 American Journal of Plant Sciences, 2013, 4, 1853-1862 http://dx.doi.org/10.4236/ajps.2013.49228 Published Online September 2013 (http://www.scirp.org/journal/ajps) Proteomics-Based Analysis of Phalaenopsis amabilis in Response toward Cymbidium Mosaic Virus and/or Odontoglossum Ringspot Virus Infection Tongfei Lai*, Yanfu Deng*, Pengchen Zhang, Zhijuan Chen, Feng Hu, Qi Zhang, Yihua Hu, Nongnong Shi# Research Centre of Plant Signaling, School of Life and Environmental Sciences, Hangzhou Normal University, Hangzhou, China. Email: #867594089@qq.com Received June 15th, 2013; revised July 20th, 2013; accepted August 21st, 2013 Copyright © 2013 Tongfei Lai et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. ABSTRACT Stress response at the protein level to viral infection in orchid plants has not been extensively investigated to date. To understand the proteomic basis of Phalaenopsis amabilis’s responses to Cymbidium Mosaic virus (CymMV), and/or Odontoglossum ring spot virus (ORSV), the total proteins were extracted from Phalaenopsis amabilis leaves infected with CymMV, ORSV, or both respectively. Differentially expressed proteins were identified by two-dimensional elec- trophoresis, and 27 of these proteins that had significant changes were further examined by mass spectrometry. Com- paring CymMV-infected leaves with mock-inoculated ones, 2 proteins were significantly up-regulated, 9 were signifi- cantly down-regulated and 1 previously undetected protein was identified. 10 proteins were significantly up-regulated, 3 significantly down-regulated and 1 previously undetected protein was identified in ORSV-infected leaves. 6 proteins were significantly up-regulated and 9 significantly down-regulated proteins were found in co-infected leaves. These identified proteins are involved in disease resistance, stress response, transcriptional regulation, energy metabolism, protein modification and the previously unknown proteins were not involved with known protein pathways. Proteins significantly up-regulated were ATP sulfurylase, down-regulated proteins included glutamate decarboxylase isozyme 2, RNA polymerase alpha subunit and chloroplastic peptide deformylase 1A were proteins with similar alteration trend after all infection treatments. Significantly up-regulated were Thioredoxin H-type and down-regulated Cytosolic phos- phoglycerate kinase I which were proteins that have been shown to be specifically regulated by the infection with CymMV. Significantly up-regulated were proteins like Rubisco large subunit, Triosephosphate isomerase, NADP-spe- cific isocitrate dehydrogenase and Cinn amoyl CoA reductase CCR2 by th e infection of ORSV. Protein regu lation in co- infected leaves followed a pattern similar to that of any of the single virus infection results. These experiments demon- strated that an exogenous pathogen infection in plants, regardless of viral type or pattern, induced a protein defense response system and a number of metabolism changes in P. amabilis with detectable protein alterations. These cellular alterations appeared to be specific responses to different pathogens. We suggest that altered proteins above might play important roles in pathogenesis and metabolic interactions between P. amabilis and viruses that infect them. Keywords: Phalaenopsis amabilis; Two-Dimensional Electrophoresis; Mass Spectrometry; Cymbidium Mosaic Virus; Odontoglossum Ringspot Virus 1. Introduction Phalaenopsis amabilis belongs to the genus Phalaenop- sis of the family Orchidaceae. It has a single short stem, succulent, fleshy leaves, beautiful shape, co lorful fl owers, long lasting flowering period and a high ornamental value. It is one of the four most famous Orchidaceae along with Cattleya hybrida, Vanda spp. and Dendrobium nobile in the world [1]. How eve r, P. amabilis is v e r y su s- ceptible to infection by Cymbidium mosaic virus (Cym- MV) and Odontoglossum ringspot virus (ORSV). Cym- MV-infected P. amabilis grows poorly and appears to have leaves with chlorotic stripes and sunken gray-white or necrotic ring spots as well as smaller and fewer flo- *These authors contributed equally to this work. #Corresponding author. Copyright © 2013 SciRes. AJPS  Proteomics-Based Analysis of Phalaenopsis amabilis in Response toward Cymbidium Mosaic Virus and/or Odontoglossum Ringspot Virus Infection 1854 wers with a shortened flowering period. ORSV-infected P. amabilis is not easily distinguished from the unin- fected healthy plants. Therefore, the virus-carrying plants being sold in commercial and local markets appear fre- quently and jeopardize and reduce their ornamental and economic value [2,3]. Molecular detection is the most effective and best way to identify ORSV and CymMV infected plants, which will also often combine toge ther to infect P. amabilis. At present, there have been some reports on the ad- verse effects of viral infection in P. amabilis. Liao et al. made a CymMV capsid protein gene that silenced the virus in P. amabilis and improved its resistance to Cym- MV [4]. Qin et al. found that expression of exogenous lipid transfer protein