Paper Menu >>
Journal Menu >>
 Engineering, 2012, 5, 106-109 doi:10.4236/eng.2012.410B027 Published Online October 2012 (http://www.SciRP.org/journal/eng) Copyright © 2012 SciRes. ENG MS-HRM to Detect Serum DNA Methylation of Intrauterine Growth Ret ardation Children Yan Du1,2, Youxia Zhou2, Qian Wu3,4* 1Department of Pathology, Maternity and Children's Health Care Centers in Wuxi, Wuxi, China 2Research Cen ter of Learn in g Science, So utheast Uni versity, Na njing, China 3State Key Laboratory of Reproductive Medicine, Department of Hygienic Analysis and Detection, Sch oo l of P ublic Health, Nanjing Medic al University, Nan jing, China 4State Key Laboratory of Bioel ectron ics, School of Biological Science and Medical Engineering, Southeast University, Nanjing, China Email: *wuqian@njmu.edu.cn Received 2012 ABSTRACT Intrauterine growth retardation (IUGR) is also called fetal growth restriction (FGR), which is the major complications in obstetrics and one of the most important causation from high morbidity and mortality in perinatal. In this study, MS-HRM (Methylation-Sensi- tive High Resolution Melting Curve Analysis) was used to detect the methylation status of serum DNA. Gene of insulin-like gro wth factor binding protein 3 (IGFBP-3) was detected in this study. Results showed that the serum DNA methylation level of FGR fetus were lower th an that of the control grou p; different methylation levels were also fou nd between male and female fetu s; and the me- thylation level was increased with the birth weight of the newborn. Our results showed that the IGFBP-3 gene methylation level of serum DNA of newborn could be semi-quantitative detected which guide the early prevention and treatment of IUGR. It also indi- cated that the methylation status of serum DNA can be conveniently identified and quantified by inspection of the melting curves. Keywords: MS-HRM; IUGR; IGFBP-3; Serum DNA 1. Introduction Intrauterine growth retardation (IUGR) is also called fetal growth restriction (FGR), which is the major complications in obstetrics and one of the most important causation from high morbidity and mortality in perinatal. Furthermore, it is also one of the important reasons for many adult diseases such as hypertension, type II diabetes, coronary heart disease, kidney disease and so on [1]in the future. Insulin like growth factor 1(IGF-1) is a single basic protein composed of 70 amino acids; and known as one of the key factors to cellular proliferation and metabolism which plays an important role in fetal organ maturation and growth and development of children. The infant health status especially endocrine status could be indicated by serum IGF-1 level, for which it is produced by fetus itself because it cannot transfer through placental from maternal serum. Previous studies showed that the birth weight of IGF-1 knockout mice were only 60% of the normal mice [2]. Insulin like growth factor binding protein 3(IGFBP-3) is also an important protein in human serum could form a compound by binding to IGF-1 which increase the bioactivity of IGF-1. The compound could prolong the half-life of IGF-1 in serum. IGFBP3 can also promote the cell division as well as involves placenta formation and growth and development of the infant directly. It was reported that umbilical cord blood IGFBP-3 level of IUGR fetus reduced about 50% than that in normal newborn [3] . Carter et al demonstrated that the serum IGFBP3 level