Paper Menu >>
Journal Menu >>
 Engineering, 2012, 5, 153-158 doi:10.4236/eng.2012.410B040 Published Online October 2012 (http://www.SciRP.org/journal/eng) Copyright © 2012 SciRes. ENG The omp2 Gene of HPS-type Bacteria Cloning and Sequence Analy si s Isolates from Sichuan Province* Lirui Li1, Zexiao Yang1, Yin Wang1#, Qiu Jin1, Xulong Wu1, Ro ngcha ng Zhang 2 1Sichuan Agricultural University, Sichuan Laboratory of Animal Disease and Human Health, SLADHH, Ya'an ,China 2Tibet Changdu animal he al th and phytosanitary supervision, TCAHPS, Changdu, China Email: 704029604@qq.com, #576122816@qq.com, Received 2012 ABSTRACT In order to compare the homology and antigen of Haemophilus parasuis (HPS)4 outer membrane protein P2(omp2), we design a test with specific primers, using PCR amplification of isolates of Haemophilus parasuis from Sichuan Province(HP Sch2010), ompP2 gene will be cloned into the pGM-T vector, and transformed into E. coli DH5α. Identified by PCR and sequencing and analysis, the sequencing results showed that the published 4 HPS SW124 stra ins omp2 gene (1077bp), compared with the amplified 1086bp pur- pose fragment(containing omp2 genes), is relatively stable, with the nucleotide homology level 97% and amino acid homology level of 92.5 %. The variabl e region s are mainly con centrated in th e three base sequences: 40-65,110-156,180-202. Keywords: Haemophilus parasuis, virulence genes, omp2 1. Introduction Haemophilus parasuis is a principal member of the genus by the Pasteurella Division Haemophilus, considered as the main pa- thogens of the Glaser's disease [1]. Nowadays, HPS disease became one of the major bacterial diseases affecting the swine indust ry. At the sa me ti me it increase t he inci dence of infection in pathogenic porcine reproductive and respiratory syndrome virus infection caused by immunosuppression, result in major economic losses, being a serious threat to the healthy develop- ment of the pig industry worldwide. Because of the numerous serotypes of the disease, and different serotypes cannot provide effective cross-protection [2], it makes the effective prevention of Haemophilus parasuis disease very difficult. The study showed that Omp2 HPS outer membrane is the most abundant protein [3], and the immune electron microscopy technique showed