 J. Biomedical Science and Engineering, 2011, 4, 70-75 doi:10.4236/jbise.2011.41009 Published Online January 2011 (http://www.SciRP.org/journal/jbise/ JBiSE ). Published Online January 2011 in SciRes. http://www.scirp.org/journal/JBiSE Analysis of the arginine biosynthetic gene cluster argCJBDFR of Corynebacterium crenatum Haitao Jiao1, Yong Yuan2, Yo nghua Xiong2, Xuelan Chen1 1College of life sciences, Jiangxi Normal University, Nanchang, China; 2State Key Laboratory of Food Science and Technology, Nanchang University, Nanchang, China. Email: yhxiongchen@163.com, xuelanchen14@yahoo.com.cn Received 8 October 2010; revised 28 October 2010; accepted 8 November 2010. ABSTRACT Objective: Corynebacterium crenatum AS1.542, a Gram-positive bacterium and indigenous nonpatho- genic corynebacteria, is widely exploited for the in- dustrial production of amino acids. The objective of this paper is to clarify the genetic information of the arginine biosynthetic pathway, and further more contribute to the improvement of arginine produc- tion. Methods: Polymerase chain reaction (PCR) technology was employed for obtaining the arginine biosynthetic gene sequence and softwares eg. Laser- gene, BPROM, RNAshapes were used for the analysis of obtained sequences. Results: Arginine bio- synethetic gene cluster of C. crenatum, comprising argJ, argB, argD, argF, argR and part of argC, has been amplified and sequenced. The gene order has been established as argCJBDFR, with a entire length of 6.08kb. Conclusion: An internal promoter was found in the upstream of argB gene, four argBDFR ORFs are located in a same transcription unit, and the transcripiton termination of argC gene is irrele- vant with the rho-factor. Comparison with ornithine acetyltransferase (coded by argJ gene) from C. glu- tamate, ornithine acetyltransferase from C. crenatum also belongs to the monofunctional enzymes. Keywords: Corynebacte riu m crenatum; argCJBDFR Sequence; Ornithine Acetyltransferase; argR Gene 1. INTRODUCTION Arginine biosynthesis commences with the acetylation of the amino group of glutamate (Figure 1). Eight en- zymes coded by eight or nine genes take part in the process of catalyzation, resulting the biotransformation of glutamate into arginine [1]. The pathway of arginine biosynthesis can be divided into two parts according to two strategies evolved in the removal of the acetyl group. One is called the “linear” pathway, in which argE gene coded acetylornithinase catalyses the hydrolysis of N-acetylornithine into the arginine precursor ornithine and acetate; the other is called the “economic cyclic” pathway, in which acetylornithine is catalyzed into or- nithine and acetyl groups, and recycled with generation of acetylglutamate by argJ gene coded ornithine acetyl- transferase [2]. ArgJ has both acetylornithinase (coded by argE) function and N-acetylglutamate synthase (coded by argA) functions in the “cyclic” pathway. Lit- eratures showed that Enterobacteriaceae and Sulfolobus solfataricus [2,3] adopted the “linear” pathway in which the metabolic flow is controlled by arginine-mediated feedback inhibition of the