Paper Menu >>
Journal Menu >>
 J. Biomedical Science and Engineering, 2009, 2, 128-134 Published Online April 2009 in SciRes. http://www.scirp.org/journal/jbise JBiSE Analysis and expression of the polyhedrin gene of Antheraea pernyi nucleopolyhedrovirus (AnpeNPV) Jia-Xi Huang1, Hui-Ling Wu1, Yan Wu1, Shan-Ying Zhu1, Wen-Bing Wang1 Institute of Life Sciences, Jiangsu University, 301 Xuefu Road, Zhenjiang 212013, P. R. China Received Dec. 3rd, 2008; revised Jan. 1st, 2009; accepted Jan. 5th, 2009 ABSTRACT The polyhedrin (polh) gene is often used to analyse evolution of baculovirus. In this report, the polh of Antheraea pernyi nucleopolyhedro- virus (AnpeNPV) was cloned and sequenced. The Open reading frame (ORF) of the AnpeNPV consists of 738 nucleotides encoding 245 amino acids with molecular masses of 29 kDa. The deduced amino acids were significant homol- ogy with other baculoviruses, such as Attacus ricini NPV (ArNPV) and Autographa californica NPV (AcNPV). A strongly hydrophilic region was predicted at positions from 30 to 50 of the An- peNPV Polh protein by bioinformatics analysis. Expression of the polh gene of AnpeNPV in E. coli was examined by SDS–PAGE, Western blot and Mass-spectrum analysis. The result showed that the bacterium expression system was suitable for the virus gene expression. It indi- cated that the products of the polh gene ex- pressed in this system can be easier to use for raising antibodies. Keywords: Antheraea Pernyi, Insect, Baculovi- rus, NPV, Polyhedrin, Prokaryotic Expression 1. INTRODUCTION The Chinese oak silkworm Antheraea pernyi (Lepidop- tera: Saturniidae) is an economically important insect primarily for the production of tussah silk. In recent years, consumption of the silkworm pupae as food deli- cacies has also gained tremendous popularity. The jaun- dice disease of the oak silkworm caused by the infection of A.pernyi nucleopolyhedrovirus (AnpeNPV) is a major threat to the tussah industry [1]. AnpeNPV is a member of the Baculoviridae with large, enveloped, dou- ble-stranded DNA. Baculoviridae are widely known to the scientific community in the form of commercial baculovirus expression vectors (BEVs) [2,3]. Baculovi- ruses also have an established application as insecticides against agricultural and forestry pests [4,5]. Currently, the Baculoviridae comprises two genera, Nucleopolyhe- drovirus (NPV) and Granulovirus (GV) [1]. During the infection cycle, NPVs produce two structurally and functionally distinct virion phenotypes: occlusion- de- rived virus (ODV) and budded virus (BV) [6]. The oc- cluded viruses of the NPV are referred to as