that a panel of monoclonal antibodies directed against the exposed area of omp2 particles with the complement can be highly efficient immune [4]. Therefore, our laboratory cloned the Sichuan isolates of the HPS omp2 gene, and made a com- parison and analysis of its sequence, so that we can provide a close reference of tracking for Haemophilus parasuis clinical gene mutation s and develop ing new and efficient vacci nes. 2. Materials and Methods 2.1. Enzymes, Reagents and Strain Tap polymerase and other reagents purchased were bought in Bowande Biological,limited,the pGM-19T kit and DL5000 Maker, DNA recovery reagents, plasmid extraction kit were purchased from Dalian TaKaRa Company. Haemophilus para- suis clinical isolates Sch2010 (serotype 4) from infected pig in Sichuan. 2.2. Primer Design and Synthesis According to the omp2 gene sequence of SH0165 strains of Haemophilus parasuis in GenBank (access number: CP001321.1), we used biological software Oligo 6.0 and Pri- mer 5.0 to design omp2 full genome sequence of primer 1, synthesized by Shanghai Shenggong Biological Engineering Co., Ltd. The primer sequences were as following: upstream: 5′- ATGGGAAGGTAATGGC -3′, downstream :5′- GTACTCGCTAAAGCAG -3′. 2.3. Amplification of omp2 Gene by PCR We incubated TSB broth under 37℃ for 24 hours to collect bacteria as a template for PCR amplification, according to the conventional cloning methods of omp2 gene. PCR reaction system (50μL): PrimeSTAR (HS (Premix) 25μL, bacteria 2.0μL, upstream and downstream primer (20 pmol) 1.5μL, sterile ultrapure water 20μL. PCR reaction conditions: 94℃ initial denaturation for 5 mi- nutes; 94℃ for 45s, 59℃ for 1 minute, 72℃ for 45s, 30 cycles; final extension at 72℃ for 10 minutes. Observe the results by taking 5μL amplified product, using 1% agarose gel electrophoresis. 2.4. Cloning of the omp2 Gene We cut the t arg et fr agmen t fro m t he agaro se gel u nd er UV l ight, purified DNA under the Dalian TaKaRa DNA purification kit instructions. Purified products were cloned into the pGM-T *This study was supported by Grants from Program for Changjiang Scholars and Innovative Research Team in University(PCSIRT)IRT 0848, and “211-Projects” Shuangzhi Plan in Sichuan Agricultural University, Li Lirui,, Zhang Rongchang and Wang Yin , all should be considered as first authors. #Corresponding author. 