first biosynthetic step; all the other prokaryotes, including Methanogenic archaea, Neisseria gonorrboese, members of the genus Bacillus and the eukaryotic microbes, use the “cyclic” pathway in which the metabolic flow was controlled by argin- ine-mediated feedback inhibition of the second biosyn- thetic step or by orthinine-mediated feedback inhibition of the fifth step [2]. Although the argJ gene by itself would thus be able to assure both the first and the fifth steps of arginine biosynthesis in above mentioned or- ganisms, there is genetic evidence for the existence of the cloned ornithine acetyltransferase genes from Pseu- domonas aeruginosa [4], Saccbaromyces cerevisiae [5], Streptomyces coelicolor [6] and Corynebacterium glu- tamicum complementing E. coli argE but not argA mu- tants. C. crenatum AS1.542, a Gram-positive bacterium and indigenous nonpathogenic corynebacteria, is widely ex- ploited for the industrial production of amino acids. The genetic information of arginine biosynthetic pathway was analyzed and clarified in this paper with the aim to contribute to the improvement of arginine production. 2. MATERIALS AND METHODS 2.1. Reagent All the primers were synthesized by Shanghai Biotech-  H. T. Jiao et al. / J. Biomedical Science and Engineering 4 (2011) 70-75 Copyright © 2011 SciRes. JBiSE 71 glutamate ↓ argA acetylglutamate synthase acetylglutamate ↓ argB N-acetylglutamate kinase acetylglutamate phosphate ↓ argC N-acetylglutamate semialdehyde dehyogenase acetylglutamate semialdehyde ↓ argD N-acetyl-ornithine aminotransferase glutamate ↓ acetyl ornithine argJ ↓ ↓ argE N-acetylornithine aminotransferase ornithine acetylornithine aminotransferase ↓ argI,F ornithine carbamoyltransferase citrulline ↓ argG argininosuccinate synthetase ornithine succinate ↓ argH argininosuccinase arginine Figure 1. The “linear” (in E. coli) and the “alternative cyclic” (in B. stearothermophilus and C. glutamate) arginine biosynthesis pathways. nology Corporation, China. Gel extraction kit and pGEM- T-Easy vector were purchased from Promega, USA. 2.2. Bacterial Cultivation C. crenatum and C. glutamicum were cultured in a rotary shaker incubator at 150 rpm under 30°C in Luria-Bertani (LB) medium. 2.3. DNA Manipulation, Amplification, Sequencing and Analysis Chromosomal DNA of C. crenatum was isolated as de- scribed by Shengdong L [7]. Total genomic DNA (50 ng) was used as a template for PCR amplification of argCJBDFR gene cluster. The employed primers were designed using the conserved sequences of C. glutamicum ATCC 13032, C. diphtheriae gravis NCTC13129, C. efficiens YS-314 and Mycobacterium tuberculosis CDC 1551. The primers were: sense-1 (5’-TCAAGGTTGCA ATCGCAGGAGCC-3’), antisense-1 (5’-GCAACTCAC CAATAAGACCAGTGG-3’), sense-2 (5’-CCGCAGCG CCGTGTTTACACGTAACC-3’), antisense-2 (5’-GAC AAGATTGTTGTCGTGAAATATG-3’), sense-3 (5’-AT CTTTGGAATCATGCCGGAATC-3’), antisense-3 (5’- TCTTCGTCGGTGATCACCAGCGG-3’), sense-4 (5’- CATGCCAGATTCTGGCTGATCTGCAG-3’) and an- tisense-4 (5’-GCAAGAACGATGCGGTTAGTCATG-3’), respectively. The amplicons were purified using gel ex- traction kit and sub-cloned into pGEM-T-Easy