 Open Journal of Genetics, 2013, 3, 141-158 OJGen http://dx.doi.org/10.4236/ojgen.2013.33017 Published Online September 2013 (http://www.scirp.org/journal/ojgen/) Characterization of randomly amplified polymorphic DNA (RAPD) fragments revealing clonal variability in cercariae of avian schistosome Trichobilharzia szidati (Trematoda: Schistosomatidae) Anna Korsunenko1,2, Galina Chrisanfova1, Alexander Arifov1, Alexey Ryskov1, Seraphima Semyenova1 1Institute of Gene Biology, Russian Academy of Sciences, Moscow, Russia 2Center of Parasitology, A.N. Severtsov Institute of Problems of Ecology and Evolution, Russian Academy of Sciences, Moscow, Russia Email: korsanna@mail.ru Received 26 May 2013; revised 20 June 2013; accepted 8 July 2013 Copyright © 2013 Anna Korsunenko et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. ABSTRACT Recently we applied randomly amplified polymorphic DNA (RAPD) fingerprinting to detect clonal variabil- ity among individual cercariae within daughter spo- rocysts and rediae of 10 digenean trematodes (Platy- helminthes: Trematoda). The most variable RAPD patterns were obtained for Schistosomatidae repre- sentative—avian schistosome Trichobilharzia szidati. In this work, 50 polymorphic DNA fragments of ap- proximately 300 - 1500 bp from RAPD patterns of individual T. szidati cercariae were cloned and se- quenced. As a result genomic DNA sequences (total length of approximately 41,000 bp) revealing clonal variability in T. szidati cercariae were obtained and analyzed. The analysis indicated that these sequences contained tandem, inverted and dispersed repeats as well as regions homological to retroelements of two human parasites, Schistosoma mansoni and S. japoni- cum. Tandem and inverted repeats constituted 8.9% and 22.1% respectively, while the percentage of dis- persed repeats was 21.0%. The average content of these components was 41.7% with the average AT content being 59.0%. About 40% of sequences in- cluded regions ranging in length from 96 to 1005 bp which displayed amino acid homology with open reading frame pol products of S. mansoni and S. ja- ponicum retroelements: non-long terminal repeat re- trotransposons (nLTRs, 76%), long terminal repeat retrotransposons (LTRs, 14%), and Penelope-like elements (PLEs, 10%). Most of these regions (86.4%) contained frameshifts, gaps, and stop-codons. The largest portion of them was homological to nLTRs of the RTE clade (67%). The number of sequences ho- mologous to the members of CR1 lineage was 7 times smaller (9%). Homology with LTRs of Gypsy/Ty3 and BEL clades was revealed in 5% and 9% of cases re- spectively. We assume that the repetitive elements including retroelement-like sequences described in the current study may serve as the source of clonal variability detected previously in T. szidati and other digenean trematodes. Such genome regions rapidly accumulate mutations and thus may play an impor- tant functional role in the life history of the species. Keywords: RAPD Variability; Cercariae Heterogeneity; Trichobilharzia szidati Repetitive DNA 1. INTRODUCTION The vast majority of eukaryotic species reproduces bi- sexually, yet approximately one out of every 1000 mul- ticellular eukaryotic taxa is unisexual (parthenogenetic) or asexual. Parthenogenetic reproduction occurs in many phyla, especially in plants and invertebrates [1]. Among invertebrates, the Digenea (Platyhelminthes: Trematoda) have by far the most complex life cycles which usually involve free-living and parasitic stages and always in- corporates both parthenogenetic (within molluscan first intermediate host) and sexual (within vertebrate defini- tive