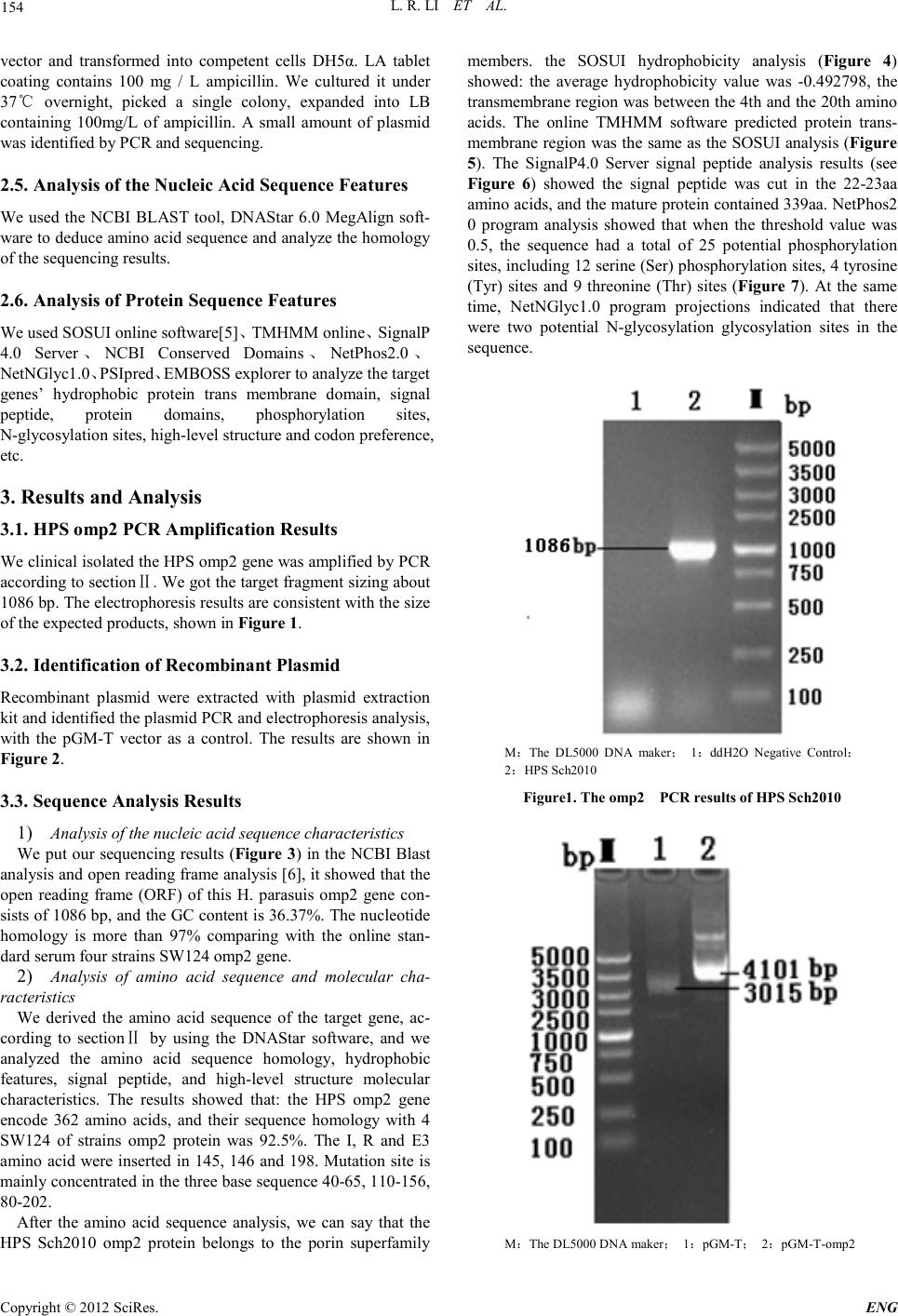 L. R. LI ET AL. Copyright © 2012 SciRes. ENG 154 vector and transformed into competent cells DH5α. LA tablet coating contains 100 mg / L ampicillin. We cultured it under 37℃ overnight, picked a single colony, expanded into LB containing 100mg/L of ampicillin. A small amount of plasmid was identified by PCR and sequenci ng. 2.5. Analysis of the Nucleic Acid Sequence Features We used the NCBI BLAST tool, DNAStar 6.0 MegAlign soft- ware to d educe amin o acid sequence an d anal yze the homology of the sequencing results. 2.6. Analysis of Protein Sequence Features We used SOSUI online software[5]、TMHMM online、SignalP 4.0 Server、NCBI Conserved Domains、NetPhos2.0 、 NetNGlyc1.0、PSIp red、EMBOSS explorer to analyze the target genes’ hydrophobic protein trans membrane domain, signal peptide, protein domains, phosphorylation sites, N-glycosylation sites, high-level structur e and cod on preference, etc. 3. Results and Analysis 3.1. HPS omp2 PCR A mplif icat ion Results We clinical isolated the HPS omp2 gene was amplified by PCR accordi ng to sectionⅡ. We got the t arget fragmen t sizing about 1086 bp. The electrophoresis results are consistent with the size of the expected products, shown in Figure 1. 3.2. Identificat ion of Recombinant Plasmid Recombinant plasmid wer e extracted with plasmid extraction kit and identified the plasmid PCR and electropho resi s anal ysis, with the pGM-T vector as a control. The results are shown in Figure 2 . 3.3. Sequence Analy sis Resul ts 1) Analysis of th e nucleic a cid sequence chara cteristics We put our sequencing results (Figure 3) in the NCBI Blast analysis and open reading frame analysis [6], it showed that the open reading frame (ORF) of this H. parasuis omp2 gene con- sists of 1086 bp, and the GC content is 36.37%. The nucleotide homology is more than 97% comparing with the online stan- dard serum four strains SW124 omp2 gene. 2) Analysis of amino acid sequence and molecular cha- racteristics We derived the amino acid sequence of the target gene, ac- cording to sectionⅡ by using the DNAStar software, and we analyzed the amino acid sequence homology, hydrophobic features, signal peptide, and high-level structure molecular characteristics. The results showed