encoding gene could enhance the plants frost resistance [5]; Specified Tyrosine phospha- tase PaPTP1 gene is related to flower development and external mechanical damage [6]. However, there are few studies on pathogenesis to P. amabilis and none of these reports used proteomics-based analysis to examine the mechanisms altered by infection with the exception of re- cent investigations with soybean [7] and cucumber [8]. Proteomics is currently a powerful tool to systematically analyze the cellular protein expression profiles and post- translational modifications under virus invasion in the host of rice [9-11], tobacco [12], soybean [13], tomato [14] and Arabidopsis thaliana [15] respectively. In this study, we utilized the two-dimensional electro- phoresis (2-DE) and mass spectrometry analysis to com- pare 2-DE gel profiles of the total proteins from CymMV and/or ORSV infected leaves in an attempt to find pro- teins that may be respons ive to viral infection or proteins related to disease resistance. The results may provide new clues to the understanding of the interactive responses in protein expression of P. amabilis when infected with viruses. 2. Materials and Methods 2.1. Inoculations and Molecular Detections Healthy P. amabilis seed lings with 5 - 6 leaves were cul- tured in a green house at 25˚C and infected with CymMV or ORSV separately, or with both CymMV and ORSV, through sap inoculation. Four weeks post-inoculation, im- mune-captured reverse-transcript polymerase chain reac- tion (IC-RT-PCR) [16] and sequencing techniques were utilized to detect the presence of infection. Self-made high-titer polyclonal antibodies [17] and two virus spe- cific primer pairs (5’-TCCGAATTCAGTCTTACACTATTACTGAC-3’ and 5’-AAGTTCGAATTATTCAGTAGGGGGTGC-3’ for CymMV; 5’-AATGGTGTTAGTGATATTCG-3’ and 5’-CCACTATGCATTATCGTATG-3’ for ORSV) were applied in the diagnosis. 2.2. Protein Extraction Total leaf proteins were extracted using tris-saturated phenol method from P. amabilis plantlets 4 weeks after virus(es)- or mock-inoculation. In detail, 2 g leaf sam- ples were collected and mixed using mortar and pestle with a small amount of quartz sand, polyvinylpyrrolidone (PVP) and 4 ml of extraction buffer (0.1 M Tris, 0.05 M vitamin C, 0.09 M sodium tetra-borate, 0.1 M EDTA-Na2, 2% β-mercaptoethanol (v/v), 1% Triton-100 (v/v) and 30% sucrose (w/v)). This mixture was ground on ice and then transferred to a 50 mL centrifuge tube. 5 mL of the extraction was mixed with 9 mL of tris-saturated phenol (pH 8.0), shaken with a shaker for 15 min and centri- fuged at 15,000 × g for 20 min at 4˚C. The lower phenol phase was then collected and precipitated by mixing with a 5-fold volume of ice-cold ammonium sulfate and me- thanol. After being stored at −20˚C for 12 h, the precipi- tated proteins were collected by centrifugation at 5000 × g for 15 min at 4˚C, and washed with ice-cold methanol and acetone for 2 - 3 times. After drying at room tem- perature for 5 min, the proteins were dissolved in 200 μL lysis buffer (7 M urea, 2 M thiourea, 1% dithiothreitol (w/v), 4% CHAPS (w/v), 1% ampholytes (v/v), pH 3 - 10) at room temperature for 2 - 3 h. The extraction was car- ried out three times with 3 days interval. All pr otei ns f r om the three extractions for each sample were mixed toge- ther for further analysis. Total protein concentration was determined using Bradford method [18]. 2.3. Two-Dimensional Gel Electrophoresis Analysis (2-DE) Protein hydration and isoelectric focusing electrophoresis: Isoelectric focusing electrophoresis was carried out on an EttanTM IPGphor IITM system (Amersham, USA). 150 μg protein lysate was mixed with 125 μL rehydration solu- tion (8 M urea, 2% CHAPS (w/v), 0.2% bromo phenol blue (w/v), 0.5% ampholytes (v/v), pH 3 - 10) and incu- bated at 20˚C for 14 h to prepare 7 cm of pH 4 - 7 IPG strips. The prepared IPG strips were then transferred to the EttanTM IPGphorIITM system and electrophoresed for 0.5 h at 300 V, 300 V/hr at Grad 1000 V, 5000 V/hr at Grad 5000 V, 5000 V/hr at 5000 V, and 1 h at 300 V. Gel strip balance: After isoelectric focusing electro- phoresis, gel strips were equilibrated with salt solution I (6 M urea, 0.075 M Tris-HCl, pH 8.8, 2% SDS (w/v), 0.4% bromo phenol blue (w/v) and 1% DTT (w/v)) and salt solution II (6 M urea, 0.075 M Tris-HCl pH 8.8, 2% SDS w/v), 0.4% bromo phenol blue (w/v), and 2.5% io- doacetamide (w/v)) by shaking the strips at room tem- perature for 15 min each. Copyright © 2013 SciRes. AJPS  Proteomics-Based Analysis of Phalaenopsis amabilis in Response toward Cymbidium Mosaic Virus and/or Odontoglossum Ringspot Virus Infection 1855 SDS-Polyacrylamide gel electrophoresis (SDS-PAGE): SDS-PAGE was carried out on 12.5% separating gel and 5% stacking gel using a DYCZ-23A small vertical elec- trophoresis apparatus (Liuyi Company, Beijing). After gel polymerization, the pre-balanced IEF gels were placed on the top of the stacking gel. Proteins were further sepa- rated in electrode solution (25 mM Tris, 90 mM glycine and 0.1% SDS (w/v)) at 120 V for 1.5 h and stained with coomassie brilliant blue