore the results from the 3D re- gion growing method must be evaluated as the evalua- tion of every single image by a pulmonologist is unvia ble. The sampling used in this work is based on the human lung, and is divided into four distinct regions (Figure 7) such as 1, 2, 3 and 4 corresponding to the superior lobes, the hilar region, the base and the diaphragmatic region, respectively. The group of images for analysis is constructed using 3 CT images of each region, totaling 12 images per exam. Since 11 complete exams were used, there were 132 Figure 6. Illustration of the lung segmentation from CT exam using 3D region growing with different thickness. Regiom of the upper lobes Hilar region Base region Diaphagmatic region Figure 7. Lung regions considered during image selection for each exam. Qualitative analyses of segmentation methods. images. These images were used in the evaluation, by the medical experts, of the CT exam lung segmentations, which means that 264 lungs have to be evaluated. This analysis is realized using qualitative metrics due to the impossibility of using quantitative metrics. A quanti- tative analysis would be unviable as the manual segmen- tation of the images by a specialist physician is extre- mely slow and tiring, considering a total of 132 images. Thus, the qualitative evaluation used in the segmen- tation of the lungs by the 3D region growing method consists in attributing grades to the segmentation ob- tained by the method. In this work, two pulmonologists were responsible for this evaluation. The attributed grades in this evaluation possessed values from 1 to 5, in which the respective Qualitative Evaluation Criteria (QEC) of the segmentation is from worst, bad, reasona- ble, acceptable to optimal. The results are considered satisfactory when they are optimal or acceptable, which represents little to no error in the results. The physicians who evaluated the results of the methods in question are professors and pulmonologists at the Walter Cantídio Hospital, and were nominated as physician 1 (P1) and physician 2 (P2). The physicians’ evaluations, for each classification performance, were presented in Table 3. The Kappa index value, obtained in the inter observer concordance is 0.684, indicating substantial concordance between the physicians concerning the 3D region grow- ing algorithm. The indexes obtained in Table 5 show the quality of the segmentation results, and the physicians 1 and 2 con- sidered 64.78% and 68.18%, respectively, of the results satisfactory, having little or no errors. However the two physicians (1 and 2) identified serious errors in the results obtained by the 3D region growing method in 29.16% and 28.55%, respectively, and these were classified in group 3 of Table 5. Bad segmentations, classified as class 2, were identified by the two physicians in 6.06% and 2.27% of the results obtained by the 3D region growing method. Table 5. Qualitative evaluation of the specialist P1 and P2. 