vector. The selected clones were subjected to sequencing of argCJBDFR gene cluster fragments with SP6 and T7 sequencing primers using ABI prism 3730 sequencer. The sequence data were compiled, aligned and ana- lyzed using Lasergene software (DNASTAR), Soft- berry’s BPROM (www.softberry.com) and RNAshapes WebServices (BiBiServ) et al. 3. RESULTS AND DISCUSSION 3.1. PCR Amplification The results of PCR amplification were shown in Figure 2. The full-length of amplified DNA, aligned with SeqMan function of Lasergene software, was 6080 bp. The full-length DNA sequence alignment of C. crenatum Figure 2. PCR amplification of tandem ar- ginine biosynthetic genes (Lane 1: DL2000 Marker; lane 2, 3 4 and 5: argCJ, argJBD, argDF and argFR, respectively).  H. T. Jiao et al. / J. Biomedical Science and Engineering 4 (2011) 70-75 Copyright © 2011 SciRes. JBiSE 72 showed a very high homology with the arginine biosyn- thetic gene cluster: argCJBDFR of C. glutamicum ATCC 13032 by blastn analysis in NCBI. This result indicated that the 6080 bp sequence of C. crenatum was the argin- ine biosynthetic gene cluster. The acession number for the sequence in Genbank is AY509864. 3.2. Sequence Analysis Analysis of the nucleotide sequence revealed the pres- ence of five intact open reading frames (ORFs): argJ, argB, argD, argF, argR, and partial of argC ORF (shown in Figure 3). Sequence analysis of the gene cluster indicated that the argB TAA termination coden was contiguous to the initiation codon of the argD gene, the distance between argD and argF was 13 bp, and argF gene was blocked off argR by 3 bp. This phenomenon suggested that the four ORFs located in the same transcription unit. Two relatively long intergenic spacers were found in the argC/argJ and argJ/argB, and a potential promoter re- gion existing in the upstream of argB gene was doped out, but there was no potential promoter existing in the upstream of argJ gene. The argB upstream sequence was shown in Figure 4. The interval between Sextama and Pribnow shown in under-double-line is 15 bp. A pair of inverse repeat sequence indicated by under-single-line is presumed to be operator region, also known as Arg box recognized by control protein. The Arg box consensus were described as TNTGA ATWWWWATTCANW in E. coli [8], CATGAATAAAA ATKCAAK in B. subtilis [9] and AWTGCATRWWYAT GCAWT in Streptomycetes [10] (where W = A or T, K = G or T, R = A or G, Y = T or C, N = any base). In Addi- tion, Binding of ArgR homologs to the sites similar to ARG boxes has been reported in Salmonella typhi- murium [13] and other Bacillus species (B. licheniformis [11] and B. stearothermophilus [12]). The most popular base of Arg boxes from the strains mentioned above wereA and T, whereas G and C in C. crenatum (as shown in Figure 4). The difference might be related with the control of arginine biosynthesis of corynebacterium and the distance of cognation. A stem-loop structure was found in the downstream of argC gene using RNAshapes tool. Two primary charac- ters of rho-independent terminator were appeared: a re- verse repeat sequence and the reverse repeat sequence