polyhedra. Polyhedrin is the major protein component of the poly- hedra [7]. The polh gene is not essential for viral devel- opment, and normally deletion of the polh gene is not interfering with viral replication in cultured cells. How- ever, in per os infectivity, the polyhedra or occlusion bodies are required for the oral infection of insects [8]. Baculovirus entry into host cells involves that ODVs are released from the occlusion body by the alkaline envi- ronment within the midgut lumen of the larva and sub- sequently initiate primary infection of the mature co- lumnar epithelial cells of the midgut [6]. In order to explore effective propagation and infectiv- ity of the polyhedra, this paper analysised the nucleotide sequence and promoter (prmoter-Ap) of the polh gene of AnpeNPV by bioinformatics tools, and further prokary- otic expression for AnpeNPV polyhedrin (polh-Ap). 2. MATERIALS AND METHODS 2.1 Materials The Wild-type AnpeNPV strain was maintained in our laboratory. Restriction Enzymes, T4 DNA ligase, PCR reagents pMD18-T and DNA purification kit were pur- chased from TaKaRa Company (China, Dalian); primers and other reagents were bought from Shanghai Sangon Bio-technology Corpotation. The vectors for expression, and Escherichia coli strain DH5α and BL21 were kept in our laboratory. 2.2. Aplification of the AnpeNPV polh Gene AnpeNPV genomic DNA was isolated using the method described by previously [9,10] and about 15-20 ng DNA was used as template for standard PCR. The specific primers were designed based on the sequence of ORF (GenBank: EU195295). The polh-Ap forward primer (5' CCG GAA TTC ATG CCA GAT TAC TCA TAC CGG 3′)containing an EcoR I restriction site (underlined), and the reverse primer (5′ CCC AAG CTT CTA GTA CGC GGG GCC AGT 3′) containing a Hind III restriction site (underlined). The PCR conditions were 1 cycle at 94 °C for 5 min; 30 cycles at 94 °C for 45 s, 62°C for 45 s, and 72 °C for 1 min; and 1 cycle at 72 °C for 10 min. The PCR product was examined by electrophoresis in 1% agarose gel with the ethidium bromide staining. SciRes Copyright © 2009  129 J. X. Huang et al. / J. Biomedical Science and Engineering 2 (2009) 128-134 SciRes Copyright © 2009 JBiSE 2.3. Cloning and Construction of Expresion Plasmid The PCR products were ligated into pMD18-T vector using T4 DNA ligase and then transformed into E. coli (DH5a), and sequenced, respectively. The recombinant plasmid pMD-polh-Ap was digested with EcoR I and Hind III, and was purified to ligate with the Pet28a vector digested with EcoR I and Hind III, and transformed into E. coli (BL21). 