host) reproduction. There are some stages in dige- nean life cycle (parthenitae) reproducing via diploid par- thenogenesis—mother sporocyst and either daughter sporocysts or rediae [2,3]. The reproductive function in parthenitae is performed by actively functioning ger- OPEN ACCESS  A. Korsunenko et al. / Open Journal of Genetics 3 (2013) 141-158 142 minal masses [4]. Numerous free-swimming larvae (cer- cariae) are formed in daughter sporocysts or rediae after undergoing parthenogenetic reproduction. Since partheni- tae are the result of diploid parthenogenesis with only one parent involved, all cercariae forming within sporo- cyst or redia might have been expected to represent a group of genetically identical individuals—the clone. Fun- ctionally diploid parthenogenesis can be viewed as a simple cell division. Acquirement of such type of repro- duction is an essential adaptation developed by trematodes which allowed them to be evolutionary successful [2,3]. In general, genetic variation was described previously for different invertebrate clonal systems using a variety of molecular approaches [5]. Among digeneans, the vari- able occurrence of W1 and W2 repetitive elements was detected within and among daughter sporocyst genera- tions of Schistosoma mansoni cultured in vitro [6,7]. Since an unexpected heterogeneity was found even among clonal cercariae arising from monomiracidial snail infec- tions the mitotic recombination events were suggested to occur during the parthenogenetic reproduction of schis- tosomes. The clonal variability was also detected in S. mansoni [8] and S. japonicum [9] using multi-locus mi- crosatellite analysis. In addition, RAPD variability with- in daughter sporocysts was determined in Microphallus pygmaeus and M. pseudopygmaeus [10]. RAPD-PCR is known as a DNA polymorphism analy- sis based on the amplification of random DNA segments with single primers of arbitrary nucleotide sequence, usually 1 - 12 bp in length [11,12]. It detects DNA polymorphisms produced by point mutations, insertions or deletions in the genome. RAPD assay has a distinct advantage of not requiring any specific nucleotide se- quence information for amplification and can be em- ployed across species using universal primers. This method is targeted primarily to abundant sequences with- in the genome, and usually generates a population of amplification products that can be characteristic of a specific organism. Because RAPD technique can reveal considerable polymorphism even between closely related organisms it has been used successfully for identification and differentiation of various parasite species [13]. In addition, RAPD-derived sequences obtained via cloning of PCR products can be used to design more specific and sensitive primers to develop locus-specific, or SCAR (Sequence-Characterized Amplified Regions), markers for parasite diagnosis. For example, using RAPD-derived sequences we developed a specific primer pair to detect three European causative agents of cercarial dermatitis in humans (T. franki, T. szidati, and T. regenti) during the prepatent period and after cercariae shedding [14]. Since RAPD assay is a method suitable for DNA polymorphism detection in organisms containing even small amounts of sample material we recently used this technique to reveal clonal heterogeneity between indi- vidual cercariae within sporocysts and rediae of 10 dige- nean trematodes from Schistosomatidae, Strigeidae, Gor- goderidae, Bucephalidae, Diplostomatidae, Plagiorchii- dae, Halipegidae, Notocotylidae, and Echinostomatidae families [15]. Here, we characterized RAPD fragments revealing clonal