mostly composed with G and C. The argC terminator region was shown in Figure 5. The downstream of rest genes have no fixed features, which implied that the transcription termination of the rest genes were relevant with the rho-factor. The C. crenatum argJ sequence, compared with blastp software in NCBI, shares 98.5, 70, 79 and 53% identical amino acids with the ornithine acetyltransferases (OATase) of C. glutamicum [2], C. diphtheriae gravis [14], C. efficiens [15] and M. tuberculosis [16], respec- tively. Sakanyan et al. reported that the OATase of C. glutamicum had only acetylorthine amidohydrolase function but no transacetylation by cloning of argJ gene for heterologous complementation of argA deficiency in E. coli [2]. The CLUSTAL alignment indicated that the similarity covered the whole argJ gene sequence be- tween C. crenatum and C. glutamicum. The result indi- cated that C. crenatum argJ coded OATase belongs to monofunctional enzymes. The C. glutamicum ArgJ molecular mass is 39.8 kDa, approximately 3 kDa less than the other known bacterial bifuctional OATases. Sakanyan et al held that the miss- ing 11-12 amino acids at the N-terminus was connected with the transacetylation deficiency via CLUSTAL alignment among C. glutamicum, B. sterothermophilus, B. subilis and Neisseria gonorrboeae [2]. In the present Figure 3. Genetic map of the 6.08 Kb stretch of C. crenatum A.S1.542 DNA. The long ar- rowheads indicated the orientation and location of the ORFs. Figure 4. Promoter sequence of arg B gene. 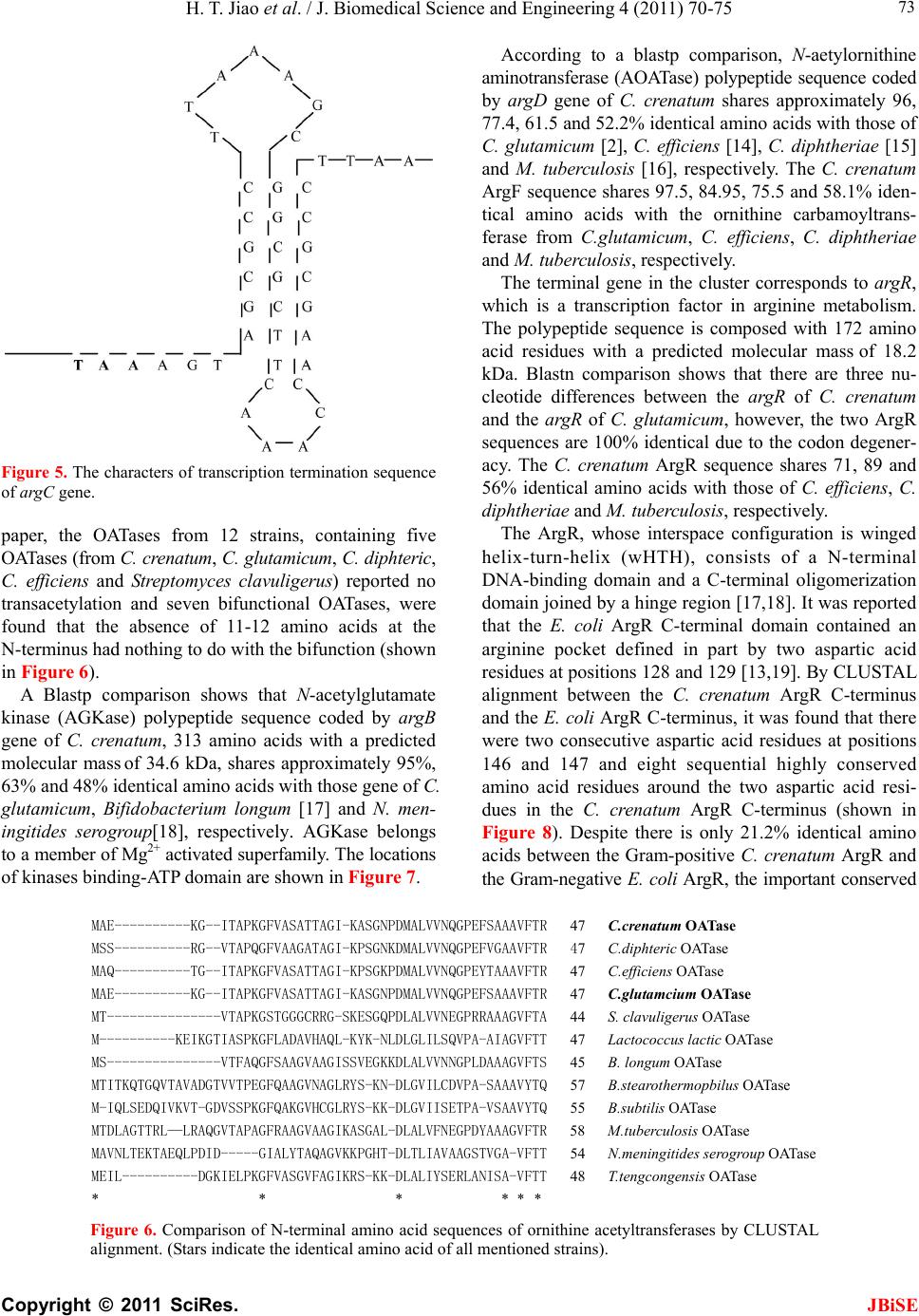 H. T. Jiao et al. / J. Biomedical Science and Engineering 4 (2011) 70-75 Copyright © 2011 SciRes. JBiSE 73 Figure 5. The characters of transcription termination sequence of argC gene. paper, the OATases from 12 strains, containing five OATases (from C. crenatum, C. glutamicum, C. diphteric, C. efficiens and Streptomyces clavuligerus) reported no transacetylation and seven bifunctional OATases, were found that the absence of 11-12 amino acids at the N-terminus had nothing to do with the bifunction (shown in Figure 6). A Blastp comparison shows that N-acetylglutamate kinase (AGKase) polypeptide sequence coded by argB gene of C. crenatum, 313 amino acids with a predicted molecular mass of 34.6 kDa, shares approximately 95%, 63% and 48% identical amino acids with those gene of C. glutamicum, Bifidobacterium longum [17] and N. men- ingitides serogroup[18], respectively. AGKase belongs to a member of Mg2+ activated superfamily. The locations of kinases binding-ATP domain are shown in Figure 7. According to a blastp comparison, N-aetylornithine aminotransferase (AOATase) polypeptide sequence coded by argD gene of C. crenatum shares approximately 96, 77.4, 61.5 and 52.2% identical amino acids with those of C. glutamicum [2], C. efficiens [14], C. diphtheriae [15] and M. tuberculosis [16], respectively. The C. crenatum ArgF sequence shares 97.5, 84.95, 75.5 and 58.1% iden- tical amino acids with the ornithine carbamoyltrans- ferase from C.glutamicum, C. efficiens, C. diphtheriae and M. tuberculosis, respectively. The terminal gene in the cluster corresponds