decreased remarkable in IUGR animal model [4]. Therefore, the growth and development of infants was affected by IGFBP3 directly or indirectly. Researches have shown that adverse intrauterine enviro nment might lead t o epigenetic chan ges which regulati ng the gene expression [5], such as passive smoking, unreasonable diet, and uterine artery blood supply deficiency. The methylation status of maternal IGFBP3 had been shown to be related with IUGR, while few reports revealing the potential relationship between IUGR and the methylation status of IGFBP3. The previous studies showed that hyper methylation of the promoter of IGFBP3 gene was highly associated with tumor [6], and the gene methylation alteration might be used as a pot ential di agno sis marker in colo rectal cancer [ 7] . It could be inferred that the methylation status of IGFBP3 promoter has certain relation with growth and development. However, no report clearly indicated whether the changes of methylation status would affect the development of fetus, infants and even adult. Investigation of the methylation status especially the methylation level of IGFBP3 is significantly important to IUGR. The most popular approaches of methylation detection rely on treatment of DNA samples with sodium bisulfate and subsequent amplification by PCR [8,9]. However, MSP (Meth ylation Specific PCR) is a non-quantitative measurement, which limited its field of application. MS-HRM (Methylation- Sensitive High Resolution Melting Curve Analysis), has been shown a simple and cost-effective post-PCR technique for routine clinical diagnosis, which is capable of analyzing methylation in a semi-quantitative manner [10]. All of the CpGs flanked by the primers binding to the target sequence can *Corresponding author.  Y. DU ET AL. Copyright © 2012 SciRes. ENG 107 be scanned by HRM regardless of the methylation status of CpGs in the primer-binding site, with no post-PCR handling and no separation step, characteristics that improve analysis time [11]. HRM might be a good choice for the detection of IGFBP3 methylation status in this study. The aim of th is stud y was to det ect the seru m DNA meth yla- tion level of IUGR infants. Results generated by HRM were also validated with traditional MSP assays. The serum IGFBP-3 was me asu red with ELISA. 2. Materials and Methods 2.1. Participants Serum samples were obtained from new-born baby between June, 2009 to July, 2011 in Maternity and Children's Health Care Centers in Wuxi. All the baby was born between 37 weeks to 40 weeks of gestation ages and the birth weight were ten percent lower than the newborn at the same gestation age. Mothers were at the age of 21-35 years old in this study, no basic disease, the weight gain during pregnancy is normal, and the gestational hypertension, diabetes, preeclampsia, polyhy- dramnios and pregnancy bile acid increased disease were all excluded. Fifty IUGR infants (thirty male and twenty female) and thirty normal controls (fourteen male and sixteen female) were selected in this experiment. This proj ect was approved by the Ethics Committee of Wuxi Maternity and Children's Health Care Centers, Southeast University and Nanjing Medical Uni- versity Clin ical Research Ethics Committee, N anjing, China. 