3D region growing Classification P1 [%] P2 [%] 1 0.00 0.00 2 6.06 2.27 3 29.16 29.55 4 48.11 50.00 5 16.67 18.18 Satisfactory 64.78 68.18 Copyright © 2013 SciRes. OPEN ACCESS 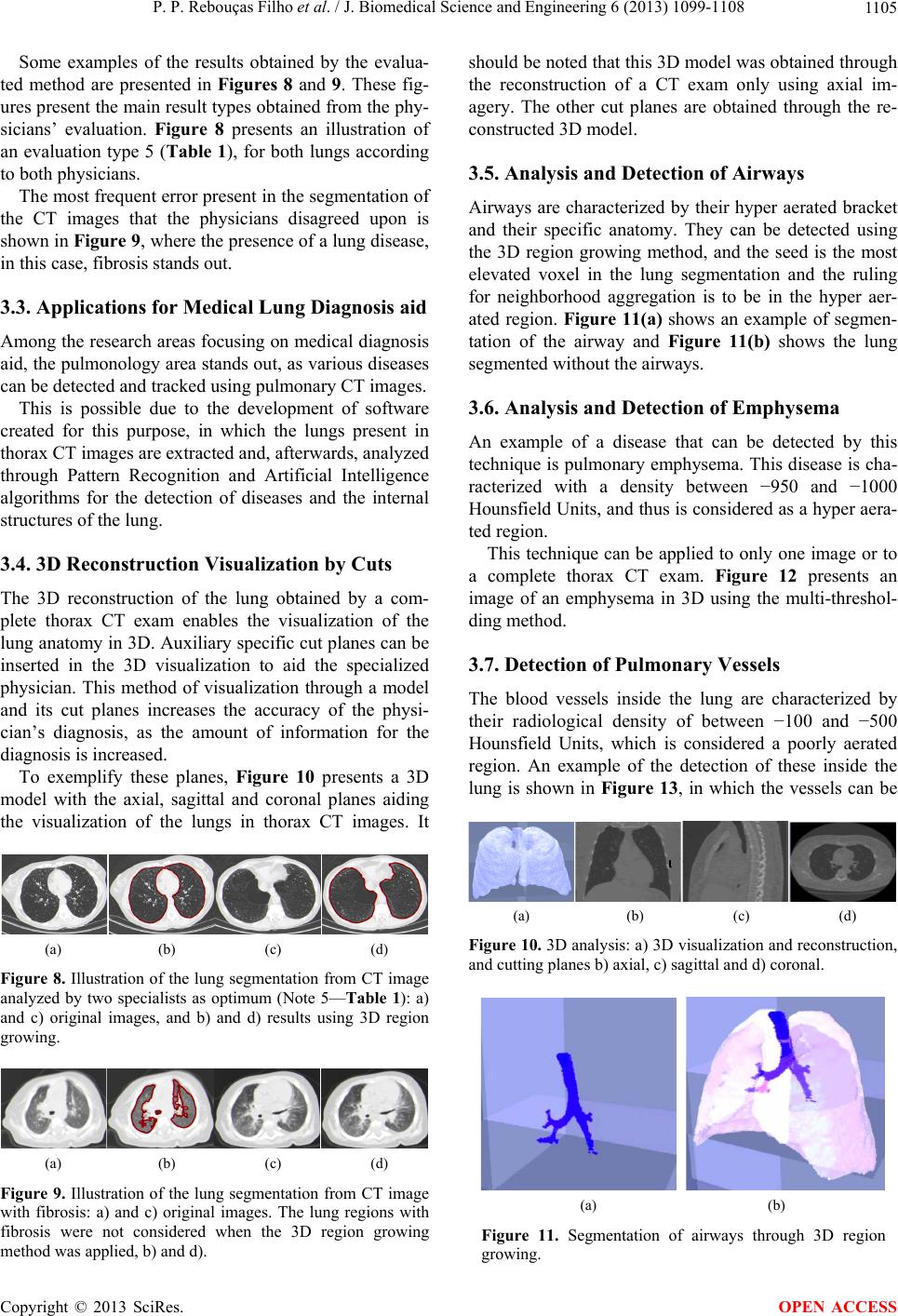 P. P. Rebouças Filho et al. / J. Biomedical Science and Engineering 6 (2013) 1099-1108 1105 Some examples of the results obtained by the evalua- ted method are presented in Figures 8 and 9. These fig- ures present the main result types obtained from the phy- sicians’ evaluation. Figure 8 presents an illustration of an evaluation type 5 (Table 1 ), for both lungs according to both physicians. The most frequent error present in the segmentation of the CT images that the physicians disagreed upon is shown in Figure 9, where the presence of a lung disease, in this case, fibrosis stands out. 3.3. Applications for Medical Lung Diagnosis aid Among the research areas focusing on medical diagnosis aid, the pulmonology area stands out, as various diseases can be detected and tracked using pulmonary CT images. This is possible due to the development of software created for this purpose, in which the lungs present in thorax CT images are extracted and, afterwards, analyzed through Pattern Recognition and Artificial Intelligence algorithms for the detection of diseases and the internal structures of the lung. 3.4. 3D Reconstruction Visualization by Cuts The 3D reconstruction of the lung obtained by a com- plete thorax CT exam enables the visualization of the lung anatomy in 3D. Auxiliary specific cut planes can be inserted in the 3D visualization to aid the specialized physician. This method of visualization through a model and its cut planes increases the accuracy of the physi- cian’s diagnosis, as the amount of information for the diagnosis is increased. To exemplify these planes, Figure 10 presents a 3D model with the axial, sagittal and coronal planes aiding the visualization of the lungs in thorax CT images. It (a) (b) (c) (d) Figure 8. Illustration of the lung segmentation from CT image analyzed by two specialists as optimum (Note 5—Table 1): a) and c) original images, and b) and d) results using 3D region growing. (a) (b) (c) (d) Figure 9. Illustration of the lung segmentation from CT image with fibrosis: a) and c) original images. The lung regions with fibrosis were not considered when the 3D region growing method was applied, b) and d). should be noted that this 3D model was obtained through the reconstruction of a CT exam only using axial im- agery. The other cut planes are obtained through the re- constructed 3D model. 3.5. Analysis and Detection of Airways Airways are characterized by their hyper aerated bracket and their specific anatomy. They can be detected using the 3D region growing method, and the seed is the most elevated voxel in the lung segmentation and the ruling for neighborhood aggregation is to be in the hyper aer- ated region. Figure 11(a) shows an example of segmen- tation of the airway and Figure 11(b) shows the lung segmented without the airways. 