Paper Menu >>
Journal Menu >>
 Journal of Biomaterials and Nanobiotechnology, 2011, 2, 41-48 doi:10.4236/jbnb.2011.21006 Published Online January 2011 (http://www.SciRP.org/journal/jbnb) Copyright © 2011 SciRes. JBNB 41 Simple Modifications to Standard TRIzol® Protocol Allow High-Yield RNA Extraction from Cells on Resorbable Materials Juliana Tsz Yan Lee1, Wai Hung Tsang2,3, King Lau Chow1,2,3 1Bioengineering Graduate Program, The Hong Kong University of Science and Technology, Hong Kong, China; 2Department of Biology, The Hong Kong University of Science and Technology, Hong Kong, China; 3Division of Life Science, The Hong Kong University of Science and Technology, Hong Kong, China. Email: juliana@ust.hk Received October 14th, 2010; revised November 25th, 2010; accepted December 3rd, 2010. ABSTRACT Resorbable bioceramics are attractive for medical applications such as bone substitution. Biochemical analysis on cells cultured on these biomaterials is vital to predict the impact of the materials in vivo and RNA extraction is an essential step in gene expression study using RT-qPCR. In this study, we describe simple modifications to the TRIzol® RNA ex- traction protocol widely used in biology and these allow high-yield extraction of RNA from cells on resorbable calcium phosphates. Without the modifications, RNA is trapped in the co-precipitated calcium compounds, rendering TRIzol® extraction method infeasible. Among the modifications, the use of extra TRIzol® to dilute the lysate before the RNA pre- cipitation step is critical for extraction of RNA from porous -tricalcium phosphate ( -TCP) discs. We also investigate the rationale behind the undesirable precipitation so as to provide clues about the modifications required for other re- sorbable materials with high application potential in bone tissue engineering. Keywords: Calcium Phosphate, Resorbable Materials, RNA Extraction, TRIzol, Acid Guanidium Thiocyanate - Phenol - Chloroform Extraction, TRI Reagent, TRIsure 1. Introduction Resorbable bioceramics with relatively high solubility are attractive materials to be used as bone substitutes since the implanted materials can eventually be replaced by bone newly formed in vivo [1-4]. In order to under- stand the cell-material interaction, gene expression of cells cultured on the materials is often monitored by means of RT-qPCR as several genes can be assessed at the same time. RNA extraction is an important step for RT-qPCR, especially for reliable quantification of gene expression level. TRIzol®, TRI Reagent® and TRIsureTM are commonly used commercial products from different companies for RNA extraction. They are modified ver- sions of the single-step acid guanidium thiocyanate- phenol- chloroform extraction technique developed by Chomczynski and Sacchi, which is particularly useful for processing of large numbers of samples and for isolation of RNA from minute quantities of cells or tissue samples [5,6]. Nevertheless, this method cannot be applied to re- sorbable calcium phosphate ceramics directly as ions dissolved from the ceramics would co-precipitate with the RNA in the alcohol precipitation step, which prevents the RNA from being re-dissolved into aqueous solution in subsequent steps. The silica-gel affinity based method may be a feasible alternative for extracting RNA from resorbable -tricalcium phosphate (-TCP) [7,8] but the cost is usually much higher. Besides, TRIzol® gives a yield of 2.4 to 93 times higher than that using silica based protocol as demonstrated in reports using different biological samples [9-11]. High yield in RNA extraction is of particular importance in certain biomaterial studies due to the limited supply of expensive biomaterial scaf- folds and the maximization of the number of samples that can be handled in each batch of experiment using cell culture. Furthermore, higher RNA concentration is at- tainable using