G-250. Electrophoresis gel imaging analysis: Electrophoresis images were acquired using a gel scanner (GS800 Cali- brated Densitometer, Bio-Rad) at transmission scanning mode and 300 dpi reso lution, and saved as TIF f iles. Pro- tein bands were analyzed using Image MasterTM 2D Pla- tinum software (Amersham Pharmacia Biotech, Uppsala, Sweden). To account for experimental variation, three biological replicate gels resulting from three independent experiments were analyzed for control and treatment. The amount of a protein spots was calculated based on the volume of that spot which was normalized against total volume of all valid spots. The normalized intensity of spots on three replicate 2-DE gels was averaged and analyzed using two-tailed, non-paired Student’s t-test to determine whether the relative change was statistically significant between samples using SPSS software (SPSS Inc., Chicago, IL, USA). The differentially expressed pro- tein spots whose expression level was more than 1.5 times higher or lower were manually excised for protein identification. 2.4. Protein Enzymatic Digestion and Identification by Mass Spectrometry Enzymatic digestion: The target protein spots were de- stained using 100 μL of destaining solution (25 mM am- monium bicarbonate and 50% acetonitrile (v/v)) for 40 min and reduced by incubating with 100 μL reducing solution (10 mM DTT and 25 mM ammonium bicarbon- ate) at 56˚C for 1 h. After cooled to room temperature, the reducing solution was discarded and proteins were incubated with equal volume of 25 mM ammonium bi- carbonate solution containing 55 mM iodoacetamide at room temperature for 45 min in dark. The supernatant was discarded and the proteins were dehydrated with 100% acetonitrile (v/v) and dried under a vacuum. The proteins were digested with 10 μL of trypsi n sol uti on for 1 h on i ce, and incubated with 5 μL of 5 mM ammonium bicarbon- ate solution at 37˚C for 16 h. The mixtures were then ex- tracted with 6 - 10 μL of 5% trifluoroacetic acid (v/v) at 40˚C for 1 h. After two successive extractions, the super- natants were collected and freeze-dried for future analysis. Protein identification by mass spectrometry: Proteins were identified using Reflective Tand em Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spec- trometry (MALDI-TOF-MS, 4800 MALDI TOF/TOF Analyzer, Applied Biosystems, USA) with MS Reflector Positive mode. The parameters were set as Laser Inten- sity 3880; Light Intensity 34; Mass Range 900 to 4000 Da; Focus Mass 2000 Da; Shots/Sub Spectrum 50, Total Shots/Spectrum 1000; Lo cal Noise Window Width (m/z) 20; Min Peak Width at Full Width Half Max (bins) 2.9. Mass spectrometric data were processed using Mass- LynxTM software to generate peak list files and blasted in public NCBInr and SWISS-PROT protein databases us- ing Mascot MS/MS Ion Search program on the Matrix Science public website (www.matrixscience.com). Search parameters were set as taxonomy, green plant; proteoly- tic enzyme, trypsin; max missed cleavages 1; fixed mod- ifications, carbamidomethyl (c); variable modifications, oxidation; peptide mass tolerance, 150 ppm. In addition, “mono-isotopic mass” was chosen. Only significant hits as defined by Mascot probability analysis were consid- ered. Mascot uses a probability-based Mowse score to evaluate data obtained from mass spectra. Mowse scores were reported as −10 × Log10 (P) where P is the prob- ability that the observed match between the experimental data and the database sequence is a random event. Pro- tein spots with a Mascot score greater than threshold (40) and C.I.% value were considered as validated ones and used to perform protein function analysis in UniprotKB/ SWISS-PROT and UniprotKB/TrEMBL databa ses (www.uniprot.org). Data analysis: The experimental data were collected from independent triplicates and significant differences were analyzed using SPSS 11.5 (SPSS Inc., Chicago, IL). P < 0.05 was considered as significance. 3. Results 3.1. Producing CymMV and/or ORSV Infected Plants IC-RT-PCR and sequencing successfully diagnosed the amplified band of approx. 600 bp for ORSV-cp (Figure 1(a)) and approx. 780 bp for CymMV-cp (Figure 1(b)) from the inoculated leaves respectively. The high homo- logies (98% for ORSV-cp and 99% for CymMV-cp) with the corresponding gene sequences in GenBank were shown for the consensus sequences. CymMV and/or ORSV in- fected plants of Phalaenopsis amabilis were confirmed by these results. 