 Open Journal of Genetics, 2013, 3, 159-170 OJGen http://dx.doi.org/10.4236/ojgen.2013.33018 Published Online September 2013 (http://www.scirp.org/journal/ojgen/) Evolutionarily conserved features of the retained intron in alternative transcripts of the nxf1 (nuclear export factor) genes in different organisms Ludmila A. Mamon, Sergey F. Kliver, Elena V. Golubkova* Department of Genetics, St. Petersburg State University, St. Petersburg, Russia Email: mamon@LM2010.spb.edu, mahajrod@gmail.com, *gelena@EG10217.spb.edu Received 27 April 2013; revised 30 May 2013; accepted 12 June 2013 Copyright © 2013 Ludmila A. Mamon et al. This is an open access article distributed under the Creative Commons Attribution Li- cense, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. ABSTRACT One of the features of intron-containing genes of the nxf (nuclear export factor) family in different organ- isms is the presence of an evolutionarily conserved exon-intron block: exon 110nt-intron-exon 37nt. The intron in this evolutionarily conserved block, which we call a “cassette” intron, can be excised or retained in alternative transcripts of nxf1. It corresponds to intron 10 - 11 in the genes that are orthologous to nxf1 in vertebrates, and intron 5 - 6 in the genes that are orthologous to nxf1 in Drosophilidae. The align- ment of sequences of cassette introns in nxf1 genes in vertebrates has revealed four evolutionarily con- served sequences: 1) 5’ flanking sequence, 2) a region containing СТЕ (constitutive transport element), 3) third conserved sequence, and 4) 3’ flanking sequence. Introns 5 - 6 of nxf1 in Drosophilidae have no similar conserved sequences. The results of sequence align- ment demonstrate a similarity between cassette in- trons of nxf1 in Drosophilidae in two poly(A) se- quences. The prevalence of Dm nxf1 transcripts con- taining cassette intron 5 - 6 under completely spliced transcripts in the heads of adult Drosophila melan- ogaster suggests a functional importance of tran- scripts that contain a retained intron. Evolutionary conservation, which in Drosophilidae is evident in the presence of poly(A) sequences in cassette introns of the nxf1 genes, is an adaptive feature: the poly(A) sequences are capable of mimicking the 3’-end of transcripts, promote transport from the nucleus to the cytoplasm, or are involved in NMD control. The ability to form characteristic secondary structures is a common feature of nxf1 cassette introns. Keywords: Nxf; Intron Retention; CTE; Poly(A); Drosophila; Vertebrates 1. INTRODUCTION 1.1. The nxf Gene Family and Functions of the nxf1 Gene The nxf (nuclear export factor) gene family was named after the function of the universal gene nxf1 responsible for the nuclear-cytoplasmic transport of most mRNAs [1-3]. This path of mRNA export is RanGTP-independ- ent [4]. Several proteins involved in mRNA export pathways are less conserved and presumably appeared later in the evolution of eukaryotes [5]. The blocking of the transport path enabled by the NXF1 protein results in the accumulation of polyadenylated RNAs within the nucleus [4,6,7]. Genes of the nxf family have been found in eukaryotic organisms of the Opisthokonta group, and are character- ized by evolutionary conservation [1,5,8]. Genomes of various fungi have only one gene, Mex67, which belongs to this family; animals usually have two to five paralo- gous genes (see Table 1) [1,9-11]. Plants and some pro- tozoa lack genes of the nxf family [5]. nxf1 genes are the most evolutionarily conserved in the nxf family. What is even more interesting is that nxf1 genes across different species of mammals exhibit a much greater degree of similarity than Mm nxf1 and Mm nxf2 or Hs nxf1 and Hs nxf2 exhibit between themselves (Figure 1). In the S. cerevisiae yeast, the factor Mex67 (mRNA EXport factor of 67 kDa), which is orthologous to other NXF1 proteins of the eukaryotes of the Opisthokonta group, is also involved in the nuclear-cytoplasmic trans- port of ribosomal RNAs [5,12,13]. Initially, the NXF1 protein in humans was identified as a potential cytoplasmic cofactor for Tip (tyrosine kinase interacting protein) encoded by the herpesvirus saimiri, and was named TAP (Tip-associated protein) *Corresponding author. OPEN ACCESS 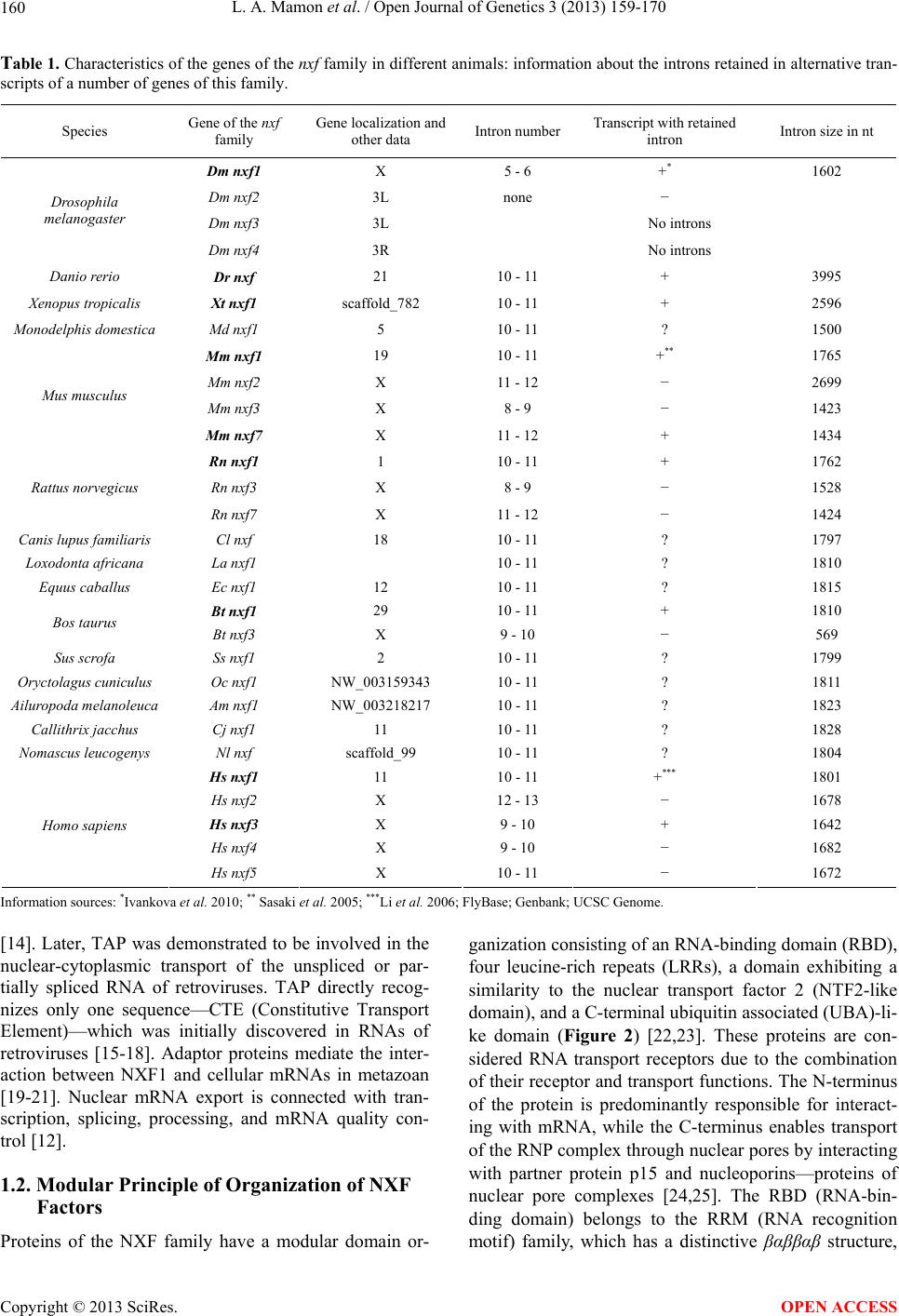 L. A. Mamon et al. / Open Journal of Genetics 3 (2013) 159-170 160 Table 1. Characteristics of the genes of the nxf family in different animals: information about the introns retained in alternative tran- scripts of a number of genes of this family. Species Gene of the nxf family Gene localization and other data Intron numberTranscript with retained intron Intron size in nt Dm nxf1 X 5 - 6 +* 1602 Dm nxf2 3L none − Dm nxf3 3L No introns Drosophila melanogaster Dm nxf4 3R No introns Danio rerio Dr nxf 21 10 - 11 + 3995 Xenopus tropicalis Xt nxf1 scaffold_782 10 - 11 + 2596 Monodelphis domestica Md nxf1 5 10 - 11 ? 1500 Mm nxf1 19 10 - 11 +** 1765 Mm nxf2 X 11 - 12 − 2699 Mm nxf3 X 8 - 9 − 1423 Mus musculus Mm nxf7 X 11 - 12 + 1434 Rn nxf1 1 10 - 11 + 1762 Rn nxf3 X 8 - 9 − 1528 Rattus norvegicus Rn nxf7 X 11 - 12 − 1424 Canis lupus familiaris Cl nxf 18 10 - 11 ? 1797 Loxodonta africana La nxf1 10 - 11 ? 1810 Equus caballus Ec nxf1 12 10 - 11 ? 1815 Bt nxf1 29 10 - 11 + 1810 Bos taurus Bt nxf3 X 9 - 10 − 569 Sus scrofa Ss nxf1 2 10 - 11 ? 1799 Oryctolagus cuniculus Oc nxf1 NW_003159343 10 - 11 ? 1811 Ailuropoda melanoleuca Am nxf1 NW_003218217 10 - 11 ? 1823 Callithrix jacchus Cj nxf1 11 10 - 11 ? 1828 Nomascus leucogenys Nl nxf scaffold_99 10 - 11 ? 