2.4. Analysis of the polh Gene The amino acid sequence was deduced with Expasy Translate tool (http://au. expasy. org/tools/dna. html) according to the AnpeNPV polh gene sequence. Align using DNAstar CLUSTAL W program. Phylogenetic tree was made by MEGA 3.1 software. In order to explore regulatory sequence in the putative promoter region, NNPP (Promoter Predication by Neural Network http://www.fruitfly.org/seq_tools/promoter.html), promoter scan and transcription factor binding sites (http://www-bimas.cit.nih.gov/molbio/proscan/) were ap- plied together to make a comprehensive prediction. 2.5. Expression of the polh Gene in E.coli A positive clone was cultured in LB medium supplement with Kanamycin (final concentration of 50μg/ ml) ove- night at 37 ℃ with shaking, then the culture was added into 100mL fresh LB medium and cultured at 37℃ with shaking to A600 about 0.6. The culture was induced with IPTG (final concentration of 8 μg/ mL) and shaked at 30 ℃ for 10 hours. SDS polyacrylamide gel was used to analyze the expression in the Mini-Protein system (Bio-Rad, USA). After electrophoresis, the gel was stained with Coomassie Brilliant Blue R250 to visualize the protein bands. Protein samples were separated on SDS-10% polyacrylamide gels and transferred to PVDF membranes. Blots were soaked in TBST buffer (10 mmol/L Tris-HCl, pH 7.6, 0.15 mol/L NaCl, 0.1% Tween 20) with 5% nonfat dried milk. The antiserum against the His-Polh fusion protein (His antibody) at a dilution of 1:2,000 monoclonal antibody was added as the first antibody, followed by addition of 1:5,000 dilu- tion horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G as the secondary antibody. Blots were visualized with the Enhanced chemiluminescence Western blot kit (Amersham). The predicted Polh protein band was cut out for Mass-spectrum analysis. 3. RESULTS 3.1. Nucleotide and Amino Acid Sequence Analysis The ORF of cloned gene has two different nucleotides from the published sequence (DQ486030), but no amino acid residues were changed. The 738 nucleotides (in- cluding the stop codon TAG) encoded a putative peptide of 235 amino acids by an Expasy Translate tool. Figure 1. Nucleotide sequence and deduced amino acid sequence of the polyhedrin gene. The predicted amino acid is represented by the one letter code designation below the nucleotide sequence. The initiate and the stop codes are framed. ATGCCAGATTACTCATACCGGCCGACCATTGGTCGCACCTATGTGTACGACAACAAGTAT M P D Y S Y R P T I G R T Y V Y D N K Y TACAAAAACTTAGGGTCCGTCATTAAAAACGCCAAGCGCAAGAAGCATTTAGTCGAACAT Y K N L G S V I K N A K R K K H L V E H GAAGAGGAAGAAAAGCATTGGGATCCTTTAGACAATTACATGGTCGCGGAAGACCCTTTC