this method even for the same yield of RNA compared with silica gel based methods as the pel- leted RNA can be dissolved in a small volume (10 µL) of buffer while a relatively large volume (100 µL) of buffer  Simple Modifications to Standard TRIzol® Protocol Allow High-Yield RNA Extraction from Cells on Resorbable Materials Copyright © 2011 SciRes. JBNB 42 is required for efficient elution of RNA from the silica gel. In order to use TRIzol® to extract RNA from cells on resorbable biomaterials, we carried out studies to devise modifications to standard protocol, which allow high quality RNA to be extracted from cells grown on -TCP. In addition, modifications that may be required for other resorbable materials were suggested based on analysis of the parameters affecting the undesirable co-precipitation. 2. Materials and Methods 2.1. Materials, Cell Culture and RNA Extraction 10mm porous α-TCP discs were synthesized by foaming method using 5% hydrogen peroxide (H2O2) solution as described [12] in the National Engineering Research Center for Biomaterials in Sichuan University, China. For cell culture study, 9 × 104 C3H10T1/2 cells (pluripo- tent mesenchymal stem cells, CCL-226™, ATCC) were plated on the -TCP disc and subjected to RNA extrac- tion using TRIzol® reagent (Invitrogen) after 6 days of culture according to the manufacturer’s instruction ex- cept drying the disc with absorbent paper and crushing the discs in TRIzol® with subsequent TCP debris re- moval by centrifugation during the homogenization step. Without further modifying the protocol, undesirable co-precipitation occurred in the RNA precipitation step. 2.2. Analysis on Precipitation We studied the chemical composition of the precipitate by energy-dispersive X-ray spectroscopy (EDX) after sputtering with gold and the crystal morphology by scan- ning electron microscopy (SEM; JSM-6390, JEOL, Ja- pan). To study the parameters affecting the undesirable co-precipitation process, different salts were added dur- ing the phase separation step of the TRIzol® protocol using cell free system as shown in Table 3 followed by studies on effects of dilution, pH and temperature (Table 4). Based on results from these experiments, further modifications to the protocol were made and RNA could be extracted eventually. 2.3. RNA Quality Check The quality and yield of the extracted RNA were as- sessed through gel electrophoresis and UV spectrometry (NanoDrop 1000, Thermo Scientific). The compatibility of the extracted RNA with RT-PCR and RT-qPCR, commonly used techniques for gene expression study, were also tested with the following procedures. DNase I treatment was performed with 500ng RNA using Ampli- fication Grade DNase I (Invitrogen) and reverse-trans- cribed (RT) into single stranded cDNA using High Ca- pacity RNA-to-cDNA Master Mix (Applied Biosystems) according to the manufacturers’ protocol with 5L and 20L reaction volumes respectively. PCR was performed with β-Actin and Col1 genes using primer pairs listed in Table 1 with an annealing temperature of 53°C for both pairs. qPCR was performed with a total reaction volume of 10 L, containing 1 L of single stranded cDNA, 5 L of TaqMan® Gene Expression Master Mix (Applied Biosystems) and 0.5 L of TaqMan® Gene Expression Assays (Applied Biosystems) in Table 2, in 96 well re- action plates (2 L of cDNA was used for the Ocn quan- tification due to the lower Ocn transcript level in the samples). 