3.2. Analysis of 2-DE Images of Different Virus Infected P. amabilis Leaves About 6 μg/μL extracted protein solution and total about 1.2 mg proteins were identified from 2 g leaf sample. Copyright © 2013 SciRes. AJPS  Proteomics-Based Analysis of Phalaenopsis amabilis in Response toward Cymbidium Mosaic Virus and/or Odontoglossum Ringspot Virus Infection Copyright © 2013 SciRes. AJPS 1856 2-DE analyses of mock-inoculated, CymMV-infected, ORSV-infected, and CymMV and ORSV co-infected P. amabilis leaves showed a highly reproducible protein ex- pression profiles. Most protein spots were concentrated in the range o f pH 5 - 7 with a molecular weigh ts of 16 - 105 kDa (Figures 2(a)-(d)). A total of 278, 263, 23 0 and 242 protein spots were detected, respectively, in mock- inoculated P. amabilis leaves (Figure 2(a)), CymMV in- fected P. amabilis leave (Figure 2(b)), ORSV infected P. amabilis leaves (Figure 2(c)) and co-infected P. amabilis leaves (Figure 2(d)). The expression of 27 proteins changed significantly after virus infection (Figure 3). In addition, compared to mock-inoculated leaves, 2 up-regulated protein spots (Protein ID 144 and 157), 9 down-regulated protein spots (Protein ID 2 2, 23, 55, 64, 81, 125 , 136, 146 and 186), 1 undetectable protein spot (Protein ID 59) and 15 un- changed protein spots were found in CymMV infected P. amabilis leaves, 10 up-regulated proteins (Protein ID 70, 100, 121, 144, 149, 150, 153, 154, 159 and 162); 3 down-regulated protein spots (Protein ID 59, 64 and 136), 1 undetectable protein spot (protein ID 186) and 13 un- changed protein spots were found in ORSV infected P. amabilis leaves, and 6 up-regulated proteins (Protein ID 100, 144, 149, 150, 159 and 162), 9 down-regulated pro- tein spots (Protein ID 22, 23, 55, 59, 64, 125, 136, 146, and 186) and 12 unchanged protein spots (Figure 3) were found in co-infected P. amabilis leaves. Among them, 4 protein spots (Protein ID 64, 136, 144 and 186) showed the same regulation tendency in all three treat- ments compared with mock-inoculated leaves. Spots 64, 136 and 186 were down-regulated and spot 144 was up- Marker 1000 750 500 1000 750 500 (bp) ORSV-cp 600 bp Healthy Mock-inoculated CymMV-inoculated CymM -cp Mock-inoculated ORSV-inoculated ORSV-inoculated Co-infected ORSV-cp Co-inoculated Co-inoculated CymMV-cp 780 bp Figure 1. Agrose-gel electrophoresis of immune-captured reverse-transcript polymerase chain reaction (IC-RT-PCR) prod- ucts from CymMV and/or ORSV inoculated leaves of Phalaenopsis amabilis. Panel A shows the amplified band of approx. 600 bp for ORSV-cp. Panel B shows the amplified band of approx. 780 bp for CymMV-cp. 16 25 35 50 71 105 (kDa) 4-7 Figure 2. Two-dimensional patterns of total proteins in Phalaenopsis amabilis from mock-inoculated leaves (Panel A), CymMV-infected leaves (Panel B), ORSV-infected leaves (Panel C) and co-infected leaves (Panel D). Protein extraction and 2D electrophoresis were performed as described in Materials and Methods. Circles with black line indicate the differentially expressed proteins that have been identified by MADLI-TOF-MS. The spots are numbered, corresponding to those in Tables 1. Electrophoresis strip: 7 cm long, IPG made, pH 4 - 7. 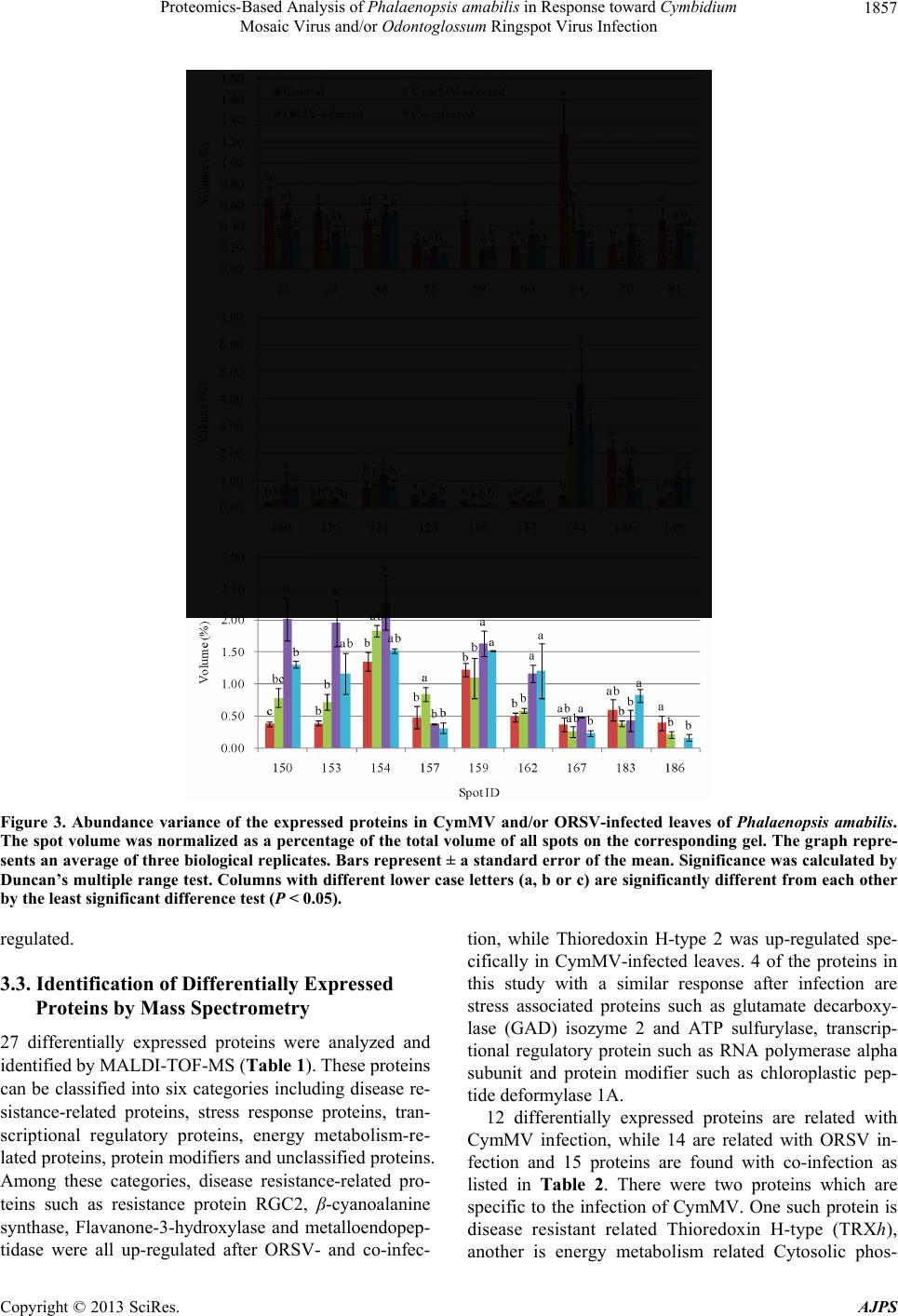 Proteomics-Based Analysis of Phalaenopsis amabilis in Response toward Cymbidium Mosaic Virus and/or Odontoglossum Ringspot Virus Infection 1857 Figure 3. Abundance variance of the expressed proteins in CymMV and/or ORSV-infected leaves of Phalaenopsis amabilis. The spot volume was normalized as a percentage of the total volume of all spots on the corresponding gel. The graph repre- sents an average of three biological replicates. Bars represent ± a standard error of the mean. Significance was calculated by Duncan’s multiple range test. Columns with different lower case letters (a, b or c) are significantly different from each other by the least significant difference test (P < 0.05). regulated. 3.3. Identification of Differentially Expressed Proteins by Mass Spectrometry 27 differentially expressed proteins were analyzed and identified by MALDI-TOF-MS (Table 1). These proteins can be classified into six categories including disease re- sistance-related proteins, stress response proteins, tran- scriptional regulatory proteins, energy metabolism-re- lated proteins, protein modifiers and unclassified proteins. Among these categories, disease resistance-related pro- teins such as resistance protein RGC2, β-cyanoalanine synthase, Flavanone-3-hydroxylase and metalloendopep- tidase were all up-regulated after ORSV- and co-infec- tion, while Thioredoxin H-type 2 was up-regulated spe- cifically in CymMV-infected leaves. 4 of the proteins in this study with a similar response after infection are stress associated proteins such as glutamate decarboxy- lase (GAD) isozyme 2 and ATP sulfurylase, transcrip- tional regulatory protein such as RNA polymerase alpha subunit and protein modifier such as chloroplastic pep- tide deformylase 1A. 12 differentially expressed proteins are related with CymMV infection, while 14 are related with ORSV in- fection and 15 proteins are found with co-infection as listed in Table 2. There were two proteins which are specific to the infection of CymMV. One such protein is disease resistant related Thioredoxin H-type (TRXh), another is energy metabolism related Cytosolic phos- Copyright © 2013 SciRes. AJPS  Proteomics-Based Analysis of Phalaenopsis amabilis in Response toward Cymbidium Mosaic Virus and/or Odontoglossum Ringspot Virus Infection 1858 Table 1. Differentially expressed protei ns in Phalaenopsis amabilis leaves after infection of CymMV and/or ORSV. Proteins ID Homologous Protein Theo. Mr (Da)/pI NCBI Accession Organism Mascot