E E E E K H W D P L D N Y M V A E D P F CTGGGGCCGGGTAAAAACCAAAAACTGACACTTTTCAAGGAAATCCGCAACGTTAAACCC L G P G K N Q K L T L F K E I R N V K P GACACAATGAAACTTATTGTCAACTGGAGCGGTAAAGAATTTCTGCGCGAAACTTGGACC D T M K L I V N W S G K E F L R E T W T CGTTTTGTTGAGGATAGCTTTCCGATTGTAAACGACCAAGAGGTCATGGATGTGTTCCTC R F V E D S F P I V N D Q E V M D V F L GTCATTAACCTGCGCCCCACGCGCCCCAACAGGTGCTACAAGTTCCTGGCGCAGCACGCG V I N L R P T R P N R C Y K F L A Q H A CTCAGATGGGACTGCGACTACGTGCCGCACGAGGTAATCCGCATTGTGGAGCCATCCTAC L R W D C D Y V P H E V I R I V E P S Y GTGGGCATGAACAACGAGTACAGAATTAGCCTCGCCAAGAAAGGCGGCGGCTGCCCCATC V G M N N E Y R I S L A K K G G G C P I ATGAACATTCACAGCGAGTACACCAACTCGTTTGAATCGTTTGTAAACCGCGTAATCTGG M N I H S E Y T N S F E S F V N R V I W GAGAACTTTTACAAGCCCATTGTGTACATTGGCACGGACTCGGGTGAGGAGGAGGAAATT E N F Y K P I V Y I G T D S G E E E E I CTCATCGAGGTTTCGCTTGTGTTCAAGGTCAAGGAGTTTGCGCCCGACGCGCCACTGTTT L I E V S L V F K V K E F A P D A P L F ACTGGCCCCGCGTACTAG T G P A Y  X. Huang et al. / J. Biomedical Science and Engineering 3 (2009) 128-134 130 SciRes Copyright © 2009 JBiSE The nucleotide sequence of Polh-Ap and its deduced amino acid sequence are shown in Figure 1. This de- duced polypeptide contains 16 strongly basic, 16 strongly acidic, 113 hydrophobic and 58 hydrophilic amino acids with the calculated molecular mass of 29 kDa, and the isoelectric point was of 6.1. 3.2. Protein and Homology Analysis Using BLAST software of NCBI to search for homology in the GenBank database, the deduced amino acid se- quence showed an identity of 97%, 98%, 98%, 97%, 93% and 89% to the corresponding genes of Attacus ricini NPV(ArNPV, AAP16625),Epiphyas postvittana NPV (EppoNPV, NP_203170), Maruca vitrata MNPV (YP_950731), Rachiplusia ou MNPV (RoMNPV, NP_702998) [11], Bombyx mori NPV (BmNPV, AAA 46734) [12] and Autographa californica NPV (AcNPV, NP_054037) [13], respectively. Comparison of the de- duced amino acid sequence with that of the correspond- ing genes of many species is shown in Figure 2. This protein was demonstrated to be highly conserved in baculoviruses. The predict of secondary structure for polh-Ap by CLC Protein Workbench 3.0.3.(Figure 4). There are 4 regions  131 J. X. Huang et al. / J. Biomedical Science and Engineering 2 (2009) 128-134 SciRes Copyright © 2009 JBiSE Figure 2. Alignment of the polyhedrin genes of baculoviruses, The sequences were aligned using DNAstar CLUSTAL W program. 