2.2. Extraction of DNA a nd Sodium Bisulfate Modif ication DNA from serum was extracted with a QIAamp DNA Blood Midi Kit (Qiagen, Hilden, Germany) according to the manu- facturer’s recommen dations. D NA was extracted from 2 mL of serum and eluted in 50μl of TE buffer. About One microgram of DNA was subjected to bisulfite conversion with the EZ DNA methylation kit (Zymo Research, USA). DNA quantity was assessed spectrophotometrically. The eluted DNA was used for the HRM analysis and MSP validation. 2.3. HRM Analysis PCR amplification and HRM were performed on the ABI 7500 fast (Life Technology, USA) as adapted from the published protocol [10]. The primers were designed as outlined, not more than 1 to 2 CpG sites and were placed at or adjacent to the 5’- end. The sequences of primers for IGFBP3 were as follows: forward: GGGTTAAGGTTAGAGGTGGTGTTAT; reverse: AAAAAAATTTACAATTTACAAAAACTC. PCR was performed in a 20 μl volume containing: 1× buffer, 2 U Hotstar tTaq DNA pol ymerase (Takar a), 25 0 nM of each p rimer, 2.5 mM SYTO-9, 10 ng bisulfite treated DNA template, and 3 mM MgCl2. The cycling conditions were as follows: 1 cycle of 95°C for 5 min, 40 cycles o f 95°C for 10 s, 58°C for 20 s, and 72°C for 30 s; followed by an HRM step of 95°C for 1 min, 40°C for 1 min, 70°C for 15 s, and continuous acquisition to 95°C at 1 acquisition per 0.1°C. A standard curve with known methylation ratios was includ ed in each assay and was used t o deduce th e methylation ratio of each fetal and maternal sample. Differences between and among groups were compared using Pearson’s chi-square test for qualitative variables and using Student’s t test or analysis of varian ce for continuous variables. Methylation- sensitive PCR was performed through BioRad PCR system as described previously [12]. 2.5 μL (approxi- mately 50 ng) of bisulfite-treated DNA was amplified using 2 pmol of forward primer and reverse primer. PCR conditions were 95 °C for 10 min followed by 40 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 50 s and 1 cycle at 72 °C for 8 min. 3. Results 3.1. HRM Assay and Dilution M atr ix We validated the correlation between Tm value and methyla- tion level by analyzing different sets of methylation level stan- dards (0%, 1%, 10%, 25%, 50%, 75%, 100%) firstly. All HRM assays were able to d etect rep roduci bly 1% meth ylated DN A in a background of unmethylated DNA. Linear regression analysis (Figure 1) revealed that methylation levels were highly corre- lated with Tm value, th at Tm value can be used as the s tandards of methylation. 3.2. Methylation Level of IGFBP3 in Serum of IUGR Infants In this present study, the serum DNA methylation level of IGFBP-3 was measured by MS-HRM.The analysis of all samples was repeat ed twice b y HR M an d part sample ran d oml y selected were measured by MSP. We also tested whether varying amounts of bisulfite treated target DNA would influence HRM results. We found no differences in methylation ratios when using two different DNA amounts (50 ng versus 10 ng). Results showed that lower methylation level of found in IUGR infants compared with normal control baby (P < 0.01, T-test). It could be inferred that certain relation might exist between the IGFBP-3 methylation and the development of IUGR. Different methylation level was also found in different gender of newborn. It can b e s een t hat the methylation level of Figure 1. Correlation between Tm value and methylation level measured by MS-HRM. 