1804 Hs nxf1 11 10 - 11 +*** 1801 Hs nxf2 X 12 - 13 − 1678 Hs nxf3 X 9 - 10 + 1642 Hs nxf4 X 9 - 10 − 1682 Homo sapiens Hs nxf5 X 10 - 11 − 1672 Information sources: *Ivankova et al. 2010; ** Sasaki et al. 2005; ***Li et al. 2006; FlyBase; Genbank; UCSC Genome. [14]. Later, TAP was demonstrated to be involved in the nuclear-cytoplasmic transport of the unspliced or par- tially spliced RNA of retroviruses. TAP directly recog- nizes only one sequence—CTE (Constitutive Transport Element)—which was initially discovered in RNAs of retroviruses [15-18]. Adaptor proteins mediate the inter- action between NXF1 and cellular mRNAs in metazoan [19-21]. Nuclear mRNA export is connected with tran- scription, splicing, processing, and mRNA quality con- trol [12]. 1.2. Modular Principle of Organization of NXF Factors Proteins of the NXF family have a modular domain or- ganization consisting of an RNA-binding domain (RBD), four leucine-rich repeats (LRRs), a domain exhibiting a similarity to the nuclear transport factor 2 (NTF2-like domain), and a C-terminal ubiquitin associated (UBA)-li- ke domain (Figure 2) [22,23]. These proteins are con- sidered RNA transport receptors due to the combination of their receptor and transport functions. The N-terminus of the protein is predominantly responsible for interact- ing with mRNA, while the C-terminus enables transport of the RNP complex through nuclear pores by interacting with partner protein p15 and nucleoporins—proteins of nuclear pore complexes [24,25]. The RBD (RNA-bin- ding domain) belongs to the RRM (RNA recognition motif) family, which has a distinctive βαββαβ structure, Copyright © 2013 SciRes. OPEN ACCESS  L. A. Mamon et al. / Open Journal of Genetics 3 (2013) 159-170 161 Figure 1. Phylogenetic tree of NXF family sequences, constructed for species of the phyla Nematoda, Arthropoda, and Chordata. The tree was constructed for sequences of ubiquitous transcripts. The tree was drawn by the MrBayes program (2,000,000 generations). Numbers denote posterior clade probabilities. Figure 2. Intron-exon structure of Hs nxf1 and Dm nxf1 genes. Arrow shows cassette intron. The color of exons corresponds to color of domains in protein. characteristic of many RNA-binding proteins, such as РА В —poly(A)-binding-protein, Sex-lethal protein (Sxl), and U1A and U2B″ splicing factors [16]. The receptor function is much older: RRM have been discovered in proteins of prokaryotes [26]. While the RRM domain alone is sufficient for binding with mRNA in vitro, both RRM and LRR are necessary for exporting mRNA in vivo [16,23]. The C-terminal end of NXF proteins is represented by NTF2-like and UBA-like domains. The NTF2-like do- main is thought to be derived from a prokaryotic precur- sor [27]. The C-terminal end in proteins paralogous to NXF often differs from the corresponding parts in NXF1 proteins. Consequently, many paralogous proteins, such as Dm NXF3, Dm NXF4, Ce NXF2, Hs NXF3, Hs NXF5, Mm NXF3, and Mm NXF7 are incapable of binding with nucleoporins [1,8,11,28-30]. In most cases this is due to the loss of a UBA-like domain in paralo- gous proteins, or the absence of both domains (as in the case of the Dm NXF4 protein) [1,7,31]. The unification of the transport and receptor functions probably took place simultaneously with the origin of the eukaryotic cell. In eukaryotes, transcription and mRNA processing are physically and temporally separated from translation by the nuclear membrane, which required the formation of mechanisms for the active transport of macromolecules, including mRNAs, from the nucleus to the cytoplasm. Most transcripts begin translation soon Copyright © 2013 SciRes. OPEN ACCESS  L. A. Mamon et al. / Open Journal of Genetics 3 (2013) 159-170 162 after they exit the nucleus. The specific class of tran- scripts capable of staying for extended periods of time in the cytoplasm in a state unavailable for translation pre- sumably required specialization of transport receptors. There are paralogous genes that, unlike the universal gene nxf1, usually exhibit an organ-specific character of expression in mammals [1,8]. The products of paralo- gous genes are usually localized in the cytoplasm [8,11, 28,30,32]. Several processes of transformation of genetic mate- rial underlie the formation of gene families: duplication, separation of certain genes, or fusion of different parts of genes [33-36]. The existence of clusters of linked genes in different organisms seems to support the hypothesis that the nxf gene family was formed via duplication [1,8]. Integration of cDNA copies of transcripts into a ge- nome may also produce gene copies that contain no in- trons [36-38]. Perhaps this mechanism eventually pro- duced genes Dm nxf3 and Dm nxf4, which contain no introns in D. melanogaster. It is important to note, how- ever, that the lack of introns in genes that are orthologous to Dm nxf4 is not characteristic of all species of the ge- nus Drosophila for which it has been described (FlyBase, 2012). LLR and NTF2-like domains responsible for the in- teraction of NXFs with various partner proteins are the most highly conserved [39]. 