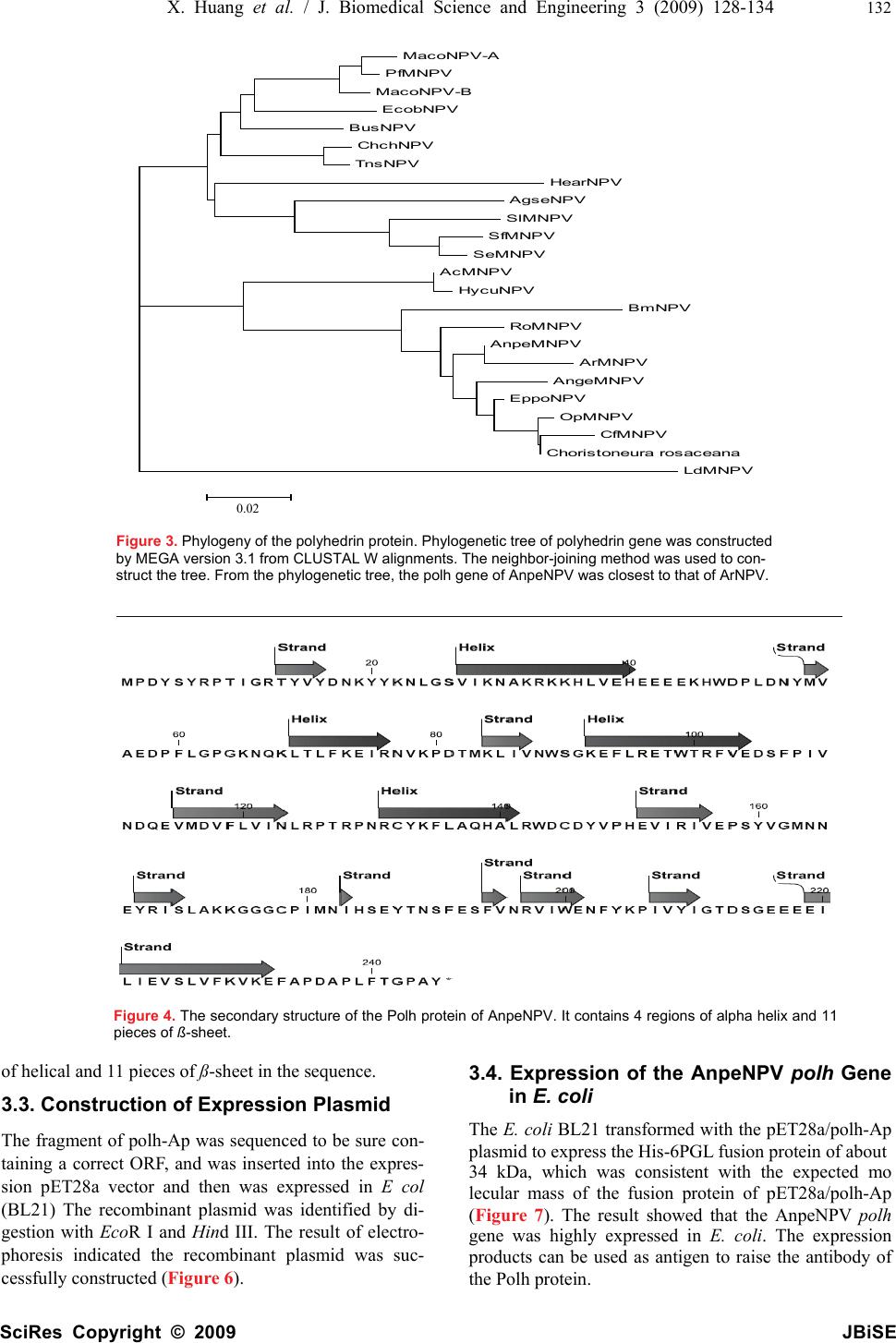 X. Huang et al. / J. Biomedical Science and Engineering 3 (2009) 128-134 132 SciRes Copyright © 2009 JBiSE MacoNPV-A PfMNPV MacoNPV-B EcobNPV BusNPV ChchNPV TnsNPV HearNPV AgseNPV SlMNPV SfMNPV SeMNPV AcMNPV HycuNPV BmNPV RoMNPV AnpeMNPV ArMNPV AngeMNPV EppoNPV OpMNPV CfMNPV Choristoneura rosaceana LdMNPV 0.02 Figure 3. Phylogeny of the polyhedrin protein. Phylogenetic tree of polyhedrin gene was constructed by MEGA version 3.1 from CLUSTAL W alignments. The neighbor-joining method was used to con- struct the tree. From the phylogenetic tree, the polh gene of AnpeNPV was closest to that of ArNPV. Figure 4. The secondary structure of the Polh protein of AnpeNPV. It contains 4 regions of alpha helix and 11 pieces of ß-sheet. of helical and 11 pieces of ß-sheet in the sequence. 3.3. Construction of Expression Plasmid The fragment of polh-Ap was sequenced to be sure con- taining a correct ORF, and was inserted into the expres- sion pET28a vector and then was expressed in E col (BL21) The recombinant plasmid was identified by di- gestion with Eco R I and Hind III. The result of electro- phoresis indicated the recombinant plasmid was suc- cessfully constructed (Figure 6). 3.4. Expression of the AnpeNPV polh Gene in E. coli The E. coli BL21 transformed with the pET28a/polh-Ap plasmid to express the His-6PGL fusion protein of about 34 kDa, which was consistent with the expected mo lecular mass of the fusion protein of pET28a/polh-Ap (Figure 7). The result showed that the AnpeNPV polh gene was highly expressed in E. coli. The expression products can be used as antigen to raise the antibody of the Polh protein. 0.02 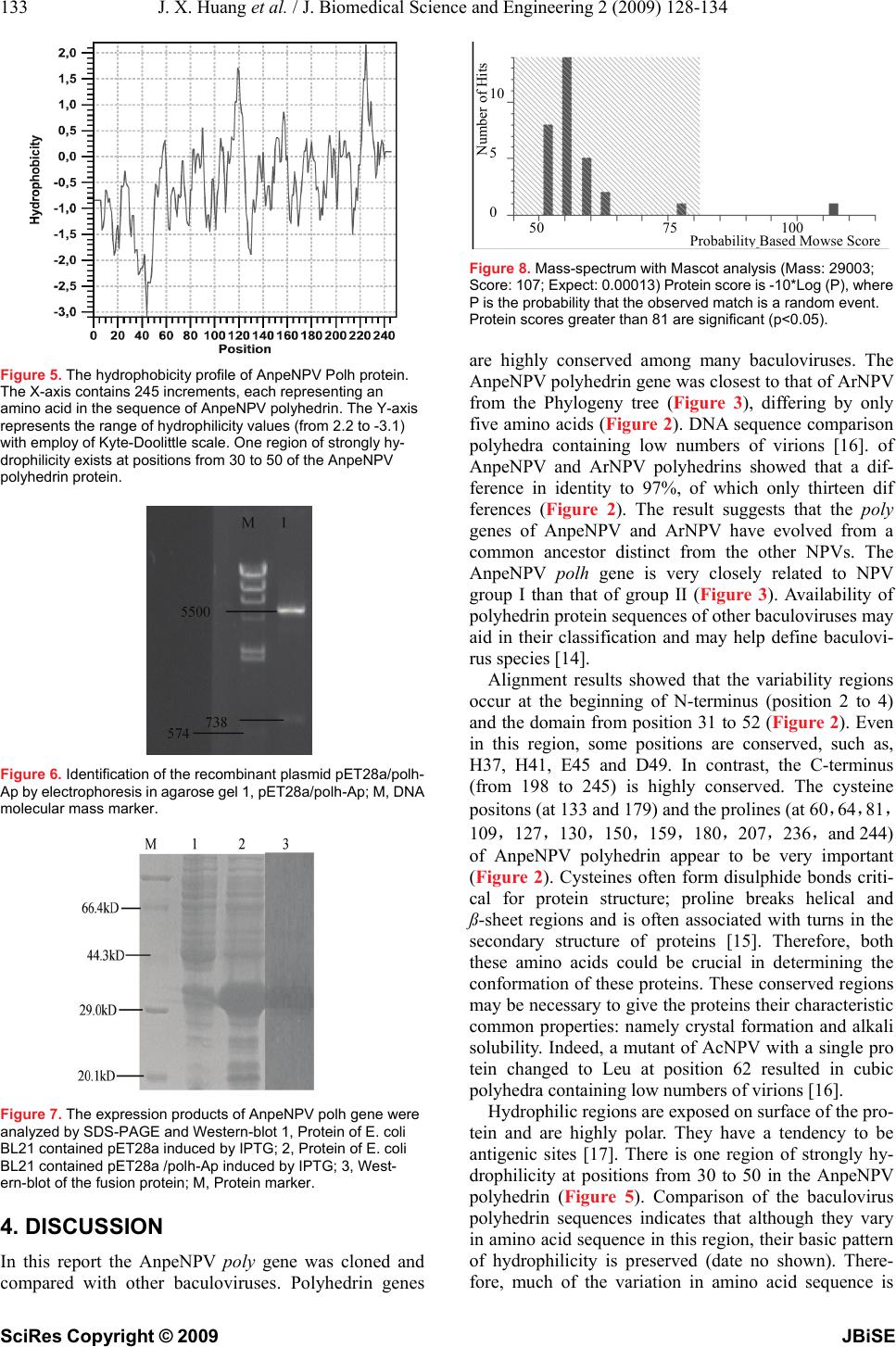 133 J. X. Huang et al. / J. Biomedical Science and Engineering 2 (2009) 128-134 SciRes Copyright © 2009 JBiSE Figure 5. The hydrophobicity profile of AnpeNPV Polh protein. The X-axis contains 245 increments, each representing an amino acid in the sequence of AnpeNPV polyhedrin. The Y-axis represents the range of hydrophilicity values (from 2.2 to -3.1) with employ of Kyte-Doolittle scale. One region of strongly hy- drophilicity exists at positions from 30 to 50 of the AnpeNPV polyhedrin protein. Figure 6. Identification of the recombinant plasmid pET28a/polh- Ap by electrophoresis in agarose gel 1, pET28a/polh-Ap; M, DNA molecular mass marker. Figure 7. The expression products of AnpeNPV polh gene were analyzed by SDS-PAGE and Western-blot 1, Protein of E. coli BL21 contained pET28a induced by IPTG; 2, Protein of E. coli BL21 contained pET28a /polh-Ap induced by IPTG; 3, West- ern-blot of the fusion protein; M, Protein marker. 