3.6. Analysis and Detection of Emphysema An example of a disease that can be detected by this technique is pulmonary emphysema. This disease is cha- racterized with a density between −950 and −1000 Hounsfield Units, and thus is considered as a hyper aera- ted region. This technique can be applied to only one image or to a complete thorax CT exam. Figure 12 presents an image of an emphysema in 3D using the multi-threshol- ding method. 3.7. Detection of Pulmonary Vessels The blood vessels inside the lung are characterized by their radiological density of between −100 and −500 Hounsfield Units, which is considered a poorly aerated region. An example of the detection of these inside the lung is shown in Figure 13, in which the vessels can be (a) (b) (c) (d) Figure 10. 3D analysis: a) 3D visualization and reconstruction, and cutting planes b) axial, c) sagittal and d) coronal. (a) (b) Figure 11. Segmentation of airways through 3D region growing. Copyright © 2013 SciRes. OPEN ACCESS 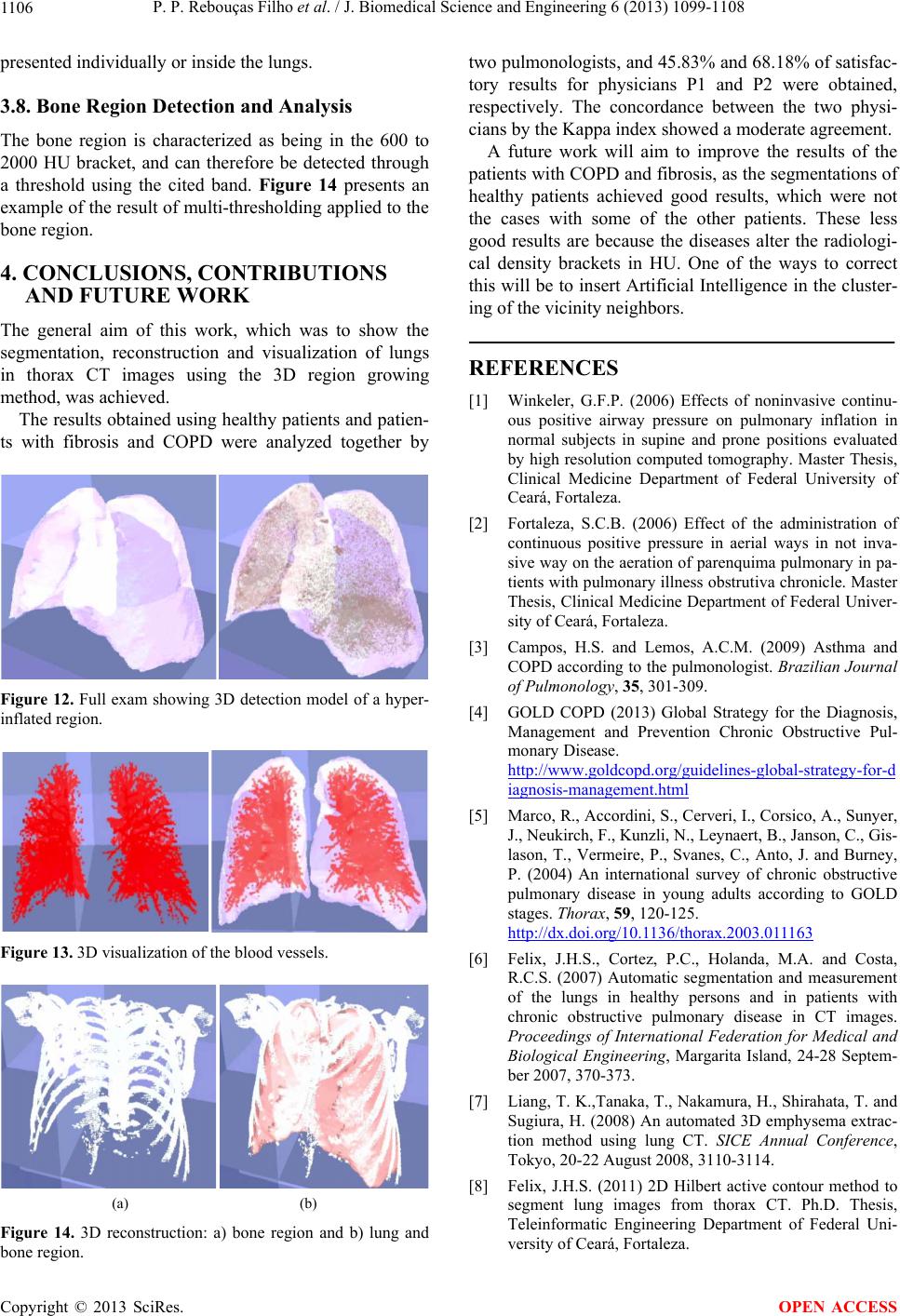 P. P. Rebouças Filho et al. / J. Biomedical Science and Engineering 6 (2013) 1099-1108 1106 presented individually or inside the lungs. 3.8. Bone Region Detection and Analysis The bone region is characterized as being in the 600 to 2000 HU bracket, and can therefore be detected through a threshold using the cited band. Figure 14 presents an example of the result of multi-thresholding applied to the bone region. 4. CONCLUSIONS, CONTRIBUTIONS AND FUTURE WORK The general aim of this work, which was to show the segmentation, reconstruction and visualization of lungs in thorax CT images using the 3D region growing method, was achieved. The results obtained using healthy patients and patien- ts with fibrosis and COPD were analyzed together by Figure 12. Full exam showing 3D detection model of a hyper- inflated region. Figure 13. 3D visualization of the blood vessels. (a) (b) Figure 14. 3D reconstruction: a) bone region and b) lung and bone region. two pulmonologists, and 45.83% and 68.18% of satisfac- tory results for physicians P1 and P2 were obtained, respectively. The concordance between the two physi- cians by the Kappa index showed a moderate agreement. A future work will aim to improve the results of the patients with COPD and fibrosis, as the segmentations of healthy patients achieved good results, which were not the cases with some of the other patients. These less good results are because the diseases alter the radiologi- cal density brackets in HU. One of the ways to correct this will be to insert Artificial Intelligence in the cluster- ing of the vicinity neighbors. REFERENCES [1] Winkeler, G.F.P. (2006) Effects of noninvasive continu- ous positive airway pressure on pulmonary inflation in normal subjects in supine and prone positions evaluated by high resolution computed tomography. Master Thesis, Clinical Medicine Department of Federal University of Ceará, Fortaleza. [2] Fortaleza, S.C.B. (2006) Effect of the administration of continuous positive pressure in aerial ways in not inva- sive way on the aeration of parenquima pulmonary in pa- tients with pulmonary illness obstrutiva chronicle. Master Thesis, Clinical Medicine Department of Federal