that: the HPS omp2 gene encode 362 amino acids, and their sequence homology with 4 SW124 of strains omp2 protein was 92.5%. The I, R and E3 amino acid were inserted in 145, 146 and 198. Mutation site is mainly con centrated in the three base seq uence 40 -65, 110-156, 80-202. After the amino acid sequence analysis, we can say that the HPS Sch2010 omp2 protein belongs to the porin superfamily members. the SOSUI hydrophobicity analysis (Figure 4) showed: the average hydrophobicity value was -0.492798, the transmembrane region was bet ween the 4th and the 20th amino acids. The online TMHMM software predicted protein trans- membrane region was the sa me as the SOSUI analysis (Figure 5). The SignalP4.0 Server signal peptide analysis results (see Figure 6) showed the signal peptide was cut in the 22-23aa amino acids, and the matu re protein con tained 339 aa. N etPho s2 0 program analysis showed that when the threshold value was 0.5, the sequence had a total of 25 potential phosphorylation sites, including 12 serine (Ser) phosphorylation sites, 4 tyrosine (Tyr) sites and 9 threonine (Thr) sites (Figure 7). At the same time, NetNGlyc1.0 program projections indicated that there were two potential N-glycosylation glycosylation sites in the sequence. M:The DL5000 DNA maker; 1:ddH2O Negative Control; 2:HPS Sch2010 Figur e1. The omp2 PCR results of HPS Sc h2 010 M:The DL5000 DNA maker; 1:pGM-T; 2:pGM-T-omp2  L. R. LI ET AL. Copyright © 2012 SciRes. E NG 155 Figure 2. Th e electophores is id ent i f i cat i on results. Figure 3. The underscore represents the primer position; the red font repre sents Sc h2010 HPS omp2 target gene bands. PSIpred procedures prediction results showed that the sec- ondary structure composition in th i s HPS Sch2010 omp2 amino acid sequence, the proportion of α-helix (Alpha-the Helix) was 28.45%, β-sheet (Beta-Strand) was 29.01%, β-turn was 23.76%, and random coil was only 9.94%. 3) Codo n pr e f e r e nc e analys i s Using the CHIPS program, the HPS Sch2010 omp2 gene analysis results showed that its Nc value was 35.731, suggest- ing that some of its frequency of codon usage has certain dif- ferences; after CUSPS analysis, we found the gene sequence encoding the same amino acid with varying degrees of differ- ence in codon usage frequency, such as those 4 Ala codon pre- fer GCA and GCT, 4 Pro codon prefer CCA, 2 Gln codon use only the CAA, Gl y pr efer GGT , and K prefer AAA. 15 codons in the sequence, including the TGC, TGT, GGG, ATA CTG, CCC and other appeared with zero frequency; TAA only ap- peared at the stop codon. 