variability between individual cercariae within daughter sporocysts of the avian schistosome T. szidati since the most variable RAPD patterns were obtained previously for this Schistosomatidae representative [15]. The analysis of polymorphic DNA fragments included sequence homology search using available nucleotide and amino sequences of the avian (Trichobilharzia spp.) and mammalian (S. mansoni and S. japonicum) schisto- somes, search for tandem, inverted and dispersed repeats as well as AT/GC ratio calculation. Such information may contribute in determining structural genome organi- zation of the avian schistosomes of Trichobilharzia spp. which are the most frequent causative agents of cercarial dermatitis in humans. RAPD-derived sequences of T. szidati obtained in the current study can be also valuable in cytogenetic studies. Our data may help to further in- vestigate the mechanisms underlying clonal variability detected in digenean trematodes. 2. MATERIALS AND METHODS In total, 250 cercariae of T. szid ati were isolated from 47 daughter sporocysts sampled from nine naturally infected snails of Lymnaea stagnalis. Four snails (Lsm1, Lsm2, Lsm7, Lsm20) were collected from Russian (Moscow) and five snails (NLst4, NLst9, NLst10, NLst11, Stag1) from Belarusian (Naroch Lake) freshwater ponds. The parasite species identification was resolved using inter- mediate host-specificity combined with ITS2 rDNA, the amplification reactions were performed by using the primers its3Trem and ITS4 Trem [16]. Pieces of individual sporocysts were isolated from snail hepatopancreas, and, after washing in distilled wa- ter, fixed in 70% ethanol. The parthenitae were dissected with a preparation needle to isolate three to six cercariae. DNA was extracted from individual cercariae as de- scribed previously [17]. Since DNA amount extracted from individual cercaria is limited, the most intensely stained and unambiguous RAPD profiles were obtained only when the DNA sam- ple was used for no more than three primers. Among tested random primers, those which detected the largest number of polymorphic markers were selected for this study. RAPD-PCR was performed with each of the cho- sen 10-mer primers, P29 (5’-CCGGCCTTAC-3’), OPA09 (5’-ACCGGACACT-3’), and OPA10 (5’-GTGATCGCAG-3’), in 25 µl volume containing 10 ng of total DNA, 75 mMTris-HCl (pH 8.8), 20 mM Copyright © 2013 SciRes. OPEN ACCESS 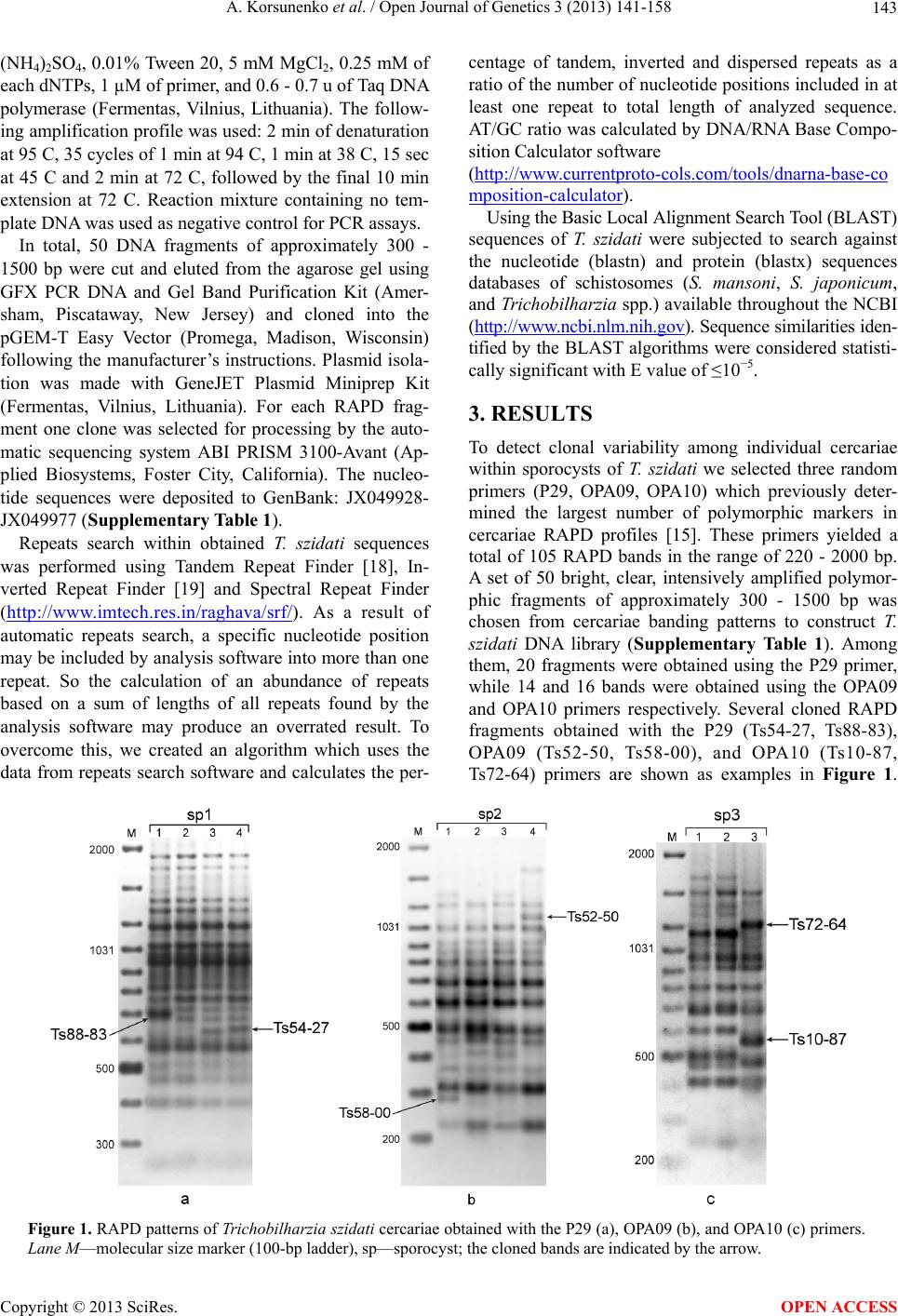 A. Korsunenko et al. / Open Journal of Genetics 3 (2013) 141-158 Copyright © 2013 SciRes. 143 centage of tandem, inverted and dispersed repeats as a ratio of the number of nucleotide positions included in at least one repeat to total length of analyzed sequence. AT/GC ratio was calculated by DNA/RNA Base Compo- sition Calculator software (http://www.currentproto-cols.com/tools/dnarna-base-co mposition-calcu lator). (NH4)2SO4, 0.01% Tween 20, 5 mM MgCl2, 0.25 mM of each dNTPs, 1 µM of primer, and 0.6 - 0.7 u of Taq DNA polymerase (Fermentas, Vilnius, Lithuania). The follow- ing amplification profile was used: 2 min of denaturation at 95 C, 35 cycles of 1 min at 94 C, 1 min at 38 C, 15 sec at 45 C and 2 min at 72 C, followed by the final 10 min extension at 72 C. Reaction mixture containing no tem- plate DNA was used as negative control for PCR assays. Using the Basic Local Alignment Search Tool (BLAST) sequences of T. szidati were subjected to search against the nucleotide (blastn) and protein (blastx) sequences databases of schistosomes (S. mansoni, S. japonicum, and Trichobilharzia spp.) available throughout the NCBI (http://www.ncbi.nlm.nih.gov). Sequence similarities iden- tified by the BLAST