Paper Menu >>
Journal Menu >>
 Engineering, 2012, 5, 139-141 doi:10.4236/eng.2012.410B036 Published Online October 2012 (http://www.SciRP.org/journal/eng) Copyright © 2012 SciRes. ENG Functional analysis of OAT gene in Aspergillus Niger* Chuang Zhou1, X ihong He1, Hao Liu1, Hao Liu2 1MOE key laborator y of Industrial Fermentation Microbiology, College of Biot echnology, Tianjin University of Sci ence & Techn ology, Tianjin, China 2College of Biotechnology, Tianjin University of Science & Technology, Tianjin, China Email: liuhao1227@gmail.com Received 2012 ABSTRACT Mitochondrial oxaloacetate tran sport er protein encoded by OAT gene transports oxaloacetate from cytoplasm into mitochondria. To investigate the primary effects of OAT gene on relative metabolism in Aspergillus niger, a oat-deleted mutant was derived from wild-type A. niger ATCC1015 using the double-crossover chromosome replacement technique. Then batch fermentation was per- formed to evaluate the mutant. Compared with the wild type, the mutant showed lower organic acids production, with the experi- mental data that the production of pyruvate and 2-ketoglutarate decreased by 31.6% and 35.7%, respectively, and by contrast, the mutant showed higher glycerol formation. The results suggest that OAT gene plays significant role s on metabolism in A. niger. Keywords: Aspergillus Niger; Oxaloacetate Transporter; Organic Acids; Glycerol Formation 1. Introduction The filamentous fungus Aspergillus niger was widely applied to indu strial enzymes and organic acid s production [1]. Despite decades of study, the production process is not well understood in details yet [2]. The release of the genomic sequences of As- pergillus niger ATCC1015 in 2006 [3-4] provided a very good opportunity to characterize the function of genes in A. niger. Additionally, the deletion of non-homologous end-joining gene Ku80 showed high recombination frequency, which provided a useful tool for gene targeting in A. niger [5-7]. In the fugus strain A. niger, citrate can be synthesed from oxaloacetate and acetyl-CoA by citrate synthase. The progress of citrate synthesis constantly accompanied with oxaloacetate consumption, so it is importan t to ensure ad equate oxaloacet ate for the production of citric acid [8]. Formers’ researches veri- fied that oxaloacetate tran spo rter delivers oxaloacetate fr o m t h e cytoplasm to the mitochondria in A. niger [3]. In this study, oxaloacetate transporter OAT null mutant was generated, and the effect of deletion of OAT on the metabolites of A. niger was investigated. 2. Materials and Methods 2.1. Strains Aspergillus niger strain ATCC 1015 used in this study was kindly provided by Dr Sun Ji Bin, Tianjin Institute of Industrial Biotechnology, CAS. Escherichia coli DH5α was used for routine cloning experi- ments. Agrobacterium tumefaciens AGL1 was used for the trans- formation of A. niger. 