4. Discussion In recent years, HPS has grown widely popular in China. It is an important pathogen of infected pigs, with numerous of dif- ferent serotypes. Its cross-protective immunity is no good, which causes serious economic losses to the pig industry all over the world. Among the Haemophilus parasuis virulence factors, outer membrane protein (OMP) was the most re- searched one. Out er me mbr ane protein is one of the main viru- lence factors of HPS[7-10] .Experimentally infected pigs with HPS produce omp antibody[11] , while Tadjine M[ 12] prepared HPS monoclonal antibody react with 15 serotypes of HPS OMP (major outer membrane protein) and its wild-types, which indi- cate the outer membrane protein may have stronger immune protective effect. M eantime, Mcvicker thought that the HPS virulence strains and non-virulent strain[13] can be distin- guis hed by omp2. This experi ment u sed a pair of self-designed specific p ri mers, which were successfully amplified by PCR to be used in clon- ing analysis of the Sch omp2 . The gene sequence analysis showed that the HPS omp2 gene fragment had the highest ho- mology of 99%, with the serum type 2 SW140 omp2 gene in GenBank (access number FJ416461.1), and their amino acid homology was 97.5%; it also had the gene homology of 97% with the serum type 4 SW124 omp2 in GenBank (access num- ber for FJ685761.1), and amino acid homology of 92.5%. This showed that the omp2 virulence gene of Sch HPS was relatively stable but there we re also certain mutations. It had higher ho- mology with standard serotype 2 than with the standard sero- type 4. And after the secondary structure prediction and amino acid mutation site analysis we found that section 145, 146 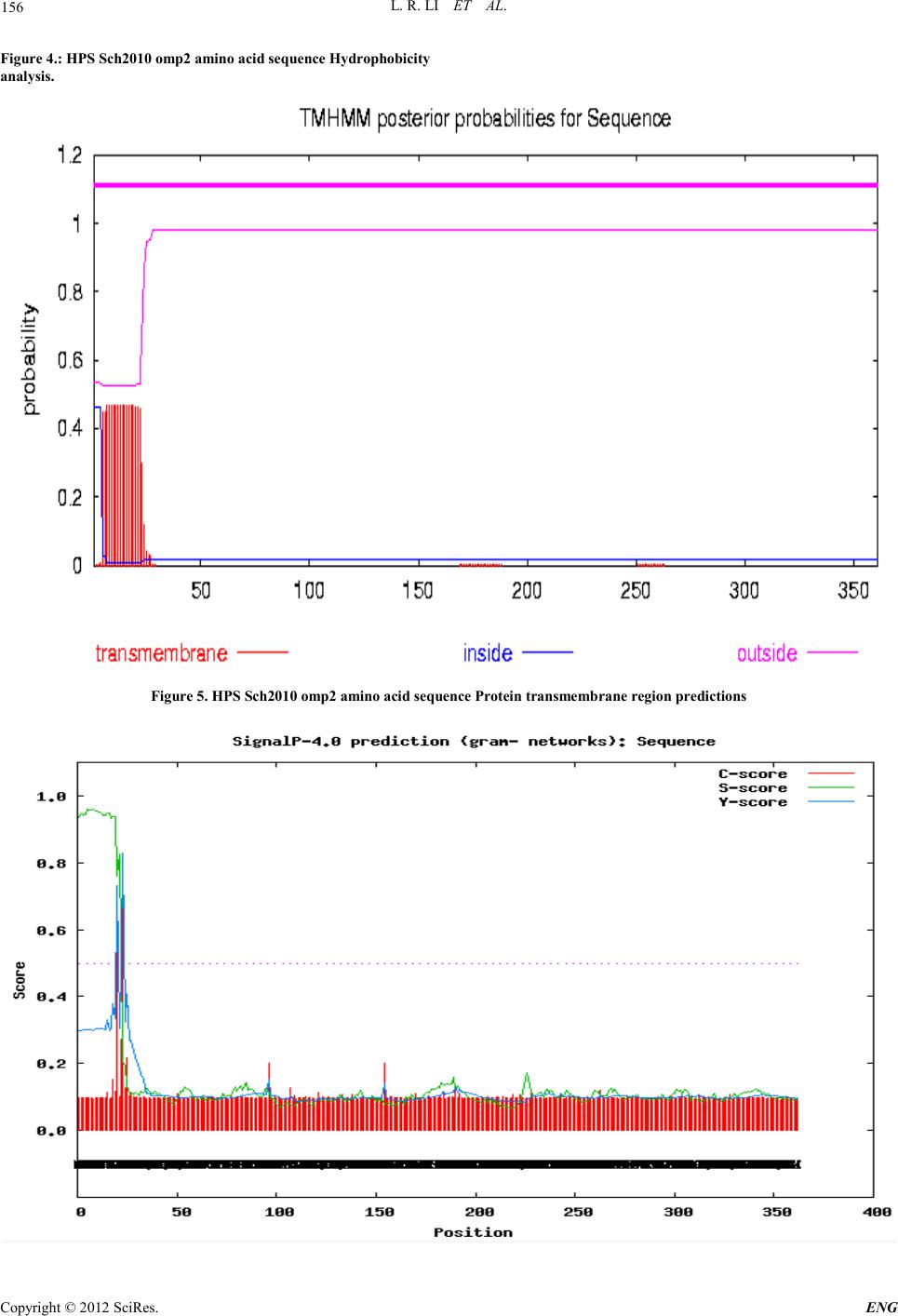 L. R. LI ET AL. Copyright © 2012 SciRes. ENG 156 Figure 4.: HPS Sch2010 omp2 am ino acid sequen ce Hydrophobicity analysis. Figure 5. HPS Sch2010 omp2 amino acid sequence P rotein transmembrane region predictions 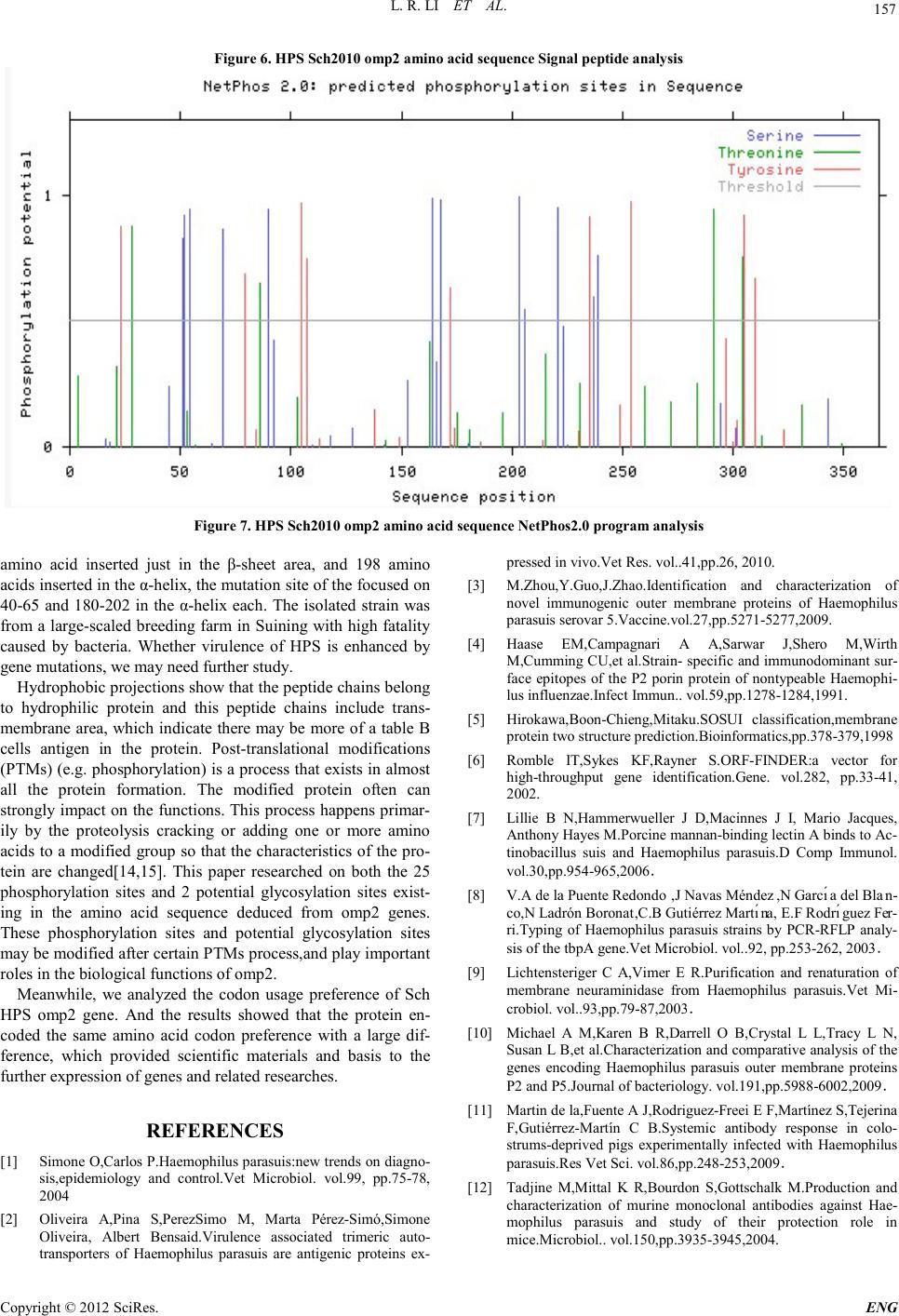 L. R. LI ET AL. Copyright © 2012 