algorithms were considered statisti- cally significant with E value of ≤10−5. In total, 50 DNA fragments of approximately 300 - 1500 bp were cut and eluted from the agarose gel using GFX PCR DNA and Gel Band Purification Kit (Amer- sham, Piscataway, New Jersey) and cloned into the pGEM-T Easy Vector (Promega, Madison, Wisconsin) following the manufacturer’s instructions. Plasmid isola- tion was made with GeneJET Plasmid Miniprep Kit (Fermentas, Vilnius, Lithuania). For each RAPD frag- ment one clone was selected for processing by the auto- matic sequencing system ABI PRISM 3100-Avant (Ap- plied Biosystems, Foster City, California). The nucleo- tide sequences were deposited to GenBank: JX049928- JX049977 (Supplementary Table 1). 3. RESULTS To detect clonal variability among individual cercariae within sporocysts of T. szidati we selected three random primers (P29, OPA09, OPA10) which previously deter- mined the largest number of polymorphic markers in cercariae RAPD profiles [15]. These primers yielded a total of 105 RAPD bands in the range of 220 - 2000 bp. Repeats search within obtained T. szidati sequences was performed using Tandem Repeat Finder [18], In- verted Repeat Finder [19] and Spectral Repeat Finder (http://www.imtech.res.in/raghava/srf/). As a result of automatic repeats search, a specific nucleotide position may be included by analysis software into more than one repeat. So the calculation of an abundance of repeats based on a sum of lengths of all repeats found by the analysis software may produce an overrated result. To overcome this, we created an algorithm which uses the data from repeats search software and calculates the per- A set of 50 bright, clear, intensively amplified polymor- phic fragments of approximately 300 - 1500 bp was chosen from cercariae banding patterns to construct T. szidati DNA library (Supplementary Table 1). Among them, 20 fragments were obtained using the P29 primer, while 14 and 16 bands were obtained using the OPA09 and OPA10 primers respectively. Several cloned RAPD fragments obtained with the P29 (Ts54-27, Ts88-83), OPA09 (Ts52-50, Ts58-00), and OPA10 (Ts10-87, Ts72-64) primers are shown as examples in Figure 1. Figure 1. RAPD patterns of Trichobilharzia szidati cercariae obtained with the P29 (a), OPA09 (b), and OPA10 (c) primers. Lane M—molecular size marker (100-bp ladder), sp—sporocyst; the cloned bands are indicated by the arrow. OPEN ACCESS  A. Korsunenko et al. / Open Journal of Genetics 3 (2013) 141-158 144 For each RAPD fragment one clone containing DNA insert was randomly selected for sequencing. In total, 50 sequences ranging in size from 281 to 1505 bp were ob- tained. There was no similarity between the sequences. The analysis indicated the prevalence of AT-bases (59.0%). Repeats search revealed a number of tandem, inverted, and dispersed repetitive components within these sequences (Supplementary Table 2). For example, one of the sequences (Ts54-27) contained a microsatellite repeat (CA)10, while a 309-bp-long minisatellite repeat was identified within another sequence (Ts59-00). This repeat consisted of nine 35-bp monomers. Interestingly, Ts49-66 included a 167-bp region similar to Sau3A minisatellite repeat of T. ocellata and T. regenti. Supple- mentary Table 3(a) gathers the information concerning sizes of homology regions, their identity/similarity (I/S) levels, and significance. In analyzed T. szidati sequences tandem and inverted repeats constituted 8.9% and 22.1% respectively, while the percentage of dispersed repeats was 21.0%. Average repeats content turned out to be 41.7% of total sequence length. Additionally, BLAST