4. DISCUSSION In this report the AnpeNPV poly gene was cloned and compared with other baculoviruses. Polyhedrin genes Figure 8. Mass-spectrum with Mascot analysis (Mass: 29003; Score: 107; Expect: 0.00013) Protein score is -10*Log (P), where P is the probability that the observed match is a random event. Protein scores greater than 81 are significant (p<0.05). are highly conserved among many baculoviruses. The AnpeNPV polyhedrin gene was closest to that of ArNPV from the Phylogeny tree (Figure 3), differing by only five amino acids (Figure 2). DNA sequence comparison polyhedra containing low numbers of virions [16]. of AnpeNPV and ArNPV polyhedrins showed that a dif- ference in identity to 97%, of which only thirteen dif ferences (Figure 2). The result suggests that the poly genes of AnpeNPV and ArNPV have evolved from a common ancestor distinct from the other NPVs. The AnpeNPV polh gene is very closely related to NPV group I than that of group II (Figure 3). Availability of polyhedrin protein sequences of other baculoviruses may aid in their classification and may help define baculovi- rus species [14]. Alignment results showed that the variability regions occur at the beginning of N-terminus (position 2 to 4) and the domain from position 31 to 52 (Figure 2). Even in this region, some positions are conserved, such as, H37, H41, E45 and D49. In contrast, the C-terminus (from 198 to 245) is highly conserved. The cysteine positons (at 133 and 179) and the prolines (at 60,64,81, 109,127,130,15 0,159,180,207,236,and 244) of AnpeNPV polyhedrin appear to be very important (Figure 2). Cysteines often form disulphide bonds criti- cal for protein structure; proline breaks helical and ß-sheet regions and is often associated with turns in the secondary structure of proteins [15]. Therefore, both these amino acids could be crucial in determining the conformation of these proteins. These conserved regions may be necessary to give the proteins their characteristic common properties: namely crystal formation and alkali solubility. Indeed, a mutant of AcNPV with a single pro tein changed to Leu at position 62 resulted in cubic polyhedra containing low numbers of virions [16]. Hydrophilic regions are exposed on surface of the pro- tein and are highly polar. They have a tendency to be antigenic sites [17]. There is one region of strongly hy- drophilicity at positions from 30 to 50 in the AnpeNPV polyhedrin (Figure 5). Comparison of the baculovirus polyhedrin sequences indicates that although they vary in amino acid sequence in this region, their basic pattern of hydrophilicity is preserved (date no shown). There- fore, much of the variation in amino acid sequence is N umber of Hits 10 5 0 50 75 100 Probabilit y Based Mowse Score  