to argR, which is a transcription factor in arginine metabolism. The polypeptide sequence is composed with 172 amino acid residues with a predicted molecular mass of 18.2 kDa. Blastn comparison shows that there are three nu- cleotide differences between the argR of C. crenatum and the argR of C. glutamicum, however, the two ArgR sequences are 100% identical due to the codon degener- acy. The C. crenatum ArgR sequence shares 71, 89 and 56% identical amino acids with those of C. efficiens, C. diphtheriae and M. tuberculosis, respectively. The ArgR, whose interspace configuration is winged helix-turn-helix (wHTH), consists of a N-terminal DNA-binding domain and a C-terminal oligomerization domain joined by a hinge region [17,18]. It was reported that the E. coli ArgR C-terminal domain contained an arginine pocket defined in part by two aspartic acid residues at positions 128 and 129 [13,19]. By CLUSTAL alignment between the C. crenatum ArgR C-terminus and the E. coli ArgR C-terminus, it was found that there were two consecutive aspartic acid residues at positions 146 and 147 and eight sequential highly conserved amino acid residues around the two aspartic acid resi- dues in the C. crenatum ArgR C-terminus (shown in Figure 8). Despite there is only 21.2% identical amino acids between the Gram-positive C. crenatum ArgR and the Gram-negative E. coli ArgR, the important conserved MAE----------KG--ITAPKGFVASATTAGI-KASGNPDMALVVNQGPEFSAAAVFTR 47 C.crenatum OATase MSS----------RG--VTAPQGFVAAGATAGI-KPSGNKDMALVVNQGPEFVGAAVFTR 47 C.diphteric OATase MAQ----------TG--ITAPKGFVASATTAGI-KPSGKPDMALVVNQGPEYTAAAVFTR 47 C.efficiens OATase MAE----------KG--ITAPKGFVASATTAGI-KASGNPDMALVVNQGPEFSAAAVFTR 47 C.glutamcium OATas e MT---------------VTAPKGSTGGGCRRG-SKESGQPDLALVVNEGPRRAAAGVFTA 44 S. clavuligerus OATase M----------KEIKGTIASPKGFLADAVHAQL-KYK-NLDLGLILSQVPA-AIAGVFTT 47 Lactococcus lactic OATase MS---------------VTFAQGFSAAGVAAGISSVEGKKDLALVVNNGPLDAAAGVFTS 45 B. longum OATase MTITKQTGQVTAVADGTVVTPEGFQAAGVNAGLRYS-KN-DLGVILCDVPA-SAAAVYTQ 57 B.stearothermopbilus OATase M-IQLSEDQIVKVT-GDVSSPKGFQAKGVHCGLRYS-KK-DLGVIISETPA-VSAAVYTQ 55 B.subtilis OATase MTDLAGTTRL—LRAQGVTAPAGFRAAGVAAGIKASGAL-DLALVFNEGPDYAAAGVFTR 58 M.tuberculosis OATase MAVNLTEKTAEQLPDID-----GIALYTAQAGVKKPGHT-DLTLIAVAAGSTVGA-VFTT 54 N.meningitides serogroup OATase MEIL----------DGKIELPKGFVASGVFAGIKRS-KK-DLALIYSERLANISA-VFTT 48 T.tengcongensis OATase * * * * * * Figure 6. Comparison of N-terminal amino acid sequences of ornithine acetyltransferases by CLUSTAL alignment. (Stars indicate the identical amino acid of all mentioned strains).  H. T. Jiao et al. / J. Biomedical Science and Engineering 4 (2011) 70-75 Copyright © 2011 SciRes. JBiSE 74 ### C. crenatum-AGKase AVRGGVSAAHVIDGRIAHSVLLELLTMGG IGTMVL 293 C. glutamicum-AGKase AVRGGVSAAHVIDGRIAHSVLLELLTMGG IGTMVL 293 C. efficiens-AGKase AVRGGVNAAHVIDGRIAHSVLLELLTMGG IGTMVL 293 C. diphteric-AGKase AVI HGVSAAHV IDGRVAHSVLLELLTSGGVGTMVV 293 E. coli-AGKase VNADQAATALAATLGADLILLSDVSGILDGKGQRIA 194 M. bovis-AGKase LRAVIGGVPSAHIIDGRVTHCVLVELFTDAGTGTKVV 292 Figure 7. AGKase shows feature of