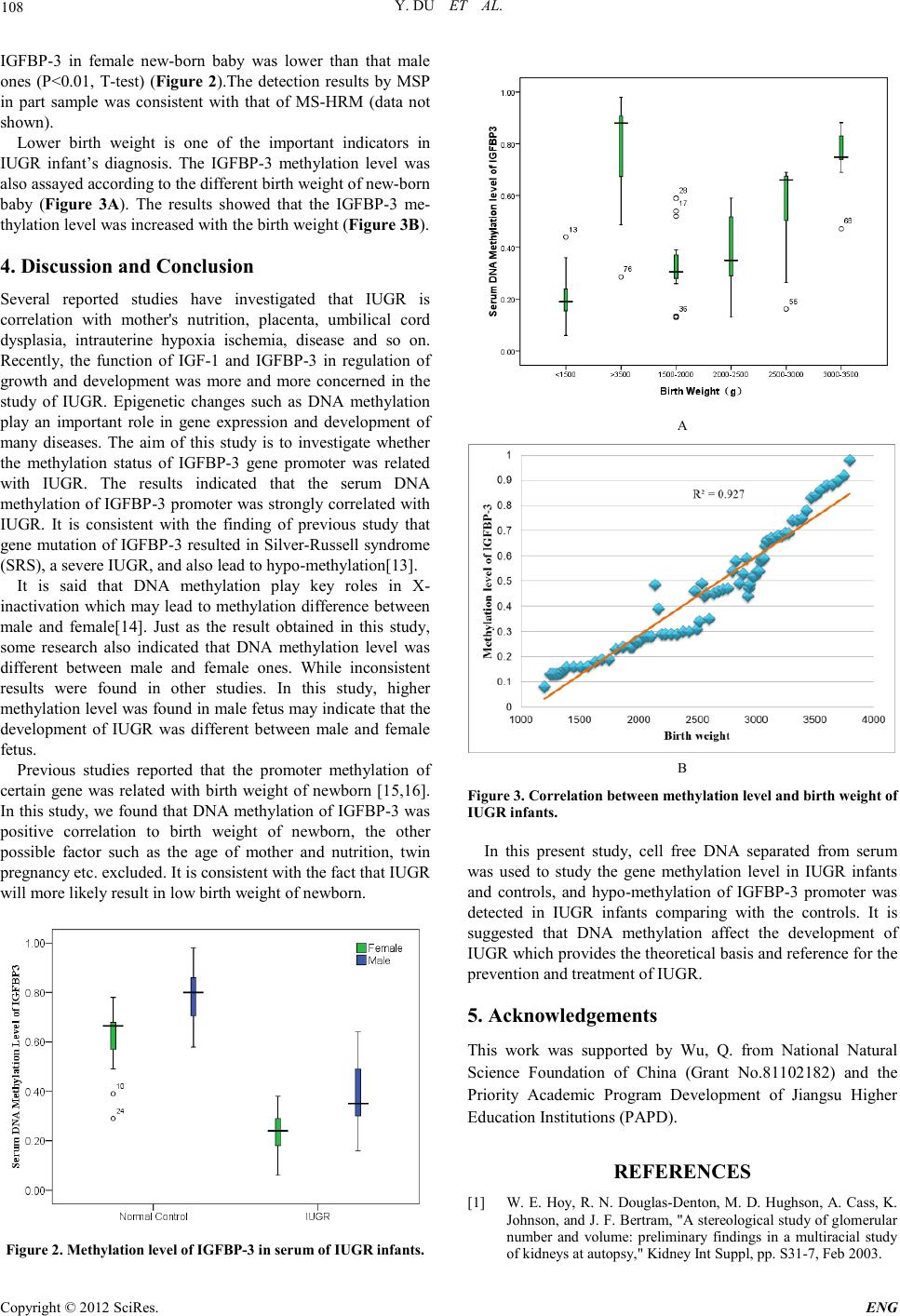 Y. DU ET AL. Copyright © 2012 SciRes. ENG 108 IGFBP-3 in female new-born baby was lower than that male ones (P<0.01, T-test) (Figure 2).The detection results by MSP in part sample was consistent with that of MS-HRM (data not shown). Lower birth weight is one of the important indicators in IUGR infant’s diagnosis. The IGFBP-3 methylation level was also assayed according to the different birth weight of new-born baby (Figure 3A). The results showed that the IGFBP-3 me- thylation level was increased with the birth weight (Figure 3B). 4. Discussion and Conclusion Several reported studies have investigated that IUGR is correlation with mother's nutrition, placenta, umbilical cord dysplasia, intrauterine hypoxia ischemia, disease and so on. Recently, the function of IGF-1 and IGFBP-3 in regulation of growth and development was more and more concerned in the study of IUGR. Epigenetic changes such as DNA methylation play an important role in gene expression and development of many diseases. The aim of this study is to investigate whether the methylation status of IGFBP-3 gene promoter was related with IUGR. The results indicated that the serum DNA methylation of IGFBP-3 promoter was strongly correlated with IUGR. It is consistent with the finding of previous study that gene mutation of IGFBP-3 resulted in Silver-Russell syndrome (SRS) , a severe IUGR, and also lead to hypo-methylation[13]. It is said that DNA methylation play key roles in X- inactivation which may lead to methylation difference between male and female[14]. Just as the result obtained in this study, some research also indicated that DNA methylation level was different between male and female ones. While inconsistent results were found in other studies. In this study, higher methylati on l evel was foun d in male fet us may ind icate th at th e development of IUGR was different between male and female fetus. Previous studies reported that the promoter methylation of certain gene was related with birth weight of newborn [15,16]. In this study, we found that DNA methylation of IGFBP-3 was positive correlation to birth weight of newborn, the other possible factor such as the age of mother and nutrition, twin pregnancy etc. exclu ded. It is consistent with the fact th at IUGR will more likely result in low birth weight of newborn. Figure 2. Methylation level of IGFBP-3 in serum of I UG R infants. A B Figure 3. Correlation between methyla tio n le ve l and birth weight of IUGR infants. In this present study, cell free DNA separated from serum was used to study the gene methylation level in IUGR infants and controls, and hypo-methylation of IGFBP-3 promoter was detected in IUGR infants comparing with the controls. It is suggested that DNA methylation affect the development of IUGR which pro vides t he theo retical basi s an d reference for t he prevention and treatment of IUGR. 5. Acknowledgements This work was supported by Wu, Q. from National Natural Science Foundation of China (Grant No.81102182) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). REFERENCES [1] W. E. Hoy, R. N. Douglas-Denton, M. D. Hughson, A. Cass, K. John son, and J. F. B ertram, "A stereologi cal stud y of glomerular number and volume: preliminary findings in a multiracial study of kidneys at autopsy," Kidney Int Suppl, pp. S31-7, Feb 2003.  