algorithms (blastn and blastx) were used to find T. szidati sequence homology with nucleotide and amino acid sequences of the avian (Trichobilharzia spp.) and mammalian (S. mansoni, S. japonicum) schistosomes available throughout the NCBI. Based on nucleotide sequence similarity search (blastn) all the sequences revealed regions ranging from 35 to 643 bp homological to numerous parts of S. mansoni and S. japonicum genome. Similarity with available Tricho- bilharzia spp. sequences was either not found or had no significance (E > 10−5) except for the case of Ts49-66 described above. Translated nucleotide sequence similar- ity search (blastx) detected 22 regions (fragments) within 19 T. szidati sequences which reveal homology with open reading frame pol products (reverse transcriptase, endonuclease, and integrase) of S. mansoni and S. ja- ponicum retroelements from three major subclasses: long terminal repeat retrotransposons (LTRs), non-long ter- minal repeat retrotransposons (nLTRs), and Penelope- like elements (PLEs) (Supplementary Table 3(b)). All these regions had no similarity between each other at amino acid level, consequently, they represented non- overlapping regions homological to retroelements. Among them, three homology regions were found in Ts82-32, two regions were detected in Ts49-66, while the other sequences revealed one region each (Supplemen- tary Table 3(b)). Nineteen of 22 detected fragments were found to be disrupted by gaps (sequences, con- taining these regions: Ts49-66, Ts51-24, Ts51-49, Ts53-51, Ts64-57, Ts72-64, Ts74-83, Ts76-85, Ts82-32), stop-codons (sequences, containing these regions: Ts48-65, Ts49-66, Ts51-49, Ts52-50, Ts64-57, Ts69-102, Ts72-64, Ts74-83, Ts76-85, Ts82-32), and frameshifts (sequences, containing these regions: Ts49-66, Ts51-24, Ts53-51, Ts71-00) (Supplementary Table 3(b)). Our data indicated the predominance of regions (n = 13) homological to the following nLTRs of the RTE clade: SjCHGCS20 and SjR2 (one region in Ts51-82, Ts71-00, Ts82-32, Ts85-90 and two in Ts49-66) as well as SR3, Perere-3, and SjCHGCS19 (Ts52-50, Ts64-57, Ts72-64, Ts74-83, Ts76-85, Ts82-32, Ts88-83) (Supple- mentary Table 3(b)). Lengths of homology subregions varied from 45 to 546 bp with I/S level ranging from 26/50% to 77/86%. Moreover, a 55-bp fragment homo- logical to untranslated region of Perere-3 was found in Ts47-78 (Supplementary Table 3(a)). In contrast, regions homological to the members of CR1 clade were found to be 2.5 times lower as compared to RTE. These regions demonstrating homology with Perere, Perere-4, -7, and Perere-2, -5, -6 were detected in Ts53-51 and Ts51-24 respectively (Supplementary Table 3(b)). Lengths of homology subregions were 177 - 405 bp, while values of I/S varied from 33/55% to 55/74%. Furthermore, a 132-bp-long region homological (I/S = 48/75%) to Per- ere-8 was revealed in Ts48-65 (Supplementary Table 3(b)). Previously, the truncated copy of Perere-8 was identified as CR1 member [20] whereas by examining a full-length copy of this element the apurinic/apyrimidinic endonuclease domain was detected which is characteris- tic of the more modern lineages of nLTRs [21]. The most lengthy homology region (1005 bp) was de- tected in Ts51-49 (Supplementary Table 3(b)). This re- gion revealed homology (I/S = 54/68%) with SjCHGCS3- retrotransposon from Gypsy/Ty3 clade of LTRs. Leng- ths of subregions homological to the other members of this lineage (Saci-5, SjCHGCS1, and SjCHGCS4) ranged from 702 to 966 bp, while I/S varied from 27/42% to 57/71%. Two other T. szidati sequences (Ts48-00, Ts83- 28) contained regions homologous to BEL representa- tives