2.2. Strain Growth A. niger strains were grown at 30 ℃ on potato dextrose agar (PDA) slant for sporulation and stock [9]. Screenin g of transformants was performed on fungal com- plete medi a (CM) containing 200 μg/mL hygromycin B and 200 μM cefotaxim [10]. E. coli DH5α was grown at 37 °C on LB medium. A. tumefa- ciens AGL1 was grown at 28°C on LB medium. The b atch cultivation medium composed of 50 g/L glucose, 5 g /L NH4NO3, 2 g/L KH2PO4, 1 g/ L MgS O4·7 H2O, 1 g /L NaCl, 0.1 g/L CaCl2·2H2O, and 1 ml/L trace element s olution. Trace element solutio n consisted of 0.8 g/L ZnSO4·7H2O, 0.8 g/L CuSO4·5H2O, 0.8 g /L FeSO4·7H2O. 2.3. OAT Gene Deletion E-mail address: liuhao@tust.edu.cnThe relative sequences of OAT gene were obtained from the genome sequences d atabase DOE Joint Genome Institute (http://genome.jgi.doe.gov/Aspni5/ Aspni5.home.html). The plasmid p44 containing hph gene was derived from pCAMBIA1300. The oatΔ strain was constructed using the standard one-step gene rep lacement strateg y in which hph gene was used as s elective marker [11]. Briefly, about 1 kb of 5’ UTR and 3’ UTR frag ments were amplified by PCR using the primer pairs oat-upF/oat-upR and oat-downF /oat-downR, respectively. The two fragments Were ligated to flank the hy- gromycin resistance cassette in p44, sequentially. The gene replace ment con struct was introduced int o A.niger ATCC 1015 via Agrobacterium tumefaciens-mediated transformation. CM containing 200 μg/mL hygromycin was used for selecting transformants. Correct gene replacement event was confirmed by PCR using primer pairs oat-upF1/oat-downR1. The se- quences of the pi mers are presented in Table 1. * Project supported by the National Nature Science Foundation of China (No.31 270125,31170134) and the Nati onal High Technology Re se arch a nd Development Program of China (863 Program) (No.2011AA02A213)  C. ZHOU ET AL. Copyright © 2012 SciRes. ENG 140 Table 1. PCR pimer. Primer name sequnece oat-upF CTGTAAGCTTC AAGCCAGACAG ATCCGTTTG oat-upR CTGGCTGCAG oat-downF TGCAGACTTTCGTATAGATAC GTCTGGATCC oat-downR GTATTCTGCCTCACACGGTAAG GTCAGGTACC oat-upF1 CAGTGTGGTGATGACGTTACAG GTCCGAGACCTTCTGGCATTGTAG oat-downR1 CTTCATGCACATTGGCAGCTGTC 2.4. Cultivation Conditions For inoculation of batch cultivations, spores of A. niger incu- bated at 30 °C on PDA solid medium for 5–7 days were har- vested with 0.05% (w/w) Tween-80. All strains were cultivated in 5L-Braun bioreactors with a working volume of 4 L. The bioreactors were equipped with two Rushton four-blade disc turbines used for culture broth mi xi n g . No b affles were incorporated in the bioreactors thereby reducing the surface area available for wall growth. Subse- quently, Spores were added t o the bioreactors to an initial con- centration of 5 × 108/L. The temperature was maintained at 30 °C. The initial aeration rate, rotate speed and pH were 0.3 vvm, 200 rpm and 2.5, respectively. After germination (ap- proximately 16 h), the culture condition described above were gradual ly adjusted to 1 vvm, 600 rpm and pH 4.5 and kep t con- stantly throughout the rest of the fermentation time. The pH was controlled by automatic addition of 1 M NaHCO3 or 2 M HCl. 2.5. Biomass Determination The biomass was determined by the use of 0.45 μm nitrocellu- lose filter. Initially, the filters were pre-dried in a microwave oven at 150 W for 10 min, and then weighed. A known weight of cell culture was filtered, and the cells were washed with distilled water. Finally, the filter was d ri ed in th e micro-wave at 150 W for 15 min and the dry weight was det ermined . 