Univer- sity of Ceará, Fortaleza. [3] Campos, H.S. and Lemos, A.C.M. (2009) Asthma and COPD according to the pulmonologist. Brazilian Journal of Pulmonology, 35, 301-309. [4] GOLD COPD (2013) Global Strategy for the Diagnosis, Management and Prevention Chronic Obstructive Pul- monary Disease. http://www.goldcopd.org/guidelines-global-strategy-for-d iagnosis-management.html [5] Marco, R., Accordini, S., Cerveri, I., Corsico, A., Sunyer, J., Neukirch, F., Kunzli, N., Leynaert, B., Janson, C., Gis- lason, T., Vermeire, P., Svanes, C., Anto, J. and Burney, P. (2004) An international survey of chronic obstructive pulmonary disease in young adults according to GOLD stages. Thorax, 59, 120-125. http://dx.doi.org/10.1136/thorax.2003.011163 [6] Felix, J.H.S., Cortez, P.C., Holanda, M.A. and Costa, R.C.S. (2007) Automatic segmentation and measurement of the lungs in healthy persons and in patients with chronic obstructive pulmonary disease in CT images. Proceedings of International Federation for Medical and Biological Engineering, Margarita Island, 24-28 Septem- ber 2007, 370-373. [7] Liang, T. K.,Tanaka, T., Nakamura, H., Shirahata, T. and Sugiura, H. (2008) An automated 3D emphysema extrac- tion method using lung CT. SICE Annual Conference, Tokyo, 20-22 August 2008, 3110-3114. [8] Felix, J.H.S. (2011) 2D Hilbert active contour method to segment lung images from thorax CT. Ph.D. Thesis, Teleinformatic Engineering Department of Federal Uni- versity of Ceará, Fortaleza. Copyright © 2013 SciRes. OPEN ACCESS  P. P. Rebouças Filho et al. / J. Biomedical Science and Engineering 6 (2013) 1099-1108 1107 [9] Sluimer, I., Prokop, M. and van Ginneken, B. (2005) Toward automated segmentation of the pathological lung in CT. IEEE Transactions on Medical Imaging, 24, 1025- 1038. http://dx.doi.org/10.1109/TMI.2005.851757 [10] Felix, J.H.S., Cortez, P.C., Holanda, M.A., Albuquerque, V.H.C., Colaço, D.F. and Alexandria, A.R. (2007) Lung and chest wall structures segmentation in CT images. I ECCOMAS Thematic Conference on Computational Vi- sion and Medical Image Processing (VipIMAGE’07), O’Porto, 17-19 October 2007. [11] Hu, S., Hoffman, E.A. and Reinhardt, J.M. (2001) Auto- matic lung segmentation for accurate quantitation of vo- lumetric X-ray CT images. IEEE Transactions on Medi- cal Imaging, 20, 490-498. http://dx.doi.org/10.1109/42.929615 [12] Silva, A.C., Carvalho, P.C.P., Nunes, R.A. and Gattass, M. (2006) Segmentation and reconstruction of the pulmo- nary parenchyma. VI Workshop on Medical Informatics (VI Workshop de Informática Médica), Vila Velha, 29-30 May 2006, 73-82. [13] Born, S., Iwamaru, D., Pfeifle, M. and Bartz, D. (2009) Three-step segmentation of the lower airways with ad- vanced leakage-control. Second International Workshop on Pulmonary Image Analysis: Extraction of airways from CT (EXACT’09), Pasadena, 11-13 July 2009, 239- 250. [14] Irving, B., Taylor, P. and Todd-Pokropek, A. (2009) 3D segmentation of the airway tree using a morphology based method. Second International Workshop on Pul- monary Image Analysis: Extraction of