SciRes. E NG 157 Figure 6. HPS Sch2010 omp2 amino acid sequence Signal peptide analysis Figure 7. HPS Sch2010 omp2 amino acid sequence NetPhos2.0 program analysis amino acid inserted just in the β-sheet area, and 198 amino acids inserted in the α-helix, the mutation site of the focused on 40-65 and 180-202 in the α-helix each. The isolated strain was from a large-scaled b reeding far m in Suining with high fatality caused by bacteria. Whether virulence of HPS is enhanced by gene mutation s , we may need further study. Hydrophobic projections show that the peptide chains belong to hydrophilic protein and this peptide chains include trans- membrane area, which indicate there may be more o f a table B cells antigen in the protein. Post-translational modifications (P TM s ) (e.g. phosphorylation) is a process that exists in almost all the protein formation. The modified protein often can strongly impact on the functions. This process happens primar- ily by the proteolysis cracking or adding one or more amino acids to a modified group so that the characteristics of the pro- tein are ch anged[14,15]. This paper researched on both the 25 phosphorylation sites and 2 potential glycosylation sites exist- ing in the amino acid sequence deduced from omp2 genes. These phosphorylation sites and potential glycosylation sites may be modified after certain P TMs process,and play important roles in the biological functions of omp 2. Meanwhile, we analyzed the codon usage preferen ce of Sch HPS omp2 gene. And the results showed that the protein en- coded the same amino acid codon preference with a large dif- ference, which provided scientific materials and basis to the further expression of genes and related researches. REFERENCES [1] Simone O,Carlos P.Haemophilus parasuis:new trends on diagno- sis,epidemiology and control.Vet Microbiol. vol.99, pp.75-78, 2004 [2] Oliveira A,Pina S,PerezSimo M, Marta Pérez-Simó,Simone Oliveira, Albert Bensaid.Virulence associated trimeric auto- tran sporters of Haemophilus parasuis are antigenic proteins ex- pressed in vivo. Vet Res. vol..41,pp.26, 2010. [3] M.Zhou,Y.Guo,J.Zhao.Identification and characterization of novel immunogenic outer membrane proteins of Haemophilus parasuis serovar 5.Vac cine.vol. 27,pp .5271-5277,2009. [4] Haase EM,Campagnari A A,Sarwar