Score/Threshold*Sequences Matched Disease resistant proteins 149 Resistance protein RGC2 47,007/5.95 gi|34485415 Lactuca saligna 42/40 9 150 Beta-cyanoalanine synthase 38176.3/6.86 gi|11559260 Solanum tuberosum 44/40 6 157 Thioredoxin H-type 2 13038.7/5.56 gi|586099 Nicotiana tabacum 46/40 4 159 Flavanone-3-hydroxylase 20922.5/5.09 gi|20513407 Sequoia sempervirens 51/40 6 162 Metalloendopeptidase 93138.8/6.78 gi|42568277 Arabidopsis thaliana 68/40 19 Stress resp onse proteins 22 Vacuolar proton -ATPase 68409.5/5.23 gi|11527563Hordeum vugare subsp. vugare 82/40 15 23 V-type proton ATPase catalytic subunit A 68791.9/5.29 gi|137460 Daucus carota 105/40 20 64 Glutamate decarboxylase isozyme 2 55895.4/5.58 gi|3252854 Nicotiana tabacum 41/40 7 121 Cinnamoyl CoA reductase CCR2 36606.8/6.11 gi|12407990 Arabidopsis thaliana 56/40 8 144 ATP sulfurylase 52262.9/8.73 gi|4033353 Brassica juncea 60/40 12 167 Heat shock protein 17a.11 12046.1/5.67 gi|7768333 Triticum aestivum 50/40 5 Proteins related to transcriptional regulation 136 RNA polymerase alpha subunit 38922.4/6.36 gi|11467223Zea mays 50/40 7 183 RNA recognition motif-containing protein 60758.7/9.2gi|30682379Arabidopsis thaliana 48/40 11 Proteins related to energy metabolism 55 ATP synthase subunit alpha,mitochondrial 55229.6/5.7 gi|114419 Arabido psis thaliana 43/40 7 59 ATP synthase beta subunit 53445.9/5.22 gi|14718074Hippeastrum papilio 109/40 12 60 Rubisco large subunit 48780.6/6.57 gi|1518390 Solaria atropurpurea 195/40 16 70 Rubisco large subunit 51850.2/6.34 gi|16973388Tetrastigma hookeri 41/40 7 81 Cytosolic phosphoglycerate kinase 1 42642.6/5.7 gi|3738259 Populus nigra 51/40 10 125 Ribulose-1,5-bisphosphate carboxylase 51539.5/5.91 gi|4176743 Llligera luzonensis 122/40 12 153 Triosephosphate isomerase 27588.3/6.6 gi|553107 Oryza sativa 47/40 7 154 NADP-specific isocitrate dehydrogenase 45703.1/6.13 gi|20260384Arabidopsis thaliana 50/40 8 Proteins related to protein modification 48 Chaperoning 60 beta subunit 63769.6/6.21 gi|15222729Arabidopsis th aliana 70/40 14 120 Cycling-dependent protein kinase 38105.4/5.07 gi|15220145Arabidopsis th aliana 42/40 6 186 Peptide deformylase 1A, chloroplast 28895.3/8.82 gi|17433051Arabidopsis th aliana 51/40 7 Others proteins 100 Oxygen-evolvin g enh ancer prot ein 34718.7/8.73 gi|131388 Triticum aestivum 52/40 9 137 Unkown protein 36053.7/6.25 gi|15220679Arabidopsis th aliana 44/40 7 146 Hypothetical protein 27335.5/5.55 gi|2244768 Arabidopsis thaliana 46/40 3 *: specified for non-model plant—Phalaenopsis amabilis. Copyright © 2013 SciRes. AJPS  Proteomics-Based Analysis of Phalaenopsis amabilis in Response toward Cymbidium Mosaic Virus and/or Odontoglossum Ringspot Virus Infection 1859 Table 2. Proteins potentially related with Phalaenopsis amabilis—CymMV and/or ORSV interactions. CymMV-infected leaves ORSV-infected leaves CymMV and ORSV-infected leaves Vacuolar proton-ATPase (22*) V-type proton ATPase catalytic subunit A (23) ATP synthase subunit alpha, mitochondrial (55) ATP synthase beta subunit (59) Glutamate decarboxylase isozyme 2 (64) §Cytosolic phosphoglycerate kinase 1 (81) Ribulose-1,5-bisphosphate carboxylase (125) RNA polymerase alpha subunit (136) ATP sulfuryl a s e (144) Hypothetical protein (146) §Thioredoxin H-type 2 (157) Peptide deformylase 1A, chloroplastic (186) ATP synthase beta subunit (59) Glutamate decarboxylase isozyme 2 (64) ¡Rubisco large subunit (70) Oxygen-envolvin g enhancer prot ein (100) ¡Cinnamoyl CoA reductase CCR2 (121) RNA polymerase alpha subunit (136) ATP sulfuryl a s e (144) Resistance protein RGC2 (149) Beta-cyanoalanine synthase (150) ¡Triosephosphate isomerase (153) ¡NADP-specific isocitrate dehy drogenase (154) Flavanone-3-hydroxylase (159) Metalloendopeptidase (162) Peptide deformylase 1A, chloroplastic (186) Vacuolar proton-ATPase (22) V-type proton ATPase catalytic subunit A ( 23) ATP synthase subunit alpha,mitochondrial (55) ATP synthase beta subunit (59) Glutamate decarboxylase isozyme 2 (64) Oxygen-envolvin g enhancer prot ein (100) Ribulose-1,5-bisphosphate carboxylase (125) RNA polymerase alpha subunit (136) ATP sulfuryl a s e (144) Hypothetical protein (146) Resistance protein RGC2 (149) Beta-cyanoalanine synthase (150) Flavanone-3-hydroxylase (159) Metalloendopeptidase (162) Peptide deformylase 1A, chloroplastic (186) *: protein ID no.; §: proteins specific to CymMV-infection; ¡: proteins specific to ORSV-infection. phoglycerate kinase I (PGK). The previously undetected protein is an energy metabolism related ATP synthase beta subunit. There were four proteins which were spe- cifically regulated when infected with ORSV. These are proteins, an energy metabolism related Rubisco large subunit, a Triosephosphate isomerase (TPI) and a NADP- specific isocitrate dehydrogenase (IDH), and stress re- sponse related Cinnamoyl CoA redu ctase 2 (CCR2). The previously undetected protein is a protein modification related Peptide deformylase 1A and is Chloroplastic. T here were no proteins uniquely regulated with co-infection, and no protein was altered more significantly than that found in any single infection. There were also no detect- able novel proteins detected in co-infected leaves. The pattern of expressed proteins in co-infected leaves fol- lowed that of any of the single virus infection results (Figure 3 and Table 2). 