1.3. Interaction of NXF1 Proteins with RNA Nuclear mRNAs are exported as RNP-complexes [40]. NXF1 interacts with mRNA by means of adaptor pro- teins, which are part of mRNP complexes during tran- scription, processing, and transport [25,41-43]. During mRNA export, NXF1 partners with proteins E1B-AP5, RAE1, and members of evolutionarily conserved fami- lies of proteins—Yra1p in yeast and REF in mammals [24,44,45], as well as components of the TREX (TRan- scription-EXport) complex [40,43]. Most genes of eukaryotes have introns that are then deleted from pre-mRNA during splicing. As a rule, cel- lular mRNAs with introns do not leave the nucleus [16,46]. Translation of incompletely spliced mRNA due to the presence of a premature termination codon may produce truncated proteins, which are potentially delete- rious to a cell. In the nucleus, transcripts undergo quality control, and normally only completely spliced mRNAs are capable of export [47,48]. Genes that correspond to transcripts with intron reten- tion are not uncommon in humans [49]. The expression of intron-containing messages has been shown to occur in a variety of diseases including several cancers [50], and also as a response to vascular injury in rats [51]. Transcripts with retained introns may serve as sources of alternative protein products with an independent function that may, among other things, influence the function of a full-length product [52]. If mRNAs with retained introns are abundant in the cytoplasm, intron retention is probably regulated by fac- tors involved in both splicing and mRNA export. Some of the retroviruses have been shown to export unspliced RNA by means of cis-acting RNA elements, termed con- stitutive transport elements (CTEs), which interact di- rectly with cellular export proteins [53]. Export of mRNA with retained introns in simple retroviruses is carried out with the help of NXF1 (TAP), which binds directly to the CTE sequence in the genome of retrovi- ruses [15-18,24,53,54]. Microinjection experiments per- formed on Xenopus oocytes have demonstrated that TAP (Hs NXF1) directly interacts with the CTE, allowing the export of CTE-containing RNAs [15]. Moreover, TAP remains bound to CTE-containing RNAs in polyriboso- mes and may be present inside the nucleus or in the nu- clear rim, as well as in the cytoplasm [55]. 2. RESULTS AND DISCUSSIONS 2.1. Alternative Intron-Retaining Transcripts of nxf1 Genes Both the domain structure and the intron-exon structure of the NXF family proteins are evolutionarily conserved. Among the known nxf genes, most have an intron-exon structure. Mex67 in fungi, nxf4 in some species of the genus Drosophila, and Dm nxf3 in D. melanogaster, have no introns. A feature specific to nxf1 genes is the existence of al- ternative transcripts with a retained intron flanked by evolutionarily conserved exons: 110 nt upstream and 37 nt downstream of the respective intron. This intron from the evolutionarily conserved block, which can be excised or retained in alternative transcripts of nxf1, we named a “cassette” intron. Such an evolutionarily conserved exon- intron block is characteristic of most of the nxf family genes that exhibit an intron-exon structure. There exist nxf genes in which the sequences of exons 110 nt and 37 nt are not separated by an intron, and are represented by exon 147 nt: Dm nxf2 and Ce nxf2, for example. Tran- scripts of nxf1 genes with a cassette intron between ex- ons 110 nt and 37 nt have been shown for nxf1 genes in M. musculus [8], H. sapiens [56], and D. melanogaster [57]. Such nxf1 transcripts have been found in other spe- cies, as well. ESTs (expressed sequence tags) include a portion of one of the aforementioned exons and a portion of the cassette intron (Genbank, UCSC) (see Table 1). We have demonstrated that the transcript with cassette intron 5 - 6 in the head tissues of adult fruit flies exceeds the universal, completely spliced transcript of the gene Dm nxf1, in its relative content (Figure 3) [57]. Usually, Copyright © 2013 SciRes. OPEN ACCESS  L. A. Mamon et al. / Open Journal of Genetics 3 (2013) 159-170 163 intron-containing transcripts stay in the nucleus [46,48]. Even if they enter the cytoplasm, they are subject to the cytoplasmic mRNA quality control mechanism (surveil- lance) termed NMD (nonsense-mediated mRNA decay) [58,59]. The large number of transcripts of nxf1 genes with a retained intron in different organisms raises the following questions: 1) How do intron-containing transcripts manage to exit the nucleus, bypassing the mRNA quality control mechanism? 2) What makes the intron-containing transcripts im- mune to NMD in the cytoplasm? 3) What evolutionarily conserved features characterize cassette introns in alternative intron-containing tran- scripts of nxf1 genes? Analyzing specific features of the intron retained in alternative transcripts of nxf1 genes may help answer these questions. The existence of a sequence with a length of about 100 nt, which resembles the CTE of the MPMV virus in cassette introns 10 - 11, represented in genes Hs nxf1 and Mm nxf1, has been observed [56]. This sequence is highly conserved in cassette introns of nxf1 in various animals (Figure 4). Hs nxf1 mRNA with the retained intron is exported from the nucleus and is represented in a polysomic fraction [56]. Due to the pres- ence of a premature stop-codon at the beginning of the cassette intron 10 - 11, translation results in a truncated protein. A similar truncated protein has been discovered in human cells [56]. The CTE sequence is an important element affecting the expression of intron-retaining mRNAs in mammals [60,61]. The CTE has not been discovered in the corresponding introns of paralogous genes of the nxf family in mice and humans. When comparing introns 5 - 6 of nxf1 genes in different Figure 3. Northern blot analysis of to- tal RNA from dif- ferent tissues of the adult Drosophila female. 