J,Shero M,Wirth M,Cumming CU,et al.Strain- specif ic and immunodomina nt sur- face epitopes of the P2 porin protein of nontypeable Haemophi- lus influenzae.Infect Immun.. vol.59,pp.1278-1284,1991. [5] Hirokawa,Boon-Chieng,Mitaku.SOSUI classification,membrane protein two str ucture prediction.Bioinformatics,pp.378-379,1998 [6] Romble IT,Sykes KF,Rayner S.ORF-FINDER:a vector for high-throughput gene identification.Gene. vol.282, pp.33-41, 2002. [7] Lillie B N,Hammerwueller J D,Macinnes J I, Mario Jacques, Anthony Hayes M.Porcine mannan-binding lecti n A bind s to Ac- tinobacillus suis and Haemophilus parasuis.D Comp Immunol. vol.30,pp.954-965,2006. [8] V.A de la Puente Redondo,J Navas Méndez,N Garcı́a del Blan- co,N Ladrón Boronat,C.B Gutiérrez Martı́na, E.F Rodrı́guez Fer- ri.Typing of Haemophilus parasuis strains by PCR-RFLP analy- sis of the tbpA gene.Vet Microbiol. vol..92, pp.253-262, 2003. [9] Lichtensteriger C A,Vimer E R.Purification and renaturation of membrane neuraminidase from Haemophilus parasuis.Vet Mi- crobiol. vol..93,pp.79-87,2003. [10] Michael A M,Karen B R,Darrell O B,Crystal L L,Tracy L N, Susan L B,et al.Characterization and comparative analysis of the genes encoding Haemophilus parasuis outer membrane proteins P2 and P5.Journal of bacteri ology. vol.191,p p.5988-6002,2009. [11] Martin de la,Fuente A J,Rodriguez-Freei E F,Ma rt ín ez S, Tej eri n a F,Gutiérrez-Martín C B.Systemic antibody response in colo- strums-deprived pigs experimentally infected with Haemophilus parasuis.Res Vet Sci. vol.86,pp.248-253,2009. [12] Tadjine M,Mittal K R,Bourdon S,Gottschalk M.Production and characterization of murine monoclonal antibodies against Hae- mophilus parasuis and study of their protection role in mice.Microbiol.. vol. 150,pp.3935-3945,2004.  L. R. LI ET AL. Copyright © 2012 SciRes. ENG 158 [13] Mcvicker J,Tabatabai L,MurielV S.Haemophilus parasuis-novel proteins-hopes for vaccine.Respiratory diseases of livestock. vol.22,pp. 2005. [14] Nikolaj B,Thomas S P,Ramneek G, Steen G,Søren B.Prediction of post-translational glycosylation and phosphorylation of pro- teins from the amino acid sequence. Proteomics. vol.4,pp.1633-1649,2004. [15] Mannm M,Jensen O N.Proteomic analysis of post-translational modi fications.Nature Biotec hnology. vol.. 21,pp.255-261,2003. |