4. Discussion Although proteomics has been widely used in examining protein function and expression profiles, it is seldomly used for analyzing interactions between virus and its host horticulture plant. Ventelon-Debout et al. (2004) applied 2-DE and mass spectrometry and found 19 and 13 up- or down-regulated proteins, respectively, in rice yellow mottle virus (RYMV) inoculated rice variety IR64 and virus-resistance variety Azucena [10]. These proteins are related to metabolism, stress resistance and translation. Abiotic stress response pathway is also activated by RYMV in both cultivars through up-regulating salt-in- duced protein (SalT), heat shock proteins (HSPs) and superoxide dismutase (SOD). Brizard et al. (2006) iden- tified that rice-RYMV interaction protein complex is in- volved in plant defense, metabolism, translation, protein synthesis and transport, and demonstrated that virus is able to recruit many proteins from its hosts to ensure its development [9]. Casado-Vela et al. (2006) revealed dif- ferential expression of peptidases, endoglucanase, chiti- nase and proteins in the ascorbate-glutathione cycle in TMV-infected, asympto matic tomato fruits, and sugg es ted that pathogenesis-related proteins and antioxidant enzy- mes may play a role in the protection against TMV infec- tion [14]. Zhao and Liu et al. (2013) recently reported that the rubisco small subunit was involved in the toba- movirus movement and silencing of NbRbCS was able to co mpr omise Tm-22-dependent resistance [12]. 4.1. Proteins Specifically Regulated by the Infection with CymMV TRXh is an intracellular small acidic protein (12 kDa) with redox activity and has multiple functions in plants. In arabidopsis, TRXh5 is involved in oxidative stress re- sponse and anti-fungal response [19]. In tobacco plants, tobacco mosaic virus could strongly induce the expres- sion of NtTRXh [20]. In this study, CymMV infection also dramatically induces TRXh expression in P. ama- bilis leaves, in consistence with th e previous result. PGK is involved in glycolysis and tricarboxylic acid (TCA) cycle and plays important roles in energy produc- tion [21]. PGK expression was significantly decreased only in CymMV-infected P. amabilis leaves, indicating that the energy metabolism of the plant host was dam- aged severely after CymMV infection. 4.2. Proteins Specifically Regulated by ORSV Infection It has been reported that sclerotinia virus infection could reduce Rubisco gene expression in sunflower even before its induced defense responses [22]. Similarly, our results showed that ORSV infection enhanced Rubisco expres- sion in leaves, implying that the modified Rubisco struc- Copyright © 2013 SciRes. AJPS  Proteomics-Based Analysis of Phalaenopsis amabilis in Response toward Cymbidium Mosaic Virus and/or Odontoglossum Ringspot Virus Infection 1860 ture is associated with different viral pathogens. TPI is involved in glycolysis and TCA cycle and plays important roles in energy production [21]. IDH is a rate- limiting enzyme in TCA cycle and plays important roles in energy metabolism, amino acid synthesis and vitamin synthesis [23,24]. TPI and IDH expression was signifi- cantly increased in ORSV-infected P. amabilis leaves, indicating that the host accelerates the transformation of energy and increases carbohydrate intermediates mainly through TCA cycle to protect itself from the harm caused by virus infection. CCR2 is the entry point for the lignin-specific branch of the phenylpropanoid pathway and one of the proteins related to plant physical defenses. CCR expression regu- lates lignin content. In this study, CCR2 expression was strongly up-regulated only in ORSV infected leaves, which may be related to the induction of specific patho- gen. The detailed mechanism of CCR2 up-regulation is unclear. 4.3. Proteins Regulated by All Infection Treatment with Similar Alteration Trend ATP sulfurylase is involved in cysteine and methionine biosynthesis [25]. Its expression was significantly up-re- gulated in all infected P. amabilis leaves, indicating that viruses could greatly induce ATP sulfurylase expression, thus promoting thiol generation to participate in the for- mation of sulfur-containing amino acids to meet the de- mand of P. amabilis for cysteine, methionine and other sulfur-containing amino acids, which have been found to be abundant in plant in response to diseases. GAD isozyme 2 is a 5-pyridoxal phosphate dependent enzyme. Its expression and activit y are increased in ma ize under stress from abscisic acid, methyl jasmonate, NaCl or cold. This up-regulation might occur at both transcrip- tional or post-transcriptional levels [26]. Ginseng GAD (PgGAD) gene expression is up- regulated under stress of non-biological factors such as temperature, osmotic pres- sure, hypoxia, and mechanical damage, and down-regu- lated under oxidative stress (H2O2) [27]. In