X. Huang et al. / J. Biomedical Science and Engineering 3 (2009) 128-134 134 SciRes Copyright © 2009 JBiSE neutral and does not alter the overall nature of the pro- teins. This region therefore presents a potential antigenic site which may be useful for production of antibodies capable of differentiating or identifying different bacu- loviruses [18]. Ultimately predicted antigenic determi- nants from proteins of pathogenic organisms might also be useful in the production of synthetic vaccines [17]. The polh gene of baculovirus is a very late gene which expressed in late stage of virus infection. It is not an essential gene in virion development and could be de- leted for foreign gene expression [19,20]. Some evi- dences showed that the level of the foreign gene expres- sion was related to genetic codes of the gene. To test the polh gene expression in another system, we constructed a bacterium expression system to express the Polh pro- tein. The result indicates that this gene is suitable for E. coli expression system. It might be helpful to produce the virus proteins to raise antibodies. ACKNOWLEDGEMENTS This work was supported by the 973 National Basic Research Program of China (2005CB121005); The Six-Field Top programs of Jiangsu Provice; National Natural Science Foundation of Jiangsu Education Communitte(06KJD180043); Innovation Foundation for Graduate Students of Jiangsu Province. REFERENCES [1] Q. Fan, S. Li, L. Wang, B. Zhang, B. Ye, Z. Zhao, Cui, L. (2007). The genome sequence of the multinucleocapsid nucleo- polyhedrovirus of the Chinese oak silkworm Antheraea pernyi. Virology 366(2), 304-315. [2] O.A. Lihoradova, I. D. Ogay, A. A. Abdukarimov, S. S. Azi- mova, D. E. Lynn, Slack, J. M. (2007). The Homingbac bacu- lovirus cloning system: An alternative way to introduce foreign DNA into baculovirus genomes. J Virol Methods 140 (1-2), 59-65. [3] Z. M. Nie, Z. F. Zhang, D. Wang, P. A. He, C. Y. Jiang, L. Song, F. Chen, J. Xu, L. Yang, L. L. Yu, J.Chen, Z. B. Lv, J. J. Lu, X. F. Wu, Zhang Y. Z. (2007) Complete sequence and organization of Antheraea pernyi nucleopolyhedrovirus, a dr-rich baculovirus. BMC Genomics 8, 248-261. [4] S. P. Cook, R. E. Webb, J. D. Podgwaite, Reardon, R. C. (2003) Increased mortality of gypsy moth Lymantria dispar (L.) (Lepi- doptera: Lymantriidae) exposed to gypsy moth nuclear polyhe- drosis virus in combination with the phenolic gycoside salicin. J Econ Entomol 96(6), 1662-1667. [5] Moscardi, F. (1999) Assessment of the application of baculovi- ruses for control of Lepidoptera. Annu Rev Entomol 44, 