1—ovaries, 2—heads. The sizes (in kb) of molecu- lar weight RNA markers are indi- cated on the left (Ivankova et al., 2010). species of Drosophila, we have not discovered extended homologous sequences (such as cassette introns in genes orthologous to the nxf1 of vertebrates). However, we have found a common feature of introns 5 - 6 of the nxf1 gene in different species of Drosophila: the presence of two poly(A) sequences, each with a length of around 100 nt (Figure 5). This feature accounts for the difference of cassette introns 5 - 6 of nxf1 genes in Drosophilidae from cassette introns 10 - 11 of nxf1 genes in vertebrates. The cassette introns of nxf1 in Drosophilidae form complex secondary structures. Poly(A) sequences of in- trons 5 - 6 of nxf1 in Drosophilidae is usually located in loops (Figure 6). Because the same RNA sequence can form several secondary structure variants, Figure 6 de- picts only the most probable structures of introns 5 - 6 of nxf1 genes in some Drosophila species. The choice of species was made in accordance with the relationships between Drosophila species, taking into consideration the phylogenetic tree constructed with the divergence of sequences of intron 5 - 6 (Figure 7). What happens to the poly(A) RNA sequence? The presence of a poly(A) tail is an important element in RNA export [62]. Export efficiency depends on the length of this sequence. The poly(A) sequence is respon- sible for binding with corresponding proteins [63]. The poly(A) sequence is believed to be a recognition target for export factors [62]. Taking into account the secondary structure of cassette introns of nxf1 genes in Drosophilidae, poly(A) se- quences are probably open for interaction with proteins that may recognize them. Poly(A) sequences may not only serve as nuclear export markers, but may also pro- tect the corresponding transcript from degradation in the cytoplasm. It has been demonstrated that the protein PABPC1 (cytoplasmic poly(A)-binding protein) sup- presses NMD in D. melanogaster [64]. 2.2. Specific Features of Retained Introns in Alternative Transcripts of nxf Genes Cassette introns of nxf1 genes in different species of mammals form complex secondary structures, as well as cassette introns in Drosophilidae nxf1 (Figure 8). It is possible that this feature of cassette introns determines the fate of intron-retaining transcripts in the cytoplasm. It is also possible that cassette introns of nxf1 genes have an independent function both inside the intron-retained transcript, and as the product of splicing of the pre- mRNA of the nxf1. RNAs transcribed from introns are known to participate in a number of processes related to post-transcriptional control of gene expression [65]. Cassette introns of nxf1 genes in vertebrates have four evolutionarily conserved regions (Figure 4). The first, with a length of around 130 nt, is the 5’-end of cassette introns (Figure 4). This sequence includes 17 complete Copyright © 2013 SciRes. OPEN ACCESS 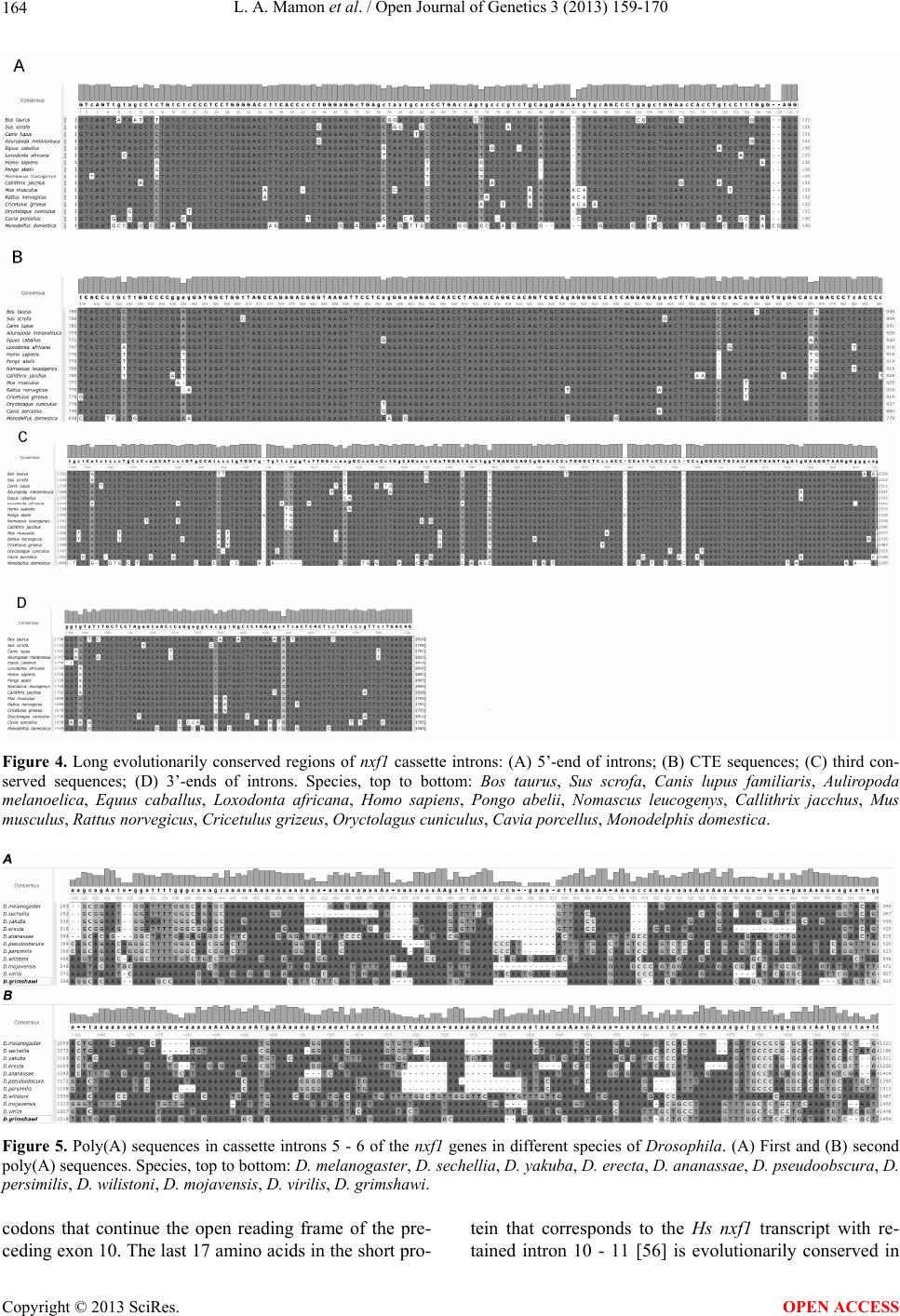 L. A. Mamon et al. / Open Journal of Genetics 3 (2013) 159-170 Copyright © 2013 SciRes. 164 Figure 4. Long evolutionarily conserved regions of nxf1 cassette introns: (A) 5’-end of introns; (B) CTE sequences; (C) third con- served sequences; (D) 3’-ends of introns. Species, top to bottom: Bos taurus, Sus scrofa, Canis lupus familiaris, Auliropoda melanoelica, Equus caballus, Loxodonta africana, Homo sapiens, Pongo abelii, Nomascus leucogenys, Callithrix jacchus, Mus musculus, Rattus norvegicus, Cricetulus grizeus, Oryctolagus cuniculus, Cavia porcellus, Monodelphis domestica. Figure 5. Poly(A) sequences in cassette introns 5 - 6 of the nxf1 genes in different species of Drosophila. (A) First and (B) second poly(A) sequences. Species, top to bottom: D. melanogaster, D. sechellia, D. yakuba, D. erecta, D. ananassae, D. pseudoobscura, D. persimilis, D. wilistoni, D. mojavensis, D. virilis, D. grimshawi. codons that continue the open reading frame of the pre- ceding exon 10. The last 17 amino acids in the short pro- tein that corresponds to the Hs nxf1 transcript with re- tained intron 10 - 11 [56] is evolutionarily conserved in OPEN ACCESS  L. A. Mamon et al. / Open Journal of Genetics 3 (2013) 159-170 165 Figure 6. Secondary structures of cassette introns 5 - 6 of the nxf1 genes in five species of Drosophila. Marked loops, formed using poly(A) sequences. Figure 7. Phylogenetic tree constructed using divergences of the sequences of cassette intron 5 - 6 of nxf1 genes in eleven species of Drosophila. truncated proteins corresponding to the intron-containing transcripts in other vertebrates (Figure 9). The first stop-codon in intron 10 - 11 is duplicated by the next one, 12 nt down. Evolutionary conservation of the first region (Figure 4(a)) suggests the functional significance of the truncated NXF1. The only species that is not present in the list is the M. musculus. Deleting one of the five nu- cleotides (C) at the position 40 - 44 of Mm nxf1 intron shifts the open reading frame compared to other species, and the C-terminus of the supposed truncated protein corresponds to the 80th full codon in intron 10 - 11 of Mm nxf1. The first conserved region of cassette introns (Figure 4(a)) is partially complimentary to the fourth conserved region (Figure 4(d)), which is at the 3’-end of the intron. In secondary RNA structures the first and fourth regions form a “stem”, thus “closing” the secondary structure of cassette introns 10 - 11 (Figure 8). Copyright © 2013 SciRes. OPEN ACCESS  L. A. Mamon et al. / Open Journal of Genetics 3 (2013) 159-170 166 The second evolutionarily conserved sequence (Fig- ure 4(b)) contains CTE and has been identified by Li with co-authors [56]. Whereas this sequence does not contain an ORF (open