3. Results and Discussion 3.1. Overview Figure 1 shows the standard TRIzol® RNA extraction procedures with modifications required for extraction of RNA from cells on resorbable materials. Using the stan- dard TRIzol® RNA extraction protocol as the basis, RNA extraction was started with the porous TCP discs con Table 1. Primers used in RT-PCR. Gene (Gene bank ID) Primer sequence Genomic amplicon cDNA amplicon 5' ATGGATGACGATATCGCTG 3' Mouse β-Actin (NM_007393.2) 5' ATGAGGTAGTCTGTCAGGT 3' 1110 bp 569 bp 5' CCCAGAACATCACCTATCAC 3' Mouse Col1 (NM_00742.3) 5' TTGGTCACGTTCAGTTGGTC 3' 639 bp 510 bp Table 2. Taqman® Gene Expression Assays used in RT-qPCR. Gene Gene symbol Assay ID Amplicon lengthReference sequence -Actin Actb Mm02619580_g1 143bp NM_007393.3 Cbfa1 Runx2 Mm01269515_mH 74bp NM_009820.4 Col1 Col1a1 Mm00801666_g1 89bp NM_007742.3 Alpl Alpl Mm00475834_m1 65bp NM_007431.2 Ocn Bglap-rs1 Mm00649782_gH 89bp NM_031368.4 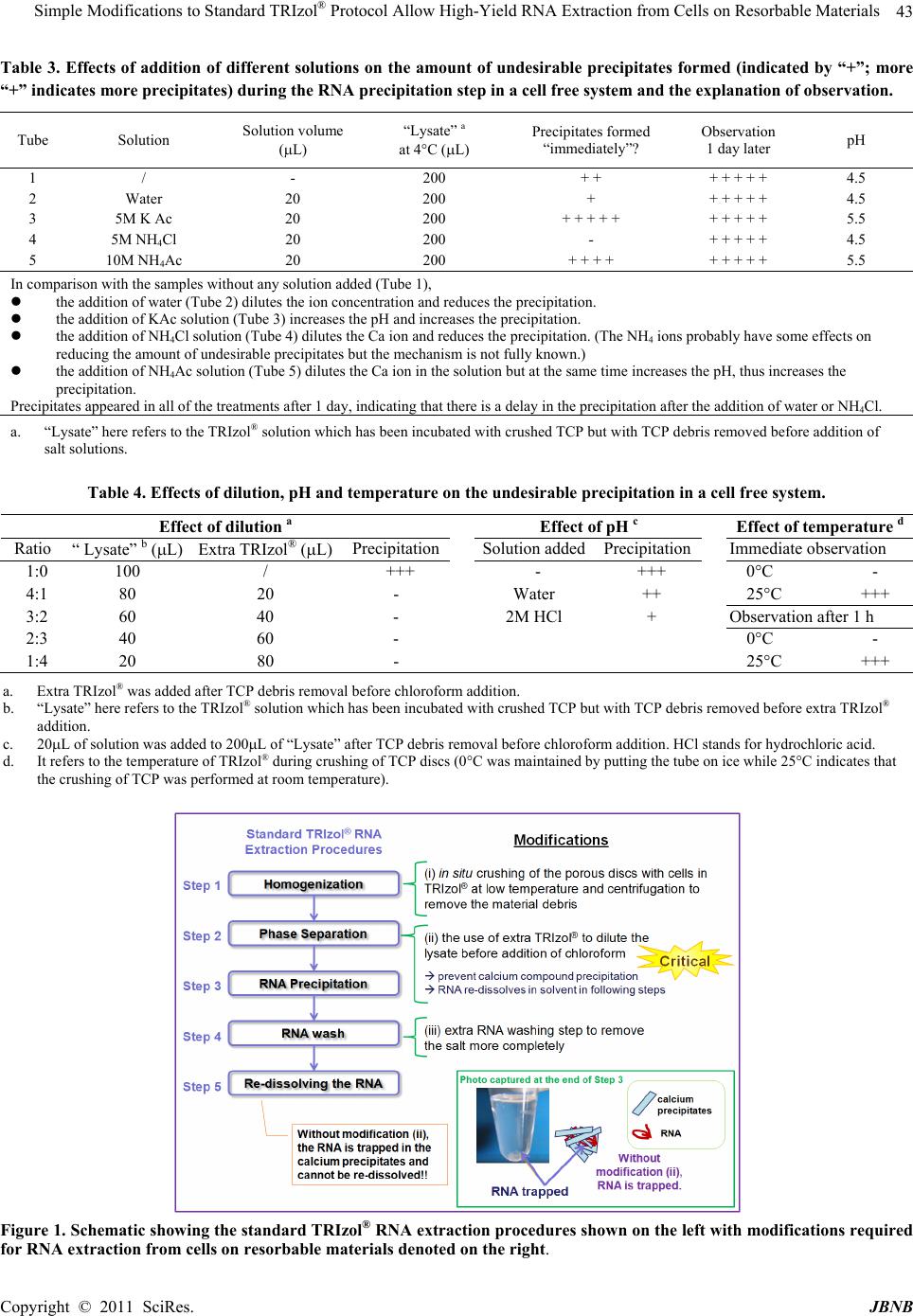 Simple Modifications to Standard TRIzol® Protocol Allow High-Yield RNA Extraction from Cells on Resorbable Materials Copyright © 2011 SciRes. JBNB 43 Table 3. Effects of addition of different solutions on the amount of undesirable precipitates formed (indicated by “+”; more “+” indicates more precipitates) during the RNA precipitation step in a cell free system and the explanation of observation. Tube Solution Solution volume (L) “Lysate” a at 4°C (L) Precipitates formed “immediately”? Observation 1 day later pH 1 / - 200 + + + + + + + 4.5 2 Water 20 200 + + + + + + 4.5 3 5M K Ac 20 200 + + + + + + + + + + 5.5 4 5M NH4Cl 20 200 - + + + + + 4.5 5 10M NH4Ac 20 200 + + + + + + + + + 5.5 In comparison with the samples without any solution added (Tube 1), the addition of water (Tube 2) dilutes the ion concentration and reduces the precipitation. the addition of KAc solution (Tube 3) increases the pH and increases the precipitation. the addition of NH4Cl solution (Tube 4) dilutes the Ca ion and reduces the precipitation. (The NH4 ions probably have some effects on reducing the amount of undesirable precipitates but the mechanism is not fully known.) the addition of NH4Ac solution (Tube 5) dilutes the Ca ion in the solution but at the same time increases the pH, thus increases the precipitation. Precipitates appeared in all of the treatments after 1 day, indicating that there is a delay in the precipitation after the addition of water or NH4Cl. a. “Lysate” here refers to the TRIzol® solution which has been incubated with crushed TCP but with TCP debris removed before addition of salt solutions. Table 4. Effects of dilution, pH and temperature on the unde sirable precipitation in a cell free system . Effect of dilution a Effect of pH c Effect of temperature d Ratio “ Lysate” b (L) Extra TRIzol® (L) Precipitation Solution addedPrecipitation Immediate observation 1:0 100 / +++ - +++ 0°C - 4:1 80 20 - Water ++ 25°C +++ 3:2 60 40 - 2M HCl + Observation after 1 h 2:3 40 60 - 0°C - 1:4 20 80 - 25°C +++ a. Extra TRIzol® was added after TCP debris removal before chloroform addition. b. “Lysate” here refers to the TRIzol® solution which has been incubated with crushed TCP but with TCP debris removed before extra TRIzol® addition. c. 20L of solution was added to 200L of “Lysate” after TCP debris removal before chloroform addition. HCl stands for hydrochloric acid. d. It refers to the temperature of TRIzol® during crushing of TCP discs (0°C was maintained by putting the tube on ice while 25°C indicates that the crushing of TCP was performed at room temperature). Figure 1. Schematic showing the standard TRIzol® RNA extraction procedures shown on the left with modifications required for RNA extraction from cells on resorbable materials denoted on the right.  Simple Modifications to Standard TRIzol® Protocol Allow High-Yield RNA Extraction from Cells on Resorbable Materials Copyright © 2011 SciRes. JBNB 44 taining the attached cells crushed in TRIzol®. TCP debris was removed by centrifugation in Step 1 (Homogeniza- tion). Step 2 (Phase separation) and Step 3 (RNA pre- cipitation using isopropanol) were performed as de- scribed by the manufacturer. Using this procedure, unde- sirable co-precipitate appeared, making the precipitated RNA unable to be dissolved in Step 5. Thus no RNA could be extracted. Analysis of the undesirable precipita- tion which eventually leads to the suggested modifica- tions will be explained in the following. 