Y. DU ET AL. Copyright © 2012 SciRes. ENG 109 [2] K. A. Woods, C. Camacho-Hubner, D. Barter, A. J. Clark, and M. O. Savage, "Insulin-like growth factor I gene deletion causing intrauterine growth retardation and severe short stature," Acta Paediatr Supp l, vol. 423, pp. 39-45, Nov 1997. [3] L. C. Giudice, F. de Zegher, S. E. Gargosky, B. A. Dsupin, L. de las Fuentes, R. A. Crystal, R. L. Hintz, and R. G. Rosenfeld, "Insulin-like growth factors and their binding proteins in the term and preterm human fetus and neonate with normal and extremes of intrauterine growth," J Clin Endocrinol Metab, vol. 80, pp. 1548-55, May 1995. [4] A. M. Carter, M. J. Kingston, K. K. Han, D. M. Mazzuca, K. Nygard, and V. K. Han, "Altered expression of IGFs and IGF-binding proteins during intrauterine growth restriction in guinea pigs," J Endocrin ol, vol. 184, pp. 179-89, Jan 2005. [5] M. Faxen, J. Nasiell, A. Blanck, H. Nisell, and N. O. Lunell, "Altered mRNA expression pattern of placental epidermal growth factor receptor (EGFR) in pregnancies complicated by preec lampsi a and/or intrau terin e growth ret ardat ion," Am J Per i- nat o l , vol. 15, pp. 9-13, Jan 199 8. [6] A. Lorente, W. Mueller, E. Urdangarin, P. Lazcoz, A. von Deimling, and J. S. Castresana, "Detection of methylation in promoter sequences by melting curve analysis-based semiquan- titative real time PCR," BMC Cancer, vol. 8, p. 61, 2008. [7] S. E. Cottrell, "Molecular diagnostic applications of DNA me- thylation technology," Clin Biochem, vol. 37, pp. 595-604, Jul 2004. [8] C. T. Wittwer, G. H. Reed, C. N. Gundry, J. G. Vandersteen, and R. J. Pryor, "High-resolution genotyping by amplicon melting analysis using LCGreen," Clin Chem, vol. 49, pp. 853-60, Jun 2003. [9] M. Frommer, L. E. McDonald, D. S. Millar, C. M. Collis, F. Watt, G. W. Grigg, P. L. Molloy, and C. L. Paul, "A genomic sequencing protocol that yields a positive display of 5-me thyl cytosi ne resid ues in individua l DNA strand s," P roc Natl Acad Sci U S A, vol. 89, pp. 1827-31, Ma r 1 1992. [10] T. K. Wojdacz and A. Dobrovic, "Methylation-sensitive high resolution melting (MS-HRM): a new app roach f or sensi tive and high-throughput assessm ent of methylation," Nucleic Acids Res, vol. 35, p. e 4 1, 2 007. [11] H. Nakagawa, R. B. Chadwick, P. Peltomaki, C. Plass, Y. Na- kamura, and A. de La Chapelle, "Loss of imprinting of the insu- lin-like growth factor II gene occurs by biallelic methylation in a core region of H19-associated CTCF-binding sites in colorectal cancer," Proc Natl Acad Sci U S A, vol. 98, pp. 591-6, Jan 16 2001. [12] J. Worm, A. Aggerholm, and P. Guldberg, "In-tube DNA me- thylation profiling by fluorescence melting curve analysis," Clin Chem, vol. 47, pp. 1183-9, 2001. [13] I. C. Beserra, M. G. Ribeiro, P. F. Collett-Solberg, M. Vaisman, and M . M . Gu i ma ra es, " IGF-I and IGF Binding Protein-3 Gener- ation Tests and Response to Growth Hormone in Children with Silver-Russell Syndrome," Int J Pediatr Endocrinol, vol. 2010, p. 546854. [14] A. D. Riggs, "X inactivation, differentiation, and DNA methyla- tion," Cytogenet Cell Genet, vol. 14, pp. 9-25, 1975. [15] A. C. Filiberto, M. A. Maccani, D. Koestler, C. Wil- helm-Benartzi, M. Avissar-Whiting, C. E. Banister, L. A. Gagne, and C. J. Marsit, "Birthweight is associated with DNA promoter methylation of the glucocorticoid receptor in human placenta," Epigenetics, vol. 6, pp. 566-72, May . [16] J. C. Ferreira, S. Choufani, D. Grafodatskaya, D. T. Butcher, C. Zhao, D. Chitayat, C. Shuman, J. Kingdom, S. Keating, and R. Weksberg, "WNT2 promoter methylation in human placenta is associated with low birthweight percentile in the neonate," Epi- genetics, vol. 6, pp. 440-9, Apr. |