reading frame), the third evolu- tionarily conserved sequence (Figure 4(c)) in introns 10 - 11 of the genes Hs nxf1 and Mm nxf1 is a long open reading frame. The supposed proteins, which correspond to the open reading frame in the third evolutionarily conserved se- quence of introns 10 - 11 of the genes Hs nxf1 and Mm nxf1, exhibit a great degree of similarity (data not pro- vided). If the open reading frame is significant, it may be preserved in other cassette introns of nxf1 genes in ver- tebrates. This, however, we did not observe when evalu- ating the third evolutionarily conserved region of nxf1 genes in other vertebrates for the presence of a long evo- lutionarily conserved ORF. In all the depicted secondary structures of cassette introns, region three is a hairpin (Figure 8), which supports the assumption of a structural role of this evolutionarily conserved region. Long non-coding RNAs often contain ORF. It is speculated that such sequences can be translated, but with very low efficiency, or only at a specific stage of development [66]. Little is known about ncRNAs that code for functionally significant short proteins [67]. Po- tentially, the possibility for synthesizing short peptides also exists for cassette introns of nxf1 genes. Short pep- tides, according to some researchers [68], may be a new class of bioactive signaling molecules. Many transcripts, including those that code for a pro- tein, also carry out regulatory functions. They are capa- ble of interactions via a specific nucleotide sequence, thus playing a structural role or serving as catalysts [66,69]. Evolutionarily conserved sequences in the cas- sette intron of nxf1 genes raise the questions of what adaptive advantages these sequences bring, and what the functional significance of intron-retained transcripts is. The evolutionary path of forming adaptive features may vary depending on the characteristics of a particular taxon. The existence of sequences that may facilitate export of transcripts containing these introns from the nucleus to the cytoplasm is a feature shared between introns 10 - 11 in nxf1 genes in vertebrates and introns 5 - 6 in nxf1 genes of different species of Drosophila. This, along with the ability of these introns to form secondary structures, Figure 8. Secondary structures corresponding to the minimum energy state of the cassette intron of the nxf1 gene in different species of mammals. A—Callithrix jacchus; B—Canis lupus familiaris; C—Monodelphis domestica; D— Loxodontha africana; E—Rattus norvegicus; F—Equus cabalus. Roman numerals denote evolutionarily conserved re- gions: I—5’-end of intron, II—CTE sequence, III—third conserved sequence, IV—3’-end of intron. Copyright © 2013 SciRes. OPEN ACCESS  L. A. Mamon et al. / Open Journal of Genetics 3 (2013) 159-170 167 Figure 9. C-terminal end of the short isoform of the NXF1 protein in mam- mals, which is presumably translated from an alternative transcript containing unspliced intron 10 - 11. suggests the functional significance of such transcripts. It is possible that the retention of the cassette intron in transcripts of nxf1 genes facilitates the synthesis of the truncated protein NXF1. We have shown that in the heads of adult flies the transcript with cassette intron 5 - 6 is more abundant than completely spliced transcripts (Figure 3) [57]. It would be interesting to test the hy- pothesis about the existence of the truncated protein Dm NXF1 specific to the brain tissues of the fruit fly. 3. CONCLUSION A great variety of alternative transcripts, including in- tron-retained and non-coding RNAs, is characteristic of the nervous system. Thus, it is not surprising that tran- scripts with cassette intron 5 - 6 in Dm nxf1 are found primarily in the head of fruit flies. Insects that are char- acterized by complex behavior are model organisms, which facilitate the study of the molecular mechanisms of the nervous system. The large number of transcripts with retained intron 5 - 6 of the Dm nxf1 gene in the head of adult fruit flies points to the functional significance of these transcripts. The existence of evolutionarily con- served sequences in the cassette introns of nxf1 genes in animals within some taxonomic groups can be regarded as the acquisition of adaptive properties of the corre- sponding intron-containing transcripts. These sequences may provide benefits in nuclear-cytoplasmic transport, resistance to NMD, and a possible involvement in the regulation of gene expression. The ability of cassette introns to form complex secondary structures suggests that these introns may have an independent, possibly structural, function and also be a source of non-coding RNAs. 4. METHODS For the analysis of nucleotide sequences we used the Unipro UGENE v1.10.0 software suite (http://ugene.unipro.ru) [70] along with third-party tools: ClustalW (http://www.clustal.org/clustal2/) [71] and T- Coffee (http://www.tcoffee.org/Projects/tcoffee/) [72] alignment algorithms. The tree structure was built using the MrBayes pro- gram (2,000,000 generations) (http://mrbayes.sourceforge.net/index.php) [73]. To view the phylogram in a more convenient form we used the FigTree v1.3.1 editor (http://tree.bio.ed.ac.uk/software/figtree/). To build the secondary structures of nucleotide se- quences we used the UNAFold v3.8 suite (http://mfold.rna.albany.edu/). Secondary structure pre- diction was based on the minimum free energy calcula- tion [74]. The following databases were used: Genbank (http://www.ncbi.nlm.nih.gov/) Flybase (http://flybase.org/) UCSC Genome (http://genome.ucsc.edu) 5. ACKNOWLEDGEMENTS