257-289. [6] X. Dai, T. M. Stewart, J. A. Pathakamuri, Q. Li, Theilmann, D. A. (2004) Autographa californica multiple nucleopolyhedrovirus exon0 (orf141), which encodes a RING finger protein, is re- quired for efficient production of budded virus. J Virol 78(18), 9633-9644. [7] S. G. Kamita, S. Maeda, Hammock, B. D. (2003) High-fre- quency homologous recombination between baculoviruses in- volves DNA replication. J Virol 77(24), 13053-13061. [8] A. M. Khurad, A. Mahulikar, M. K. Rathod, M. M. Rai, S. Kanginakudru, Nagaraju J. (2004) Vertical transmission of nu- cleopolyhedrovirus in the silkworm, Bombyx mori L. Journal of Invertebrate Pathology 87, 8–15. [9] S. Gomi, C. E. Zhou, W. Y. Yih, K. Majima, Maeda S. (1997) Deletion analysis of four of eighteen late gene expression factor gene homologues of the baculovirus, BmNPV. Virology 230, 35-47. [10] W. B. Wang, S. Y. Zhu, L. Q. Wang, F. Yu, Shen W. D. (2005) Cloning and sequence analysis of the Antheraea pernyi nucleo- polyhedrovirus gp64 gene. J Biosci 30, 605-610. [11] L. H. Robert, Bonning B.C. (2003) Comparative analysis of the genomes of Rachiplusia ou and Autographa californica multiple nucleopolyhedroviruses Journal of General Virology 84, 1827–1842. [12] S. Gomi, K. Majima, Maeda S. (1999) Sequence analysis of the genome of Bombyx mori nucleopolyhedrovirus. J Gen. Virol. 80, 1323-1337. [13] M. D. Ayres, S. C. Howard, J. Kuzio, M. Lopez-Ferber, Possee R.D. (1994) The complete DNA sequence of Auto- grapha californica nuclear polyhedrosis virus. Virology 202, 586-605. [14] E. A. van Strien, D. Zuidema, R.W. Goldbach, Vlak, J. M. (1992) Nucleotide sequence and transcriptional analysis of the polyhedrin gene of Spodoptera exigua nuclear polyhedrosis vi- rus. J Gen Virol 73 ( Pt 11), 2813-2821. [15] P. Y. Chou, Fasman, G. D. (1977) Beta-turns in proteins. J Mol Biol 115(2), 135-175. [16] E. B. Carstens, A. Krebs, Gallerneault, C. E. (1986) Identifica- tion of an amino acid essential to the normal assembly of Auto- grapha californica nuclear polyhedrosis virus polyhedra. J Virol 58(2), 684-688. [17] T. P. Hopp, Woods, K. R. (1981). Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci U S A 78(6), 3824-3828. [18] Rohrmann, G. F. (1986) Polyhedrin structure. J Gen Virol 67(8), 1499-1513. [19] R. D. Possee, S. C. Howard, (1987) Analysis of the polyhedrin gene promoter of the Autographa californica nuclear polyhedro- sis virus. Nucleic Acids Res 15(24), 10233-10248. [20] G. E. Smith, M. J. Fraser, Summers, M. D. (1983) Molecular engineering of the Autographa californica nuclear polyhedrosis virus genome:deletion mutations within the polyhedrin gene. J Virol 46, 584-593. |