3.2. Identification of the Undesirable Precipitate To identify the components of the undesirable precipi- tate, we studied the morphology and the composition of the precipitate by SEM and EDX. SEM examination shows that the precipitate is in the form of clustered crystals (Figure 2(a)). From the EDX analysis (Figures 2(b) and 2(c)), the precipitate was found to be mainly composed of Ca, O, C and N. Calcium thiocyanate (Ca(SCN)2) and calcium acetate (Ca(C2H3O2)2) may be the possible constituents of the precipitate, leading to the interpretation of having the precipitate coming from the dissolution of the calcium containing biomaterials during the TRIzol® mediated cell lysis step, rather than being the direct precipitation of components present in the TRIzol® reagent. 3.3. Parameters Affecting the Undesir able Co-Precipitation Different approaches to get rid of the undesirable co-precipitates were attempted and the key experiments leading to the modifications are highlighted in this report. In molecular biology, precipitation with ethanol is the standard method to recover nucleic acids from aqueous solutions and ammonium acetate is frequently used to reduce the co-precipitation of unwanted contaminants such as dNTPs or oligosaccharides with nucleic acids [13]. This ethanol precipitation step has high similarity to the RNA precipitation step using isopropanol in the TRIzol® protocol. And having known that most ammo- nium, potassium, acetate and chloride compounds have high solubility in aqueous solutions [14], we attempted to avoid the undesirable co-precipitation by adding different salt solutions in the phase separation step (Step 2 of Fig- ure 1). Cell-free system was used to simplify the analysis and the results together with the explanation of observa- tion are summarised in Table 3. The addition of ammo- nium chloride solution eliminates the undesirable pre- cipitation but more importantly the results revealed that dilution and pH are critical parameters in the precipita- tion process. Besides, we proposed that lower tempera- ture would reduce the precipitation by slowing down the dissolution of calcium ions from TCP. Hence, we carried out experiments to investigate the effects of these three parameters: dilution, pH and temperature. 3.3.1. Effect of Dilution From Table 3, the addition of water reduces the precipi- tation but it also dilutes other salts in the TRIzol® which may affect the phase equilibrium in the phase separation step and hence affects the normal RNA precipitation process. Thus we have experimented using TRIzol® to dilute the Ca ions after TCP debris removal. Surprisingly, the addition of as little as 1/4 volume of TRIzol® already eliminates the unwanted precipitation (Table 4), proba- bly by preventing the solution from being saturated in one of the insoluble calcium compounds. For more solu- ble materials, larger extra TRIzol® to lysate ratio may be required. 3.3.2. Effect of pH Besides, the addition of HCl can reduce the precipitates formed (Table 4). This observation indicates that lower pH increases the solubility of the insoluble compounds, thus less precipitates would be formed. Therefore we can reduce the amount of precipitates by lowering the pH during the RNA precipitation step. On the other hand, increasing the pH would lower the solubility of the cal- cium phosphate materials [15], which in turn may reduce the dissolution of Ca ions from materials that are more soluble than TCP. By using “self-made TRIzol® reagent” (a) (b) (c) Figure 2. (a) SEM photo of the precipitate, (b) EDX spectrum and (b) Atomic concentration of the precipitate in elemental analysis using EDX (average ± SD). Element Atomic % (average ± SD) O 40.59 ±0.05 C 29.53 ±0.85 N 16.78 ±0.62 Ca 11.35 ±0.27 Au 1.76 ±0.02  Simple Modifications to Standard TRIzol® Protocol Allow High-Yield RNA Extraction from Cells on Resorbable Materials Copyright © 2011 SciRes. JBNB 45 (with all the components in TRIzol® reagent except so- dium acetate) at neutral pH to lyse the cells on the re- sorbable materials and adjusting the pH after material de- bris removal by using sodium acetate (2 M, pH 4) (50 µL added per 1 mL), the undesirable co-precipitates can probably be reduced. 