Our work is supported by the Leading Scientific Schools program (SciSch-6455.2010.4; SciSch-5345.2012.4), the Russian Foundation for Basic Research (projects 09-04-00697 and 12-04-00934), Federal Tar- get Program (02.740.11.0698). REFERENCES [1] Herold, A., Suyama, M., Rodrigues, J.P., Braun, I.C., Copyright © 2013 SciRes. OPEN ACCESS  L. A. Mamon et al. / Open Journal of Genetics 3 (2013) 159-170 168 Kutay, U., Carmo-Fonseca, M., Bork, P. and Izaurralde, E. (2000) TAP (NXF1) Belongs to a multigene family of putative RNA export factors with a conserved modular architecture. Molecular and Cellular Biology, 20, 8996- 9008. doi:10.1128/MCB.20.23.8996-9008.2000 [2] Wilkie, G.S., Zimyanin, V., Kirby, R., Korey, C., Fran- cis-Lang, H., Van Vactor, D. and Davis, I. (2001) Small bristles, the Drosophila ortholog of NXF-1, is essential for mRNA export throughout development. RNA, 7, 1781-1792. [3] Herold, A., Teixeira, L. and Izaurralde, E. (2003) Ge- nome-wide analysis of nuclear mRNA export pathways in Drosophila. EMBO Journal, 22, 2472-2483. doi:10.1093/emboj/cdg233 [4] Hurt, E., Strässer, K., Segref, A., Bailer, S., Schlaich, N., Presutti, C., Tollervey, D. and Jansen, R. (2000) Mex67p mediates nuclear export of a variety of RNA polymerase II transcripts. The Journal of Biological Chemistry, 275, 8361-8368. doi:10.1074/jbc.275.12.8361 [5] Serpeloni, M., Vidal, N., Goldenberg, S., Avila, A.R. and Hoffmann, F.G. (2011) Comparative genomics of pro- teins involved in RNA nucleoplasmic export. BMC Evo- lutionary Biology, 11, 7. http://www.biomedcentral.com/1471-2148/11/7 doi:10.1186/1471-2148-11-7 [6] Segref, A., Sharma, K., Doye, V., Hellwig, A., Huber, J., Lührmann, R. and Hurt, E. (1997) Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO Journal, 16, 3256-3271. doi:10.1093/emboj/16.11.3256 [7] Herold, A., Klymenko, T. and Izaurralde, E. (2001) NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA, 7, 1768-1780. [8] Sasaki, M., Takeda, E., Takano, K., Yomogida, K., Kata- hira, J. and Yoneda, Y. (2005) Molecular cloning and functional characterization of mouse Nxf family gene products. Genom ic s, 85, 641-653. doi:10.1016/j.ygeno.2005.01.003 [9] Tan, W., Zolotukhin, A.S., Bear, J., Patenaude, D.J. and Felber, B.K. (2000) The mRNA export in Caenorhabditis elegans is mediated by Ce-NXF-1, an ortholog of human TAP/NXF and Saccharomyces cerevisiae Mex67p. RNA, 6, 1762-1772. doi:10.1017/S1355838200000832 [10] Tan, W., Zolotukhin, A.S., Tretyakova, I., Bear, J., Lin- dtner, S., Smulevitch, S.V. and Felber, B.K. (2005) Iden- tification and characterization of the mouse nuclear ex- port factor (Nxf) family members. Nucleic Acids Re- search, 33, 3855-3865. doi:10.1093/nar/gki706 [11] Jun, L., Frints, S., Duhamel, H., Herold, A., Abad-Rod- rigues, J., Dotti, C., Izaurralde, E., Marynen, P. and Froyen, G. (2001) NXF5, a novel member of the nuclear RNA export factor family, is lost in a male patient with a syndromic form of mental retardation. Current Biology, 11, 1381-1391. doi:10.1016/S0960-9822(01)00419-5 [12] Köhler, A. and Hurt, E. (2007) Exporting RNA from the nucleus to the cytoplasm. Nature Reviews Molecular Cell Biology, 8, 761-773. doi:10.1038/nrm2255 [13] Yao, W., Roser, D., Köhler, A., Bradatsch, B., Baβler, J. and Hurt, E. (2007) Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67- Mtr2. Molecular Cell, 26, 51-62. doi:10.1016/j.molcel.2007.02.018 [14] Yoon, D.W., Lee, H., Seol, W., DeMaria, M., Rosen- zweig, M. and Jung, J.U. (1997) Tap: A novel protein that interacts with tip of herpesvirus saimiri and induces lym- phocyte aggregation. Immunity, 6, 571-582. doi:10.1016/S1074-7613(00)80345-3 [15] Grüter, P., Tabemero, C., von Kobbe, C., Schmitt, C., Saavedra, C., Bachi, A., Wilm, M., Felber, B.K. and Izaurralde, E. (1998) TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Molecular Cell, 1, 649-659. doi:10.1016/S1097-2765(00)80065-9 [16] Liker, E., Fernandez, E., Izaurralde, E. and Conti, E. (2000) The structure of the mRNA nuclear export factor TAP reveals a cis arrangement of a non-canonical RNP domain and an LRR domain. EMBO Journal, 19, 5587- 5598. doi:10.1093/emboj/19.21.5587 [17] Zolotukhin, A.S., Michalowski, D., Smulevitch, S. and Felber, B.K. (2001) Retroviral constitutive transport ele- ment evolved from cellular TAP(NXF1)-binding sequences. Journal of Virology, 75, 5567-5575. doi:10.1128/JVI.75.12.5567-5575.2001 [18] Teplova, M., Wohlbold, L., Khin, N.W., Izaurralde, E. and Patel, D.J. (2012) Structure-function studies of nu- cleocytoplasmic transport of retroviral genomic RNA by mRNA export factor TAP. Nature Structural and Mo- lecular Biology, 18, 990-998. doi:10.1038/nsmb.2094 [19] Gatfield, D. and Izaurralde, E. (2002) REF1/Aly and the additional exon junction complex proteins and dispensa- ble for nuclear mRNA export. The Journal of Cell Biol- ogy, 159, 579-588. doi:10.1083/jcb.200207128 [20] Huang, Y., Gattoni, R., Stevenin, J. and Steitz, J.A. (2003) SR splicing factors serve as adapter proteins for TAP- dependent mRNA export. Molecular Cell, 11, 837-843. doi:10.1016/S1097-2765(03)00089-3 [21] Hautbergue, G.M., Hung, M.-L., Golovanov, A.P., Lian, L.