this stud y, we found that GAD2 expression in infected P. amabilis leaves was significantly decreased, possibly due to virus infec- tion-induced host allergic reactions, which could increase local accumulation of active oxygen, thus reducing GAD expression. The α subunit of RNA polymerase is the core compo- nent for gene transcription, and regulates transcription initiation [28]. RNA polymerase expression was signifi- cantly decreased in all infected P. amabilis leaves, indi- cating that viral infection reduces protein metabolism and regeneration, and disrupts normal ph ysiological activities of plants. Peptide deformylase is a metalloprotease involved in the posttranslational deformylation of N-terminal methio- nine residue of newly synthesized proteins [29]. The ex- pression of chloroplast peptide deformylase 1A was sig- nificantly reduced in all virus infected P. Amabilis leaves, especially in ORSV-infected leaves, suggesting that viral infection has significant impact on protein modification in P. amabilis, and the symptom of chlorotic stripes in infected leaves probably resulted from the dramatically reduced expression of peptide deformylase 1A in chloro- plasts. The underlying mechanism needs to be further studied. These results demonstrated that viral infection regard- less of type and pattern could induce dramatic changes in defense response system and metabolism in P. amabilis. Nevertheless, there are apparent specific responses to different pathogens. For CymMV infection, we observed differentially expressed proteins including stress response proteins, altered transcriptional regulation and changes in energy metabolism proteins, all of which, particularly down-regulation of proteins, reflect the severe damage to the host and the suscep tibility of P. ama bilis to CymMV. In terms of ORSV infection, up-regulation of most pro- teins shown above reflects the obvious disease-resistance characteristics of host P. amabilis to this viral pathogen. This result actually explains why ORSV symptoms, wh i c h usually appear faintly in P. amabilis, can be detected only by molecular techniques. It also illustrates a good symbiotic relationship between ORSV and its host P. amabilis. Transcript levels for those specifically regu- lated proteins in CymMV and/or ORSV infected leaves of P. amabilis need to be investigated further. 5. Acknowledgements This work was partially supported by China Natural Sci- ence Foundation (NSFC, 30770185) and the Key Labo- ratory Innovative Founda tion of Science and Technology Bureau in Hang zhou (20090232T05). REFERENCES [1] X. Q. Chen, Z. H. Ji and Y. F. Zheng, “The Orchids of China,” China Forestry Press, Beijing, 1998. [2] F. W. Zettler, N. J. Ko, G. C. Wisler, M. S. Elliott and S. M. Wong, “Viruses of Orchids and Their Control,” Plant Disease, Vol. 74, No. 9, 1990, pp. 621-626. doi:10.1094/PD-74-0621 [3] J. S. Hu, S. Ferreira, M. Wang and M. Q. Xu, “Detection of Cymbidium Mosaic Virus, Odontoglossum Ringspot Virus, Tomato Spotted Wilt Virus and Potyviruses Infect- ing Orchids in Hawaii,” Plant Disease, Vol. 77, No. 5, 1993, pp. 464-468. doi:10.1007/s10658-008-9293-2 [4] L. J. Liao, I. C. Pan, Y. L. Chan, Y. H. Hsu, W. H. Chen Copyright © 2013 SciRes. AJPS  Proteomics-Based Analysis of Phalaenopsis amabilis in Response toward Cymbidium Mosaic Virus and/or Odontoglossum Ringspot Virus Infection 1861 and M. T. Chan, “Transgene Silencing in Phalaenopsis Expressing the Coat Protein of Cymbidium Mosaic Virus Is a Manifestation of RNA-Mediated Resistance,” Mole- cular Breeding, Vol. 13, No. 3, 2004, pp. 229-242. doi:10.1023/B:MOLB.0000022527.68551.30 [5] X. Y. Qin, Y. Liu, S. J. Mao, T. B. Li, H. K. Wu, C. C. Chu and Y. P. Wang, “Genetic Transformation of Lipid Transfer Protein Encoding Gene in Phalaenopsis amabi- lis to Enhance Cold Resistance,” Euphytica, Vol. 177, No. 1, 2011, pp. 33-43. doi:10.1007/s10681-010-0246-4 [6] S. F. Fu, C. W. Lin, T. W. Kao, D. D. Huang and H. J. Huang, “PaPTP1, a Gene Encoding Protein Tyrosine Phosphatase from Orchid, Phalaenopsis amabilis, Is Re- gulated during Floral Development and Induced by Wounding,” Plant Molecular Biology Reporter, Vol. 29, No. 1, 2011, pp. 106-116. doi:10.1007/s11105-010-0216-y [7] H. Yang, Y. P. Huang, H. J. Zhi and D. Y. Yu, “Proteo- mics-Based Analysis of Novel Genes Involved in Re- sponse toward Soybean Mosaic Virus Infection,” Mole- cular Biology Reporter, Vol. 38, No. 1, 2011, pp. 511- 521. doi:10.1007/s11033-010-0135-x [8] H. Y. Fan, J. Chen, C. M. Lv, C. Y. Zhang and Q. Sun, “Proteomic Analysis of F2 Generation of Cucumber against the Cucumber Powdery Mildew Disease,” Acta Horticulturae Sinica, Vol. 34, No. 2, 2007, pp. 349-354. [9] J. P. Brizard, C. Carapito, F. Delalande, A. Van Dorsse- laer and C. Brugidou, “Proteome Analysis of Plant-Virus Interactome: Comprehensive Data for Virus Multiplica- tion inside Their Hosts,” Molecular and Cell Proteomics, Vol. 5, No. 12, 2006, pp. 2279-2297. doi:10.1074/mcp.M600173-MCP200 [10] M. Ventelon-Debout, F. Delalande, J. P. Brizard, H. Die- mer, D. A. Van and C. Brugidou, “Proteome Analysis of