of LTRs (SjCHGCS16, Saci-1, etc.) (Supplemen- tary Table 3(b)). Lengths of homology subregions were 96 - 252 bp with I/S varying in the range of 42/63% - 57/74%. In total collection two regions demonstrated homology with PLEs: 273-bp region homological (I/S = 33/55%) to SjPenelope-2 was detected in Ts69-102, while Ts82-32 included 123-bp region similar to SjPenelope-2, -3, and Perere-10 (Supplementary Table 3(b)). The most complex structure of homology regions was identified in Ts49-66 (Figure 2). This sequence con- tained two regions homological to RTE members (SjCHGCS20 and SjR2): located towards 5’-end subre- gion I and II formed the first region, while the second one (designated as VI) was located closer to 3’-end. These two regions were homological to different parts within amino acid sequences of SjCHGCS20 and SjR2: the first region had homology with 856 - 913 and 1029 - Copyright © 2013 SciRes. OPEN ACCESS  A. Korsunenko et al. / Open Journal of Genetics 3 (2013) 141-158 145 Figure 2. Location of homology regions in Ts49-66 nucleotide sequence of Trichobilharzia szidati. Subregion I and II formed the first region homological to SjCHGCS20 and SjR2 retrotransposons, the second region is designated as VI. Subregions III, IV, and V represented the region homological to Sau3A minisatellite repeat of T. ocellata and T. regenti. Regions without any homology are indicated in black. Reading frames are indicated as +1 and +2. 1086 aa respectively, while the second region had ho- mology with 713 - 768 and 886 - 941 aa respectively. Therefore, Ts49-66 included regions homological to dif- ferent copies of SjCHGCS20 and SjR2. Finally, subre- gions III, IV, and V (located in the central part of the sequence) represented the region homological to Sau3A minisatellite repeat of T. ocellata and T. regent i . Besides that, computer-assisted sequence similarity search revealed two regions homological at amino acid level to S. mansoni regulatory elements. One of them (288 bp) detected in Ts67-61 was homologous to zinc finger transcription factor gli2 with I/S being 69/80%, while the other one (177 bp) detected in Ts04-84 was homological to transcription initiation factor tfiid with I/S being 92/98% (Supplementary Table 3(c)). In summary, a set of RAPD-derived sequences (total length of approximately 41,000 bp) which reveal clonal variability in T. szidati cercariae were obtained and ana- lyzed. The analysis indicated that these sequences con- tained tandem, inverted and dispersed repeats. The aver- age content of these components was 41.7% with the average AT content being 59.0%. About 40% of se- quences included regions ranging in length from 96 to 1005 bp which displayed amino acid homology with pol products of S. mansoni and S. japonicum retroelements: nLTRs (76%), LTRs (14%), and PLEs (10%) (Figure 3). Most of these regions (86.4%) contained frameshifts, gaps, and stop-codons. The largest portion of them was homological to nLTRs of the RTE clade (67%). The number of sequences homologous to CR1 members was 7 times smaller (9%). Homology with LTRs of Gypsy/ Ty3 and BEL lineages was revealed in 5% and 9% of cases respectively. 