-Y. and Wilson, S.A. (2008) Mutually exclusive inter- action drive handover of mRNA from export adaptor to TAP. Proceedings of the National Academy of Sciences of the USA, 105, 5154-5159. doi:10.1073/pnas.0709167105 [22] Bear, J., Tan, W., Zolotukhin, A.S., Tabernero, C., Hud- son, E.A. and Felber, B.K. (1999) Identification of novel import and export signals of human TAP, the protein that binds to the constitutive transport element to the type D retrovirus mRNAs. Molecular and Cellular Biology, 19, 6306-6317. [23] Katahira, J., Strässer, K., Podtelejnikov, A., Mann, M., Jung, J.U. and Hurt, E. (1999) The Mex67p-mediated nu- clear mRNA export pathway is conserved from yeast to human. EMBO Journal, 18, 2593-2609. doi:10.1093/emboj/18.9.2593 [24] Bachi, A., Braun, I.C., Rodrigues, J.P., Pante, N., Rib- beck, K., von Kobbe, C., Kutay, U., Wilm, M., Gorlich, D., Carmo-Fonseca, M. and Izaurralde, E. (2000) The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing Copyright © 2013 SciRes. OPEN ACCESS  L. A. Mamon et al. / Open Journal of Genetics 3 (2013) 159-170 169 RNA substrates. RNA, 6, 136-158. doi:10.1017/S1355838200991994 [25] Stutz, F. and Izaurralde, E. (2003) The interplay of nu- clear mRNP assembly, mRNA surveillance and export. Trends in Cell Biology, 13, 319-327. doi:10.1016/S0962-8924(03)00106-5 [26] Maruyama, K., Sato, N. and Ohta, N. (1999) Conserva- tion of structure and cold-regulation of RNA-binding proteins in cyanobacteria: Probable convergent evolution with eukaryotic glycine-rich RNA-binding proteins. Nu- cleic Acids Research, 27, 2029-2036. doi:10.1093/nar/27.9.2029 [27] Mans, B.J., Anantharaman, V., Aravind, L. and Koonin, E.V. (2004) Comparative genomics, evolution and origins of the nuclear envelope and pore complex. Cell Cycle, 3, 1612-1637. doi:10.4161/cc.3.12.1316 [28] Tretyakova, I., Zolotukhin, A.S., Tan, W., Bear, J., Propst, F., Ruthel, G. and Felber, B.K. (2005) NXF family pro- tein participates in cytoplasmic mRNA trafficking. The Journal of Biological Chemistry, 280, 31981-31990. doi:10.1074/jbc.M502736200 [29] Lévesque, L., Bor, Y-С., Matzat, L.H., Jin, L., Berbero- glu, S., Rekosh, D., Hammarskjöld, M.L. and Paschal, B.M. (2006) Mutations in Tap uncouple RNA export ac- tivity from translocation through the nuclear pore com- plex. Molecular Biology of the Cell, 17, 931-943. [30] Katahira, J., Miki, T., Takano, K., Maruhashi, M., Uchi- kawa, M., Tachibana, T. and Yoneda, Y. (2008) Nuclear RNA export factor 7 is localized in processing bodies and neuronal RNA granules through interactions with shut- tling hnRNPs. Nucleic Acids Research, 36, 616-628. doi:10.1093/nar/gkm556 [31] Izaurralde, E. (2001) “Friedrich Miescher Prize Awardee Lecture Review”. A conserved family of nuclear export receptors mediates the exit of messenger RNA to the cy- toplasm. Cellular and Molecular Life Sciences, 58, 1105- 1112. doi:10.1007/PL00000924 [32] Lai, D., Sakkas, D. and Huang, Y. (2006) The fragile X mental retardation protein interacts with a distinct mRNA nuclear export factor NXF2. RNA, 12, 1446-1449. doi:10.1261/rna.94306 [33] Ohno, S. (1970) Evolution by Gene Duplication. Springer- Verlag, New York. [34] Modrek, B. and Lee, C.J. (2003) Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nature Genetics, 34,177-180. doi:10.1038/ng1159 [35] Hughes, A.L. (2005) Gene duplication and the origin novel proteins. Proceedings of the National Academy of Sciences of the USA, 102, 8791-8792. doi:10.1073/pnas.0503922102 [36] Zhou, Q., Zhang, G., Zhang, Y., Xu, S., Zhao, R., Zhan, Z., Li, X., Ding, Y., Yang, S. and Wang, W. (2008) On the origin of new genes in Drosophila. Genome Research, 18, 1446-1455. doi:10.1101/gr.076588.108 [37] Brosius, J. (1991) Retroposons—Seeds of evolution. Sci- ence, 251, 753. doi:10.1126/science.1990437 [38] Bai, Y., Casola, C. and Betran, E. (2008) Evolutionary origin of regulatory regions of retrogenes in Drosophila. BMC Genomics, 9, 241. http://www.biomedcentral.com/1471-2164/9/241 [39] Braun, I.C., Herold, A., Rode, M., Conti, E. and Izaurral- de, E. (2001) Overexpression of TAP/p15 heterodimers bypasses nuclear retention and stimulates nuclear mRNA export. The Journal of Biological Chemistry, 276, 20536- 20543. doi:10.1074/jbc.M100400200 [40] Erkmann, J.A. and Kutay, U. (2004) Nuclear export of mRNA: From the site of transcription to the cytoplasm. Experimental Cell Research, 296, 12-20. [41] Cullen, B.R. (2003) Nuclear RNA export. Journal of Cell Science, 116, 587-597. doi:10.1242/jcs.00268 [42] Huang, Y.Q. and Steitz, J.A. (2005) SRprises along a messenger’s journey. Molecular Cell, 17, 613-615. doi:10.1016/j.molcel.2005.02.020 [43] Björk, P. and Wieslander, L. (2011) Nucleocytoplasmic mRNP export is an integral part of mRNP biogenesis. Chromosoma, 120, 23-38. doi:10.1007/s00412-010-0298-1 [44] Strässer, K. and Hurt, E. (2000) Yra1p, a conserved nu- clear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO Journal, 19, 410-420. doi:10.1093/emboj/19.3.410 [45] Stutz, F., Bachi, A., Doerks, T., Braun, I.C., Séraphin, B., Wilm, M., Bork, P. and Izaurralde, E. (2000) REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA, 6, 638-650. doi:10.1017/S1355838200000078 [46] Gencheva, M., Lin, T.-Y., Wu, X., Yang, L., Richard, C., Jones, M., Lin, S.-B. and Lin, R.-J. (2010) Nuclear reten- tion of unspliced pre-mRNAs by mutant DHX16/hPRP2, a spliceosomal DEAH-box protein. The Journal of Bio- logical Chemistry, 285, 35624-35632. doi:10.1074/jbc.M110.122309 [47] Chang, D.D. and Sharp, P.A. (1989) Regulation by HIV Rev depends upon recognition of splice sites. Cell, 59, 789-795. doi:10.1016/0092-8674(89)90602-8 [48] Legrain, P. and Rosbash, M. (1989) Some cis- and trans- acting mutations for splicing target pre-mRNA to the cy- toplasm. Cell, 57, 573-583. doi:10.1016/0092-8674(89)90127-X [49] Galante, P.A., Sakabe, N.J., Kirschbaum-Slager, N. and De Souza, S.J. (2004) Detection and evaluation of intron retention events in the human transcriptome. RNA, 10, 757-765. doi:10.1261/rna.5123504 [50] Justman, Q.A. and Clinton, G.M. (2002) Herstatin, an autoinhibitor of the human epidermal growth factor re- ceptor 2 tyrosine kinase, modulates epidermal growth factor signaling pathways resulting in growth arrest. The Journal of Biological Chemistry, 277, 20618-20624. doi:10.1074/jbc.M111359200 [51] Forrest, S.T., Barringhaus, K.G., Perlegas, D., Ham- marskjöld, M.L. and McNamara, C.A. (2004) Intron re- tention generates a novel Id3 isoform that inhibits vascu- lar lesion formation. The Journal of Biological Chemistry, 279, 32897-32903. doi:10.1074/jbc.M404882200 Copyright © 2013 SciRes. OPEN ACCESS  L. A. Mamon et al. / Open Journal of Genetics 3 (2013) 159-170 Copyright © 2013 SciRes. 