airways from CT (EXACT’09), Pasadena, 11-13 July, 297-307. [15] Tschirren, J., Yavarna, T. and Reinhardt, J. (2009) Air- way segmentation framework for clinical environments. The Second International Workshop on Pulmonary Image Analysis, London, 20 September 2009, 227-238. [16] Matsuoka, S., Yamashiro, T., Washko, G. R., Kurihara, Y., Nakajima, Y. and Hatabu, H. (2010) Quantitative CT assessment of chronic obstructive pulmonary disease. Radiographics, 30, 55-66. http://dx.doi.org/10.1148/rg.301095110 [17] Chen, H., Zhang, J., Xu, Y., Chen, B. and Zhang, K. (2012) Performance comparison of artificial neural net- work and logistic regression model for differentiating lung nodules on CT scans. Expert Systems with Applica- tions, 39, 11503-11509. http://dx.doi.org/10.1016/j.eswa.2012.04.001 [18] Er, O.,Yumusak, N. and Temurtas, F. (2010) Chest disea- ses diagnosis using artificial neural networks. Expert Systems with Applications, 37, 7648-7655. http://dx.doi.org/10.1016/j.eswa.2010.04.078 [19] Er, O.,Yumusak, N. and Temurtas, F. (2012) Diagnosis of chest diseases using artificial immune system. Expert Systems with Applications, 39, 1862-1868. http://dx.doi.org/10.1016/j.eswa.2011.08.064 [20] Fernández, J.M.F., López, E.J.H., Llamas, F.S., Calvillo, A.R., Galeana, P.A.C., Pacheco, G.L., Palomar., M.G.G., Femat, R. and Velázquez, M.M. (2012) Development of an optimized multi-biomarker panel for the detection of lung cancer based on principal component analysis and artificial neural network modeling. Expert Systems with Applications, 39, 10851-10856. http://dx.doi.org/10.1016/j.eswa.2012.03.008 [21] Annangi, P., Thiruvenkadam, S., Raja, A., Xu, H., Sun., X. and Mao, L. (2010) A region based active contour method for x-ray lung segmentation using prior shape and low level features. Proceedings of 7th IEEE International Symposium on Biomedical Imaging (ISBI), Rotterdam, 14-17 April 2010. [22] Wu, Y., Wang, Y. and Jia, Y. (2013) Adaptive diffusion flow active contours for image segmentation. Computer Vision and Image Understanding, 117, 1421-1435. http://dx.doi.org/10.1016/j.cviu.2013.05.003 [23] Keshani. M., Azimifar, Z., Tajeripour, F. and Boostani, R. (2013) Lung nodule segmentation and recognition using SVM classifier and active contour modeling: A complete intelligent system. Computers in Biology and Medicine, 43, 287-300. http://dx.doi.org/10.1016/j.compbiomed.2012.12.004 [24] Tan, Y., Schwartz, L. H. and Zhao, B. (2013) Segmenta- tion of lung lesions on CT scans using watershed, active contours, and Markov random field. Medical Physics, 40, 043502. http://dx.doi.org/10.1118/1.4793409 [25] Liu, L. and Bovik, A.C. (2012) Active contours with neighborhood-extending and noise-smoothing gradient vector flow external force. EURASIP Journal on Image and Video Processing, 9, 2-6. [26] Toshiba (1997) Operation manual of system: X-ray computerized tomography for all body: Calibration data acquisition. São Paulo. [27] Gevenois, P.A. and Yernault, J.C. (1995) Can computed tomography quantify pulmonary emphysema? European Respiratory Journal, 5, 843-848. [28] Vieira, S.R., Puybasset, L., Richecoeur, J., Lu, Q., Cluzel, P., Gusman, P.B., Coriat, P. and Rouby, J.J. (1998) A lung computed tomographic assessment of positive end- expiratory pressure-induced lung over distension. Ameri- can Journal of Respiratory and Critical Care Medicine, 158, 1571-1577. http://dx.doi.org/10.1164/ajrccm.158.5.9802101 [29] Gonzalez, R. C. and Woods, R. (2010) Digital image pro- cessing. 