putative ATP-binding domain protein. (# indicates a putative ATP-binding site). ## E. coli-ArgR KNLVLDIDYNDAVVVIHTSFGAAQLITARLLDS L GKAEGI LGTI AGDDT C. crenatum-ArgR DELLVSTDHSGNI AMLRTPPGAAQYLASFI DRRVGLKE-VV GTIAGDDT Consensus L D T GAAQ A D G E GTIAGDDT E. coli-ArgR I FTTPANG FTVKALYEAILELFDQEL 156 C. crenatum-ArgR VFVLARDPLTGKELGELLSG- -RTT 171 Consensus F K L E Figure 8. Comparison of amino acids sequence of C-terminal domain of ArgR between C. cre- natum and E. coli (# indicates asparagine). region binding arginine is quite consistent. The result implied that the two genes originated by a duplication of some common ancestral gene. Although it was modified and changed by different host in far-flung evolvement course, the partial region determining function still kept highly conservative. 6. ACKNOWLEDGEMENTS The authors are thankful to the financial support from the National Natural Science Foundation of China (No. 30960012). REFERENCES [1] Gigot, D., Caplier, I., Str Vehary, S., Pavel, P., Michele, L. and Osberg, D. (1987) Amino-proximal sequences of the argF and argI ornithine carbamoyltransferases from Es- cherichia coli K-12. Archives Internationales de Physi- ologie de Biochimie et de Biophysique, 86, 913-915. [2] Sakanyan, V., Petrosyan, P., Lecocq, M., Boyen, A., Le- grain, C., Demarez, M., Hallet, J.N. and Glansdorff, N. (1996) Genes and enzymes of the acetyl cycle of arginine biosynthesis in Corynebacterium glutamicum: Enzyme evolution in the early steps of the arginine pathway. Mi- crobiology, 142, 99-108. doi:10.1099/13500872-142-1-99 [3] Unin, R. and Glansdorff, N. (1986) Biosynthesis and metabolism of arginine in bacteria. Microbiology Re- views, 50, 314-352. [4] Haas, D. and Kurer, V. (1982) N-acetylglutamate syn- thetase of Pseudomonas aeruginosa. An assay in vitro and feedback inhibition by arginine. European Journal of Biochemistry, 31, 290-295. doi:10.1111/j.1432-1033.1972.tb02531.x [5] Heimberg, H., Boyen, A., Crabeel, M. and Glansdorff, N. (1990) Escherichia coli and Saccharomyces cerevisiae acetylornithine aminotransferase: Evolutionary relation- ship with ornithine aminotransferase. Gene, 90, 69-78. [6] Hindle, Z. and Callis, R. (1994) Cloning and expression in Enterobacteria coli of a Streptomyces coelicolor A3(2) argCJB gene cluster. Microbiology, 140, 311-320. doi:10.1099/13500872-140-2-311 [7] Shengdong, L. (1999) Experiment technology of mole- cule biology. China Consonancy Medical University Publishing Company, Beijing. [8] Maas, W.K. (1994) The arginine repressor of Escherichia coli. Microbiology Reviews, 58, 631-640. [9] Miller, C.M., Baumberg, S. and Stockley, P. G. (1997) Operator interactions by the Bacillus subtilis arginine repressor/activator, AhrC: Novel positioning and DNA- mediated assembly of a transcriptional activator at cata- bolic sites. Molecular Microbiology, 26, 37-48. doi:10.1046/j.1365-2958.1997.5441907.x [10] Rodriguez-Garcia, A., Ludovice, M., Martin, J.F. and Liras, P. (1997) Arginine boxes and the argR gene in Streptomyces clavuligerus: Evidence for a clear regula- tion of the arginine pathway. Molecular Microbiology, 25, 