170 OPEN ACCESS [52] Michael, I.P., Kurlender, L., Memari, N., Yousef, G.M., Du, D., Grass, L., Stephan, C., Jung, K. and Diamandis, E.P. (2005) Intron retention: A common splicing event within the human kallikrein gene family. Clinical Chem- istry, 51, 506-515. doi:10.1373/clinchem.2004.042341 [53] Hammarskjöld, M.-L. (2001) Constitutive transport ele- ment-mediated nuclear export. Current Topics in Micro- biology and Immunology, 259, 77-93. doi:10.1007/978-3-642-56597-7_4 [54] Kang, Y. and Cullen, B.R. (1999) The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleoplasmic transport sequences. Genes and Development, 13, 1126-1139. doi:10.1101/gad.13.9.1126 [55] Jin, L., Guzik, B.W., Bor, Y.-C., Rekosh, D. and Ham- marskjöld, M.-L. (2003) Tap and NXT promote transla- tion of unspliced mRNA. Genes and Development, 17, 3075-3086. doi:10.1101/gad.1155703 [56] Li, Y., Bor, Y.-C., Misawa, Y., Xue, Y., Rekosh, D. and Hammarskjöld, M.-L. (2006) An intron with a constitu- tive transport element is retained in a Tap messenger RNA. Nature, 443, 234-237. doi:10.1038/nature05107 [57] Ivankova, N., Tretyakova, I., Lyozin, G., Avanesyan, E., Zolotukhin, A., Zatsepina, O.G., Evgen’ev, M.B. and Mamon, L.A. (2010) Alternative transcripts expressed by small bristles, the Drosophila melanogaster nxf1 gene. Gene, 458, 11-19. doi:10.1016/j.gene.2010.02.013 [58] Conti, E. and Izaurralde, E. (2005) Nonsense-mediated mRNA decay: Molecular insights and mechanistic varia- tions cross species. Current Opinion in Cell Biology, 17, 316-325. doi:10.1016/j.ceb.2005.04.005 [59] Amrani, N., Sachs, M.S. and Jacobson, A. (2006) Early nonsense: mRNA decay solves a translational problem. Nature Reviews Molecular Cell Biology, 7, 415-425. doi:10.1038/nrm1942 [60] Bor, Y., Swartz, J., Morrison, A., Recosh, D., Ladomery, M. and Hammarskjöld, M.-L. (2006) The Wilms’ tumor 1 (WT1) gene (+KTS isoform) functions with a CTE to en- hance translation from an unspliced RNA with a retained intron. Genes and Development, 20, 1597-1608. doi:10.1101/gad.1402306 [61] Coyle, J.H., Bor, Y.-C., Rekosh, D. and Hammarskjold, M.-L. (2011) The Tpr protein regulates export of mRNAs with retained introns that traffic through the nxf1 path- way. RNA, 17, 1344-1356. doi:10.1261/rna.2616111 [62] Fuke, H. and Ohno, M. (2008) Role of poly(A) tail as an identity element for mRNA nuclear export. Nucleic Acids Research, 36, 1037-1049. doi:10.1093/nar/gkm1120 [63] Mangus, D.A., Evans, M.C. and Jacobson, A. (2003) Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. BMC Genome Biology, 4, 223. doi:10.1186/gb-2003-4-7-223 [64] Behm-Ansmant, I., Gatfield, D., Rehwinkel, J., Hilgers, V. and Izaurralde, E. (2007) A conserved role for cyto- plasmic poly(A)-binding protein 1 (PABPC1) in nonsen- se-mediated mRNA. EMBO Journal, 26, 1-11. doi:10.1038/sj.emboj.7601588 [65] Nakaya, H.I., Amaral, P.P., Louro, R., Lopes, A., Fachel, A.A., Moreira, Y.B., El-Jundi, T.A., Da Silva, A.M., Reis, E.M. and Verjovski-Almeida, S. (2007) Genome map- ping and expression analyses of human intronic noncod- ing RNAs reveal tissue-specific patterns and enrichment in genes related to regulation of transcription. BMC Ge- nome Biology, 8, 43. [66] Dinger, M.E., Gascoigne, D.K. and Mattick, J.S. (2011) The evolution of RNAs with multiple functions. Bio- chimie, 93, 2013-2018. doi:10.1016/j.biochi.2011.07.018 [67] Wadler, C.S. and Vanderpool, C.K. (2007) A dual func- tion for a bacterial small RNA: SgrS performs base pair- ing-dependent regulation and encodes a functional poly- peptide. Proceeding of the National Academy of Sciences of the United States of America, 104, 20454-20459. doi:10.1073/pnas.0708102104 [68] Kageyama, Y., Kondo, T. and Hashimoto, Y. (2011) Co- ding vs non-coding: Translatability of short ORFs found in putative non-coding transcripts. Biochimie, 93, 1981- 1986. doi:10.1016/j.biochi.2011.06.024 [69] Kloc, M., Foreman, V. and Reddy, S. (2011) Binary func- tion of mRNA. Biochimie, 93, 1955-1961. doi:10.1016/j.biochi.2011.07.008 [70] Okonechnikov, K., Golosova, O. and Fursov, M. (2012) Unipro UGENE: A unified bioinformatics toolkit. Bioin- formatics, 28, 1166-1167. doi:10.1093/bioinformatics/bts091 [71] Thompson, J.D., Higgins, D.G. and Gibson, T.J. (1994) CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22, 4673-4680. doi:10.1093/nar/22.22.4673 [72] Notredame, C., Higgins, D.G. and Heringa, J. (2000) T- coffee: A novel method for fast and accurate multiple sequence alignment. Journal of Molecular Biology, 302, 205-217. doi:10.1006/jmbi.2000.4042 [73] Huelsenbeck, J.P. and Ronquist, F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics, 17, 754-755. doi:10.1093/bioinformatics/17.8.754 [74] Markham, N.R. and Zuker, M. (2008) UNAFold: Soft- ware for nucleic acid folding and hybridization. In: Keith, J.M., Ed., Methods in Molecular Biology. Bioinformatics: Structure, Function and Applications, Humana Press, To- towa, 3-31.
|