Cultivar-Specific Degradation of Oryza sativa Indica and O. sativa Japonica Cellular Suspensions Undergoing Rice Yellow Mottle Virus Infection,” Proteomics, Vol. 4, No. 1, 2004, pp. 216-225. doi:10.1002/pmic.200300502 [11] C. L. Yu, Y. Yang, X. M. Wang, C. Q. Yan and J. P. Chen, “Recent Advances in Proteomic Studies on Rice- Pathogen Interactions,” China Journal of Rice Science, Vol. 24, No. 6, 2010, pp. 647-651. [12] J. Zhao, Q. Liu, H. Zhang, Q. Jia, Y. Hong and Y. Liu, “The RuBisCO Small Subunit Is Involved in the Toba- movirus Movement and Tm-22-Mediated Extreme Re- sistance,” Plant Physiology, Vol. 161, No. 1, 2013, pp. 374-383. doi:10.1104/pp.112.209213 [13] M. Babu, A. Gagarinova, J. Brandle and A. Wang, “As- sociation of the Transcriptional Response of Soybean Plants with Soybean Mosaic Virus Systemic Infection,” Journal of General Virology, Vol. 89, No. 4, 2008, pp. 1069-1080. doi:10.1099/vir.0.83531-0 [14] J. Casado-Vela, S. Selles and R. Martinez, “Proteomic Analysis of Tobacco Mosaic Virus-Infected Tomato (Ly- copersicon esculentum M.) Fruits and Detection of Viral Coat Protein,” Proteomics, Vol. 6, No. 1, 2006, pp. 196- 206. doi:10.1002/pmic.200500317 [15] S. Golem and J. Culver, “Tobacco Mosaic Virus Induced Alterations in the Gene Expression Profile of Arabidopsis thaliana,” Molecular Plant Microbe Interaction, Vol. 16, No. 8, 2003, pp. 681-688. doi:10.1094/MPMI.2003.16.8.681 [16] T. Kühne, N. N. Shi, G. Proeseler, M. J. Adams and K. Kanyuka, “The Ability of a Bymovirus to Overcome the rym4-Mediated Resistance in Barley Correlates with a Codon Change in the VPg Coding Region on RNA1,” Journal of General Virology, Vol. 84, No. 10, 2003, pp. 2853-2859. doi:10.1099/vir.0.19347-0 [17] N. N. Shi, Y. Xu, K. F. Yang, Y. Chen, H. Z. Wang, X. B. Xu and Y. G. Hong, “Development of a Sensitive Diag- nostic Assay to Detect Cymbidium Mosaic Virus and Odontoglossum Ringspot Virus in Members of the Or- chidaceae,” The Journal of Horticultural Science and Biotechnology, Vol. 86, No. 1, 2011, pp. 69-73. http://wrap.warwick.ac.uk/id/eprint/42020 [18] M. M. Bradford, “A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding,” Analytical Bio- chemistry, Vol. 72, 1976, pp. 248-254. doi:10.1016/0003-2697(76)90527-3 [19] J. P. Reichheld, D. Mestres-Ortega, C. Laloi and Y. Meyer, “The Multigenic Family of Thioredoxin h in Arabidopsis thaliana: Specific Expression and Stress Re- sponse,” Plant Physiology and Biochemistry, Vol. 40, 2002, pp. 685-690. doi:10.1016/S0981-9428(02)01406-7 [20] T. Song, “Cloning and Functional Analyses of a Tobacco Thioredoxin Gene,” Journal of Ludong University, Vol. 23, No. 3, 2007, pp. 256-260. [21] N. J. Poysti and I. J. Oresnik, “Characterization of Si- norhizobium meliloti Triose Phosphate Isomerase Genes,” Journal of Bacteriology, Vol. 189, No. 9, 2007, pp. 3445- 3451. doi:10.1128/JB.01707-06 [22] C. C. Fritz, F. P. Wolter, V. Schenkemeyer, T. Herget and P. H. Schreier, “The Gene Family Encoding the Ribulose- (1,5)-Bisphosphate Carboxylase/Oxygenase (Rubisco) Sm all Subunit of Potato,” Gene, Vol. 137, No. 2, 1993, pp. 271- 274. doi:10.1126/science.1106974G. P. Zhu, G. B. Gold- ing and A. M. Dean, “The Selective Cause of an Ancient Adaptation,” Science, Vol. 307, No. 5713, 2005, pp. 1279-1282. [24] I. L. Jung, S. K. Kim and I. G. Kim, “The RpoS-Mediated Regulation of Isocitrate Dehydrogenase Gene Expression in Escherichia coli,” Current Microbiology, Vol. 52, No. 1, 2006, pp. 21-26. doi:10.1007/s00284-005-8006-8 [25] A. Schmidt and K. Jäger, “Open Questions about Sulfur Metabolism in Plants,” Annual Review of Plant Physiol- ogy and Plant Molecular Biology, Vol. 43, 1992, pp. 325- 349. doi:10.1146/annurev.pp.43.060192.001545 [26] Y. L. Zhuang, G. J. Ren, C. M. He, X. Y. Li, Q. M. Meng, C. F. Zhu, R. C. Wang and J. R. Zhang, “Cloning and Characterization of a Maize cDNA Encoding Glutamate Decarboxylase,” Plant Molecular Biology Reporter, Vol. 28, No. 4, 2010, pp. 620-626. doi:10.1007/s11105-010-0191-3 [27] J. H. Lee, Y. J. Kim, D. Y. Jeone, G. Sathiyaraj, R. K. Copyright © 2013 SciRes. AJPS  Proteomics-Based Analysis of Phalaenopsis amabilis in Response toward Cymbidium Mosaic Virus and/or Odontoglossum Ringspot Virus Infection Copyright © 2013 SciRes. AJPS 1862 Pulla, J. S. Shim, J. G. Yin and D. C. Yang, “Isolation and Characterization of a Glutamate Decarboxylase (GAD) Gene and Their Differential Expression in Response to Abiotic Stresses from Panax Ginseng C. A. Meyer,” Mo- lecular Biology Reporter, Vol. 37, No. 7, 2010, pp. 3455- 3463. doi:10.1007/s11033-009-9937-0 [28] R. Valentina, C. Claudia, D. P. Benedetta, D. Nunzianna, A. Angela and D. Angela, “The Ribosomal Protein L2 Interacts with the RNA Polymerase α Subunit and Acts as a Transcription Modulator in Escherichia coli,” Journal of bacteriology, Vol. 192, 2010, pp. 1882-1889. doi:10.1128/JB.01503-09 [29] K. T. Nguyen, X. B. Hu, C. Colton, R. Chakrabarti, M. X. Zhu and D. H. Pei, “Characterization of a Human Peptide Deformylase: Implications for Antibacterial Drug Design, ” Biochemistry, Vol. 42, No. 33, 2003, pp. 9952-9958. doi:10.1021/bi0346446
|