219-228. doi:10.1046/j.1365-2958.1997.4511815.x [11] Maghnouj, A. and Sousa, C.T.F. (1998) The arcABDC gene cluster, encoding the arginine deiminase pathway of Bacillus licheniformis, and its activation by the arginine repressor argR. Journal of Bacteriology, 180, 6468-6475. [12] Dion, M. and Chalier, D. (1997) The highly thermostable arginine repressor of Bacillus stearothermophilus: gene cloning and repressor-operator interactions. Molecular Microbiology, 25, 385-398. doi:10.1046/j.1365-2958.1997.4781845.x [13] Kira, S.M., Andrey, A.M. and Mikhail S.G. (2001) Con- servation of the binding site for the arginine repressor in all bacterial lineages. Genome Biology, 2, 0013.1-0013.8. [14] Cerdeño-Tárraga, A.M., Efstratiou, A., Dover, L.G., Holden, M.T., Pallen, M., Bentley, S.D., Besra, G.S.,  H. T. Jiao et al. / J. Biomedical Science and Engineering 4 (2011) 70-75 Copyright © 2011 SciRes. JBiSE 75 Churcher, C., James, K.D., De Zoysa, A., Chillingworth, T., Cronin, A., Dowd, L., Feltwell, T., Hamlin, N., Hol- royd, S., Jagels, K., Moule, S., Quail, M.A., Rabbi- nowitsch, E., Rutherford, K.M., Thomson, N.R., Unwin, L., Whitehead, S., Barrell, B.G. and Parkhill, J. (2003) The complete genome sequence and analysis of Coryne- bacterium diphtheriae NCTC13129. Nucleic Acids Re- search, 22, 6516-6523. doi:10.1093/nar/gkg874 [15] Nishio, Y., Nakamura, Y., Kawarabayasi, Y., Usuda, Y., Kimura, E., Sugimoto, S., Matsui, K., Yamagishi, A., Kikuchi, H., Ikeo, K. and Gojobori, T. (2003) Compara- tive complete genome sequence analysis of the amino acid replacements responsible for the thermostability of Corynebacterium efficiens. Genome Research, 13, 1572- 1579. doi:10.1101/gr.1285603 [16] Cole, S.T., Brosch, R., Parkhill J., Garnier, T., Churcher, C., Harris, D., Gordon, S.V., Eiglmeier, K. and Gas, T. F. S. (1998) Deciphering the biology of Mycobacterium tu- berculosis from the complete genome sequence. Nature, 393, 537-544. doi:10.1038/31159 [17] Schell, M.A., Karmirantzou, M., Snel, B., Vilanova, D., Berger, B., Pessi, G., Zwahlen, M.C., Desiere, F., Bork, P., Delley, M., Pridmore, R.D. and Arigoni, F. (2002) The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proceed- ings of the National Academy of Sciences of the United States of America, 99, 14422-14427. doi:10.1073/pnas.212527599 [18] Parkhill, J., Achtman, M., Bentley, S.D., Churcher, C., Klee, S.R., Morelli, G., Basham, D., Brown, D., Chillingworth, T., Davies, R.M., Davis, P., Devlin, K., Feltwell, T., Hamlin, N., Holroyd, S., Jagels, K., Leather, S., Moule, S., Mungall, K., Quail, M.A., Rajandream, M.A., Rutherford, K.M., Simmonds, M., Skelton, J., Whitehead, S., Spratt, B.G. and Barrell, B.G. (2000) Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature, 404, 502-506. doi:10.1038/35006655 [19] Ghochikyan, A., Karaivanova, I.M., Lecocq, M., Vusio, P., Arnaud, M.C., Snapyan, M., Weigel, P., Guével, L., Buckle, M. and Sakanyan, V. (2002) Arginine Operator Binding by Heterologous and Chimeric ArgR Repressors from Escherichia coli and Bacillus stearothermophilus. J Bacteriol, 184, 6602-6614. doi:10.1128/JB.184.23.6602-6614.2002
|