3.3.3. Effect of Temperature Since most chemical reactions are slower at lower tem- -perature, we have proposed that lower temperature would reduce the undesirable precipitation by slowing down the dissolution of Ca ions from TCP. This is con- firmed by experiments summarized in Table 4. Hence, it is better to use chilled TRIzol® and keep the TRIzol® with -TCP on ice during the crushing of materials so as to reduce the amount of Ca ions dissolved from the -TCP. 3.4. Suggestions on Modifications From the above experiments, we summarized the factors affecting the undesirable precipitation in Table 5. Based on this, we proposed the modifications required for RNA extraction from cells on porous -TCP discs as shown on the right of Figure 1. (A sample protocol was attached to the end of this article). On the other hand, TCP is only one of the promising resorbable materials, while other resorbable materials may have high application potential in the future. Different materials differ greatly in solubil- ity in aqueous solution (Table 5) [16]. Analysis in Table 5 also suggests that increasing the extra TRIzol® to lysate ratio, increasing the pH during crushing of the materials and lowering the pH during the RNA precipitation step can be used for materials which are more soluble than TCP. These aim to minimize the dissolution of ions from materials and the precipitation of the undesirable co-pre- cipitates. On the other hand, TRI Reagent® and TRIsureTM have similar extraction principles and procecures as TRIzol®. Thus similar modifications can be made to their RNA extraction protocols to extract RNA from cells on resorbable materials in high yield. 3.5. Quality of the Extracted RNA To evaluate the quality of RNA extracted using the modified procedures, we used agarose gel electrophoresis analysis and UV spectrometry to check the samples. Two distinct bands showing the 28S and 18S ribosomal RNA were observed and degradation of RNA was not detected after the agarose gel electrophoresis (Figure 3(a)). By UV spectrometry, the A260/A280 ratios were consistently (a) (b) Figure 3. (a) Agarose gel image of RNA in 2% GelRed pre-stained gel (b) Conventional RT-PCR of -Actin (A1 and A2) and Col1 (C1 and C2), M: 100bp DNA ladder. Table 5. Factors affecting the dissolution of materials in the homogenization step and the precipitation of undesirable pre- cipitates in the RNA precipitation step. Dissolution of materials Precipitation of undesirable precipitates Nature of compounds Different materials differ greatly in solubility. Relative solubility a: MCPM ~ MCPA > DCPD > DCPA >> -TCP > -TCP > TTCP >> CDHA > OCP > HA [16] Most ammonium, potassium, acetate and chloride compounds have high solubility in aqueous solutions while many calcium compounds are insoluble [14]. Volume of TRIzol® A smaller TRIzol® volume reduces the total amount of ions dissolved from the materials but the volume should be large enough for effective lysis of the cells on materials. A larger volume of extra TRIzol® reduces the concentration of ions which may contribute to the undesirable precipitation. pH For many calcium phosphates (such as OCP, -TCP, -TCP, HA and TTCP) a, a higher pH reduces the dissolution rate within pH range of 4 to 7 [15]. A lower pH increases the solubility of the undesirable precipitates, thus reduces the precipitation. Temperature A lower temperature reduces the dissolution rate. A higher temperature can probably increase the solubility of the undesirable precipitates but a low