2.6. Extracellular Meta bolites Quantification To determine the extracellular metabolites, a culture sample was taken an d immediatel y fi ltered through a 0.45 μm nitrocel- lulose filter. The filtrate was frozen and kept at -20 °C until analysis. Glucose, pyruvate, citrate, succinate, oxalate and gly- cerol concentrations were determined using an cationic- ex- change column (Aminex HPX-87H, BioRad, USA) elu ted with 5mM H2SO4 with the flow rate of 0.6 mL min- 1 at 65 °C. Me- tabolites were detected b y re fractive index and UV. 3. Results and Discussion 3.1. The Construct of p 44-oat::hph Gene Delet ion Cassette The 3’ UTR fragment (966 bp) was amplified and inserted into BamH I and Kpn I sites of p44 to construct the plasmid p44-oat-dw. Th e 5’ UTR fragment (855 bp) was amplified and inserted into Hind III and Pst I sites of p44-oat-dw to obtain the plasmid p44-oat::hph (Figure 1). The result of double digestion of p44-oat::hph to confirm the successful cloning being con- struct ed (Figure 2). 3.2. The Generation of the OAT Gene Deletion in A. Niger T-DNA fragment containing the 5’ UTR (855 bp) and 3’ UTR (966 bp) fragments of the OAT gene was transferred into the strain A. niger ATCC 1015 and the transformants were selected with hygrom yc in. The resistant colonies grown in the plates were picked out and tot al DNA were extracted fro m the transfor mants, and then used as the template for PCR amplification and examination. More than 30 transformants wer e examin ed, and two gene dele- tion candidates were obtained. When the OAT gene was re- placed by hph gene, the length of the DNA fragment amplified by PCR using primer pairs oat-upF1/ oat-downR1 is about 3.5 kb (Figure 3, Lane 3) compared to 3.0 kb obtained from the wild type strain(Figure 3, Lane 1). The length of the PCR product obtained from transformants1 (Figure 3, Lane 2) is the same as that obtained from the wild type strain, indicating that the transformants1 was not the oatΔ strain. All the data sug- gested that the transformants2 was an OAT gene deletion strain. Figure 1. Map of p 44-oat::hph. Figure 2. Double digestion of p 44-oat::hph 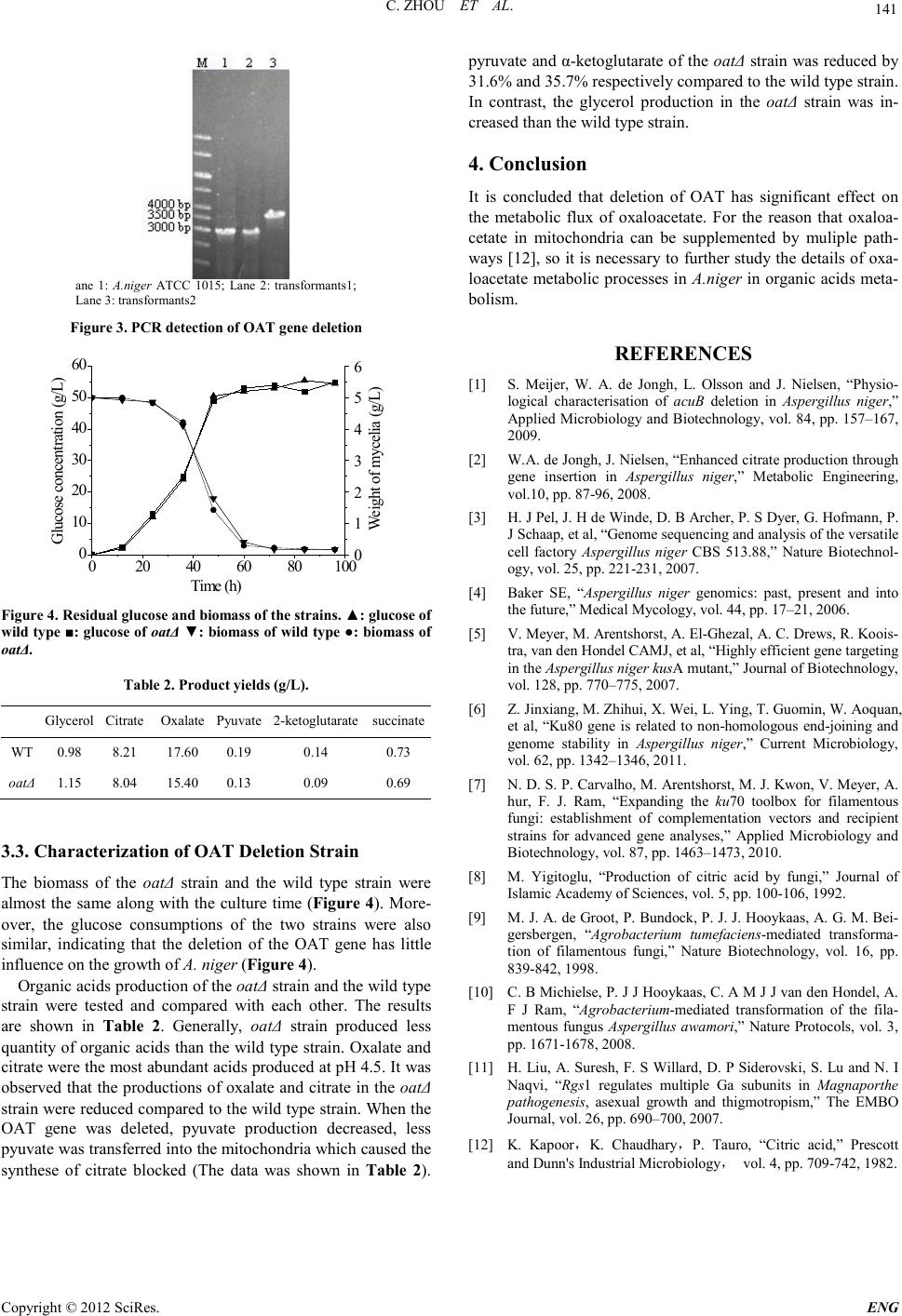 C. ZHOU ET AL. Copyright © 2012 SciRes. E NG 141 ane 1: A.niger ATCC 1015; Lane 2: transformants1; Lane 3: t r ansfor mants2 Figure 3. PC R detecti on o f OAT gene de letio n 020 40 60 80100 0 10 20 30 40 50 60 W eight of mycelia (g/L) Time (h) Glucose concentration (g/L) 0 1 2 3 4 5 6 Figure 4. Residual glucose and biomass of the strains. ▲: glucose of wild type ■: glucos e of oatΔ ▼: biomass of wi ld ty pe ●: biomass of oatΔ. Table 2. Product yields (g/L). Glycerol Citrate Oxalate Pyuvate 2-ketoglutarate succinate WT 0.98 8.21 17.60 0.19 0.14 0.73 oatΔ 1.15 8.04 15.40 0.13 0.09 0.69 3.3. Characterization of OAT Delet ion Strain The biomass of the oatΔ strain and the wild type strain were almost the same along with the culture time (Figure 4). More- over, the glucose consumptions of the two strains were also similar, indicating that the deletion of the OAT gene has little influence on the growth of A. nige r (Figure 4). Organic acids production of th e oatΔ strain and the wild type strain were tested and compared with each other. The results are shown in Table 2. Generally, oatΔ strain produced less quantity of organic acids than the wild type strai n. Oxalate and citrate wer e the most ab undant acid s