3rd Edition, Pearson Prentice Hall, New Jersey. [30] Papamarkos, N., Strouthopoulos, C. and Andreadis, I. (2000) Multithresholding of color and grey-level images through a neural network technique. Image and Vision Computing, 18, 213-222. http://dx.doi.org/10.1016/S0262-8856(99)00015-3 [31] Zahara, E., Fan, S.K.S. and Tsai, D.M. (2005) Optimal multi-thresholding using a hybrid optimization approach. Pattern Recognition Letters, 26, 1082-1095. http://dx.doi.org/10.1016/j.patrec.2004.10.003 [32] Rebouças Filho, P.P., Cortez, P.C. and Holanda, M.A. (2011) Active contour modes Crisp: New technique for segmentation the lungs in CT images. Brazilian Journal of Biomedical Engineering, 27, 259-272. [33] Cavalcante, T.S. (2010) Technique for automatic seg- mentation of the airway from thorax CT images. Master Thesis, Teleinformatic Engineering Department of Fed- Copyright © 2013 SciRes. OPEN ACCESS  P. P. Rebouças Filho et al. / J. Biomedical Science and Engineering 6 (2013) 1099-1108 Copyright © 2013 SciRes. 1108 OPEN ACCESS eral University of Ceará, Fortaleza. [34] Congalton, R.G. and Green, K. (2008) Assessing the accuracy of remotely sensed data: Principles and prac- tices. 2nd Edition, CRC Press-Taylor & Francis Group. http://dx.doi.org/10.1201/9781420055139 [35] Figueiredo, G.C. and Vieira, C.A.O. (2007) Study of the behavior of Overall Accuracy, Kappa and Tau indexes, commonly used to evaluate the classification of remote sensing images. Proceedings of XIII Brazilian Symposium of Remote Sensing, Florianópolis, 21-26 April, 5755- 5762. [36] Deng-hui, Z., Han-Kui, Z., Bin, X., Zhao-quan, H., Le, Y. and Yun-yun, C. (2010) Analysis of image fusion and classification for high resolution SAR data on-line. 2nd International Conference on Education Technology and Computer (ICETC’10), 22-24 June, Shanghai, V1-267- V1-271. [37] Can, A., Bello, M.O. and Gerdes, M.J. (2010) Quantifica- tion of subcellular molecules in tissue microarray. 20th International Conference on Pattern Recognition, Istan- bul, 23-26 August 2010, 2548-2551. http://dx.doi.org/10.1109/ICPR.2010.624 [38] Dundar, M.M., Badve, S., Bilgin, G., Raykar, V., Jain, R., Sertel, O. and Gurcan, M.N. (2011) Computerized classi- fication of intraductal breast lesions using histopatholo- gical images. IEEE Transactions on Biomedical Engi- neering, 58, 1977-1984. http://dx.doi.org/10.1109/TBME.2011.2110648 [39] Engelke, U., Pitrey, Y. and Callet, P.L. (2011) Towards an inter-observer analysis framework for multimedia qua- lity assessment. Third International Workshop on Quality of Multimedia Experience (QoMEX), Mechelen, 7-9 Sep- tember 2011, 183-188.
|