temperature is preferred to prevent RNA degradation. a. MCPM: monocalcium phosphate monohydrate; MCPA: monocalcium phosphate anhydrate; DCPD: dicalcium phosphate dihydrate; DCPA: dicalcium phosphate anhydrate; OCP: octacalcium phosphate;-TCP: -tricalcium phosphate;-TCP: -tricalcium phosphate; CDHA: calcium-deficient hydroxyapatite; HA: hydroxyapatite; TTCP: tetracalcium phosphate  Simple Modifications to Standard TRIzol® Protocol Allow High-Yield RNA Extraction from Cells on Resorbable Materials Copyright © 2011 SciRes. JBNB close to 2.0 and the A260/230 ratios were higher than 1.0, indicating that the RNA samples extracted by this me- thod are not carrying contaminating proteins and salts (Table 6). Finally, the quality of the RNA was confirmed by con- ventional RT-PCR and RT-qPCR. After DNase I treat- ment, reverse transcription and PCR amplification with primer pairs specific to the transcripts from β-Actin gene (a housekeeping gene as control) and Col1 gene (a chon- drogenic gene), specific amplicons with the correspond- ing molecular sizes were observed (Figure 3(b)). Fur- thermore, consistent Ct values from RT-qPCR analysis on β-Actin, Col1 and other bone cell differentiation- markers such as Cbfa1, Alpl and Ocn (Table 7) were noted, indicating that the RNA quality of these samples is compatible with protocols for gene expression study. In addition, this protocol allowed us to isolate 2 g of total RNA from cells grown on 10 mm -TCP disc, an amount enough for performing expression evaluation on 40 different target genes. This good yield allows the pro- tocol to be used for large scale analysis, when cell supply is limited because of the lack of large volume of bioma- terials used in the study. 4. Conclusions For RNA extraction from cells on porous -TCP, the following three modifications are sufficient to obtain high quality RNA: 1) in situ crushing of the porous discs with cells in TRIzol® at low temperature 2) the use of extra TRIzol® to dilute the lysate after TCP debris re- moval and 3) extra RNA washing step to remove the salts more completely. Without modifications, the RNA was often trapped in the undesirable precipitates which Table 6. UV absorbance readings using Nanodrop of the RNA extracted from cells on porous -TCP disc, indicating the extracted RNA is of high quality. A260/280 a A260/230 conc. (ng/mL) average 2.00 2.05 223.23 SD 0.08 0.14 40.98 a. Using other spectrometric equipments for the UV absorbance measurements often requires a dilution of the samplesand may result in readings of A260/280 deviating from 2.00 but the RNA may still be suitable for RT-PCR. In such case, the RNA quality should be confirmed by gel electrophoresis. Table 7. Ct values of -Actin, Cbfa1, Alpl, Col1 and Ocn of RT-qPCR with the RNA samples extracted using the modi- fied procedures. β-Actin Cbfa1 Alpl Col1 Ocn average 20.6 25.2 31.6 18.8 35.7 SD 0.6 0.4 0.2 0.3 0.5 prevent effective recovery and render TRIzol® extraction method incompatible with studies using -TCP. Analysis of various physical parameters affecting the formation of undesirable co-precipitates suggests that a combined protocol of increasing the extra TRIzol® to lysate ratio, increasing the pH during crushing of materi- als and lowering the pH during the RNA precipitation step would make extraction of RNA from cells cultured on other more soluble materials possible. In summary, this study introduces to the field some easy-to-perform and low cost modifications to extract RNA from cells grown on -TCP in high yield. It also offers a direction for further modifications of the proce- dures in extracting nucleic acid samples from cells cul- tured on soluble ceramic materials. The impact on bio- medical development and applications could be tremen- dous. 5. Acknowledgements We thank the constructive comments and technical as- sistances from members of the Bioengineering lab, Prof. King L. Chow lab, Prof. Yang Leng lab of the Hong Kong University of Science and Technology and the Na- tional Engineering Research Center for Biomaterials in Sichuan University. This work was supported by the Re- search Project Competition Grant of the Hong Kong University of Science and Technology (Grant no. RPC 07/08.EG02) and RGC Grant (Grant no. RGC660407). 