produced at pH 4.5. It was observed th at the producti ons of oxalat e and citrate in the oatΔ strain were reduced compared to the wild type strain. When the OAT gene was deleted, pyuvate production decreased, less pyuvate was transferred into the mitochondria which caused the synthese of citrate blocked (The data was shown in Table 2). pyruvate and α-ketoglutarate of t he oatΔ strain was reduced by 31.6% and 35.7% respectively compared to the wild type strain. In contrast, the glycerol production in the oatΔ strain was in- creased than the wild type strain. 4. Conclusion It is concluded that deletion of OAT has significant effect on the metabolic flux of oxaloacetate. For the reason that oxaloa- cetate in mitochondria can be supplemented by muliple path- way s [12], so it is necessary to further study the details of oxa- loacetate metabol ic processes in A.niger in organic acids meta- bolism. REFERENCES [1] S. Meijer, W. A. de Jongh, L. Olsson and J. Nielsen, “Physio- logical characterisation of acuB deletion in Aspergillus niger,” Applied Microbiology and Biotechnology, vol. 84, pp. 157–167, 2009. [2] W. A. de Jongh, J. Nielsen, “Enhanced citrat e producti on through gene insertion in Aspergillus niger,” Metabolic Engineering, vol.10, pp . 87-96, 2008. [3] H. J Pel, J. H de Winde, D. B Archer, P. S Dyer, G. Hofmann, P. J Sch aap , et al, “Gen ome s eq u en c in g and ana lysi s o f th e vers a t i le cell factory Aspergillus niger CBS 513.88,” Nature Biotechnol- ogy, vol. 25, pp. 221-231, 2007. [4] Baker SE, “Aspergillus niger genomics: past, present and into the future,” Medical Mycology, vol. 44, pp. 17–21, 2006. [5] V. Meyer, M. Arentshorst, A. El-Ghezal, A. C. Drews, R. Koois- tra, van den Hondel CAMJ , et al, “Highly efficient gen e targeting in the Aspergill u s niger kusA mutant ,” Jou rna l of Bi ot echn ology, vol. 128, pp. 770–775, 2007. [6] Z. Ji n xian g, M . Zhihui, X. Wei, L. Ying, T. Guomin, W. Aoquan, et al, “Ku80 gene is related to non-homologous end-joining and genome stability in Aspergillus niger,” Current Microbiology, vol. 62, pp. 1342–1346, 2011. [7] N. D. S. P. Carvalho, M. Arentshorst, M. J. Kwon, V. Meyer, A. hur, F. J. Ram, “Expanding the ku70 toolbox for filamentous fungi: establishment of complementation vectors and recipient strains for advanced gene analyses,” Applied Microbiology and Biotechnology, vol. 87 , pp. 1463–1473, 2010. [8] M. Yigitoglu, “Production of citric acid by fungi,” Journal of Islamic Academy of Sciences, vol. 5, pp. 100-106, 1992. [9] M. J. A. de Groot, P. Bundock, P. J. J. Hooykaas, A. G. M. Bei- gersbergen, “Agrobacterium tumefaciens-mediated transforma- tion of filamentous fungi,” Nature Biotechnology, vol. 16, pp. 839-842, 1998. [10] C. B Michielse, P. J J Hooykaas, C. A M J J van den Hondel, A. F J Ram, “Agrobacterium-mediated transformation of the fila- mentous fungus Aspergillus awamori,” Nature Protocols, vol. 3, pp. 1671-1678, 2008. [11] H. Liu, A. Suresh, F. S Willard, D. P Siderovski, S. Lu and N. I Naqvi, “Rgs1 regulates multiple Ga subunits in Magnaporthe pathogenesis, asexual growth and thigmotropism,” The EMBO Journal, vol. 26, pp. 690–700, 2007. [12] K. Kapoor,K. Chaudhary,P. Tauro, “Citric acid,” Prescott and Dunn's Industrial Microbiology, vol. 4, pp. 709-742, 1982. |