6. Sample Protocol for RNA Isolation from Cells Grown on Porous -TCP Discsa Materials used: 10mm porous α-TCP discs with 9 × 104 cells plated and cultured for 6 days. 1) HOMOGENIZATION At the day of analysis, wash the bioceramic disc (containing cultured cells) with PBS twice to re- move trapped cell culture medium. Briefly dry the disc on absorbent paper between washes. Briefly dry the disc on absorbent paper, gently break (not crush) the disc into smaller pieces and transfer the pieces to a 1.5 ml microcentrifuge tube. Add 0.5 ml of TRIzol® (Invitrogen) to the tube and crush the disc into powder with a steel rod with the tube placed on ice. Incubate the tube for 5 min at room temperature to lyse cells. Centrifuge the tube at 12,000 x g for 5 min at 4°C and transfer the clear orange lysate to a new tube. Repeat this step if TCP debris is observed at the bot- aAll reagents and consumables involved in RNA experiments should be free of RNase (Critical!). Besides, caution should be taken while using TRIzol® (specifically phenol) and Chloroform.  Simple Modifications to Standard TRIzol® Protocol Allow High-Yield RNA Extraction from Cells on Resorbable Materials Copyright © 2011 SciRes. JBNB 47 tom of the tube.b 2) PHASE SEPARATION Add another 0.5 ml of TRIzol® to the clear lysate and mix the content. Add 0.2 ml of chloroformc to the lysate and shake vigorously by hand for 15s and incubate the tube for 2 min at room temperature. Centrifuge the tube at 12,000 x g for 15 min at 4°C. 3) RNA PRECIPITATION Transfer the aqueous supernatant to a new tube and gently mix it with 0.5 ml of isopropanol by invert- ing the tube several times. Incubate the tube for 10 min at room temperature and centrifuge the tube at 12,000 x g for 10 min at 4°C 4) RNA WASH Remove the supernatant, add 0.5 ml of 75% ethanol to the tube and centrifuge at 7,500 x g for 5 min at 4°C. Repeat this washing step once. 5) REDISSOLVING THE RNA Remove the ethanol in the last washd and air-dry the RNA pellet (until no observable liquid droplet was found). Resuspend the RNA in RNase-free water / buffer and incubate at 60°C for 10min. Then proceed to gene expression studies after RNA quality check and quantitation. REFERENCES [1] L. L. Hench and J. M. Polak, “Third-Generation Bio- medical Materials,” Science, Vol. 295, No. 5557, 2002, pp. 1014-1017. doi:10.1126/science.1067404 [2] M. Vallet-Regí and J. M. González-Calbet, “Calcium Phosphates as Substitution of Bone Tissues,” Progress in Solid State Chemistry, Vol. 32, No. 1-2, 2004, pp. 1-31. doi:10.1016/j.progsolidstchem.2004.07.001 [3] P. Habibovic, U. Gbureck, C. J. Doillon, D. C. Bassett, C. A. van Blitterswijk and J. E. Barralet, “Osteoconduction and Osteoinduction of Low-Temperature 3D Printed Bioceramic Implants,” Biomaterials, Vol. 29, No. 7, 2008, pp. 944-953. doi:10.1016/j.biomaterials.2007.10.023 [4] C. Knabe, A. Houshmand, G. Berger, P. Ducheyne, R. Gildenhaar, I. Kranz and M. Stiller, “Effect of Rapidly Resorbable Bone Substitute Materials on the Temporal Expression of the Osteoblastic Phenotype in vitro,” Journal of Biomedical Materials Research-Part A, Vol. 84, No. 4, 2008, pp. 856-868. doi:10.1002/jbm.a.31383 [5] P. Chomczynski and N. Sacchi, “Single-Step Method of RNA Isolation by Acid Guanidinium Thiocyanate-Phenol- Chloroform Extraction,” Analytical Biochemistry, Vol. 162, No. 1, 1987, pp. 156-159. doi:10.1016/0003-2697(87)90021-2 [6] P. Chomczynski and N. Sacchi, “The Single-Step Method of RNA Isolation by Acid Guanidinium Thiocy- anate-Phenol-Chloroform Extraction: Twenty-Something Years on,” Nature Protocols, Vol. 1, No. 2, 2006, pp. 581-585. doi:10.1038/nprot.2006.83 [7] P. Niemeyer, U. Krause, J. Fellenberg, P. Kasten, A. Seckinger, A. D. Ho and H. Simank, “Evaluation of Min- eralized Collagen and α-Tricalcium Phosphate as Scaf- folds for Tissue Engineering of Bone Using Human Mes- enchymal Stem Cells,” Cells Tissues Organs, Vol. 177, No. 2, 2004, pp. 68-78. doi:10.1159/000079182 [8] U. Mayr-Wohlfart, J. Fiedler, K. Gnther, W. Puhl and S. Kessler, “Proliferation and Differentiation Rates of a Human Osteoblast-Like Cell Line (SaOS-2) in Contact with Different Bone Substitute Materials,” Journal of Biomedical Materials Research, Vol. 57, No. 1, 2001, pp. 132-139. doi:10.1002/1097-4636(200110)57:1<132::AID-JBM115 2>3.0.CO;2-K [9] M. Y. Deng, H. Wang, G. B. Ward, T. R. Beckham and T. S. McKenna, “Comparison of Six RNA Extraction Meth- ods for the Detection of Classical Swine Fever Virus by Real-Time and Conventional Reverse Transcription-PCR,” Journal of Veterinary Diagnostic Investigation, Vol. 17, No. 6, 2005, pp. 574-578. [10] L. Z. Santiago-Vázquez, L. K. Ranzer and R. G. Kerr, “Comparison of Two Total RNA Extraction Protocols Using the Marine Gorgonian Coral Pseudopterogorgia Elisabethae and its Symbiont Symbiodinium sp.,” Elec- tronic Journal of Biotechnology, Vol. 9, No. 5, 2006, pp. 598-603. [11] X. Xiang, D. Qiu, R. D. Hegele and W. C. Tan, “Com- parison of Different Methods of total RNA Extraction for Viral Detection in Sputum,” Journal of virological meth- ods, Vol. 94, No. 1-2, 2001, pp. 129-135. doi:10.1016/S0166-0934(01)00284-1 [12] H. Yuan, Z. Yang, J. D. De Bruijn, K. De Groot and X. Zhang, “Material-Dependent Bone Induction by Calcium Phosphate Ceramics: A 2.5-Year Study in Dog,” Bioma- terials, Vol. 22, No. 19, 2001, pp. 2617-2623. doi:10.1016/S0142-9612(00)00450-6 [13] J. Sambrook and D. W. Russell, “Molecular Cloning: A Laboratory Manual,” 3rd Edition, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, 2001. [14] M. M. Amiji and B. J. Sandmann, “Applied Physical Pharmacy,” McGraw-Hill, Medical Publishing Division, bFor a large number of materials being tested with cells plated in one batch, the cells may be lysed in TRIzol® at the same time and stored in a –80°C freezer after material debris removal before subsequent extrac- tion procedures which require relatively longer time. Besides, the sam- p les can be splitted into several extraction batches for easier handling i f required. cBromo-chloropropane may be used instead of chloroform as a safe r and equally efficient phase-separation agent [17]. dWhen the amount of RNA is small, the RNA pellet may be difficult to be observed by naked eye. In such case, the orientation of the tubes during centrifugation should be noted so that the position of the tiny RNA pellet is known. During this step, it is important to avoid pipet- ting the tiny precious RNA pellet together with the ethanol which would then be discarded.  Simple Modifications to Standard TRIzol® Protocol Allow High-Yield RNA Extraction from Cells on Resorbable Materials Copyright © 2011 SciRes. JBNB 48 New York, 2003. [15] E. Ferńndez, F. J. Gil, M. P. Ginebra, F. C. M. Driessens, J. A. Planell and S. M. Best, “Calcium Phosphate Bone Cements for Clinical Applications. Part I: Solution Chem- istry,” Journal of Materials Science: Materials in Medi- cine, Vol. 10, No. 3, 1999, pp. 169-176. doi:10.1023/A:1008937507714 [16] S. V. Dorozhkin and M. Epple, “Biological and Medical Significance of Calcium Phosphates,” Angewandte Che- mie-International Edition, Vol. 41, No. 17, 2002, pp. 3130-3146. doi:10.1002/1521-3773(20020902)41:17<3130::AID-AN IE3130>3.0.CO;2-1 [17] P. Chomczynski and K. Mackey, “Substitution of Chlo- roform by Bromo-Chloropropane in the Single-Step Method of RNA Isolation,” Analytical Biochemistry, Vol. 225, No. 1, 1995, pp. 163-164. doi:10.1006/abio.1995.1126 |