4. DISCUSSION The aim of this study was the molecular characterization of anonymous nuclear genome sequences produced by RAPD genotyping and revealing clonal variability be- tween individual cercariae within sporocysts of T. szi- dati. The majority of RAPD-derived sequences belonged to repetitive elements. Among them there were tandem, inverted and dispersed repeats as well as regions homo- logical to retroelements of two human parasites, S. man- soni and S. japonicum. The complete genome sequences of S. mansoni, S. japonicum, and S. haematobium were published recently [22-24]. Over a third part of their ge- nomes consists of the repetitive DNA including mobile genetic elements (MGE), predominantly retrotransposons. The average tandem, inverted and dispersed repeats con- tent (41.7%) in T. szidati sequences was comparable with total repetitive DNA constitution in S. mansoni (40.0%), S. japonicum (40.1%), and S. haematobium (43.0%) ge- nomes [22-24]. The AT-content prevalence (59.0%) was Copyright © 2013 SciRes. OPEN ACCESS  A. Korsunenko et al. / Open Journal of Genetics 3 (2013) 141-158 146 Figure 3. Distribution of homology regions detected in Tricho- bilharzia szidati nucleotide sequences by three retroelement subclasses. RTE, CR1—clades of non-long terminal repeat retrotransposons (nLTRs); Gypsy/Ty3, BEL—clades of long terminal repeat retrotransposons (LTRs); PLEs—Penelope-like elements. detected in analyzed sequences of T. szidati as previously was reported for S. mansoni, S. japonicum, and S. haematobium (65.3%, 66.5%, and 65.7% respectively) [22-24]. Almost one-half of T. szidati sequences in- cluded fragments homological to retroelements of mammalian schistosomes (nLTRs, LTRs, PLEs). The largest portion of them (76%) demonstrated homology with nLTRs. This type of MGE was shown to be pre- dominant in S. mansoni and S. japonicum genomes [22,24]. Both blood flukes are known to contain numerous fami- lies from the CR1 and RTE clades of nLTRs [21]. Like- wise, most of homology regions detected in T. szidati sequences were similar to RTE and CR1 members. The elements from R2 lineage of nLTRs also occur in S. mansoni and S. japonicum, but do not comprise a sig- nificant fraction of their genomes [21]. Homologues of R2 members were not found among analyzed T. szidati DNA fragments. This may be explained by the low fre- quency or general absence of R2 representatives in ge- nome of this avian schistosome. Additionally, we de- tected homologues of LTRs from two clades (Gypsy/Ty3 and BEL) common in mammalian schistosomes [22,24]. Among T. szidati sequences those homological to PLEs were revealed as well. PLEs are not very abundant in mammalian schistosomes, representing less than 2% of bases in the shortgun reads of their genomes [21]. Retrotransposons and other MGE colonize the ge- nomes of nearly every eukaryote and they play an im- portant role in their genome structural organization and evolution [25]. The genetic variability caused by MGE ranges from changes in single nucleotides to changes in the size and arrangement of whole genomes. Given that the majority of new MGE insertions tend to be deleteri- ous to host, different mechanisms have been developed to mitigate the reduction in host fitness. MGE commonly integrates into non-coding regions, for example, introns to increase their probability of survival because of less visibility to natural selection. However, MGE activity can result in positive benefit to their hosts providing positive selection on elements [26,27]. The genomes of most organisms carry tens to thou- sands of retrotransposon copies. The majority of them is non-functional and contains disabling mutations. The coding sequences of such copies are highly degenerate, cluttered with stop codons, frameshifts, and large indels [28]. The mammalian schistosome genome also contains defective copies of transposable elements. For example, the coding sequences of different S. mansoni retrotrans- posons found to be truncated or contain insertions of related copies [20,29]. Furthermore, retrotransposons in S. japonicum were represented by one to 793 intact cop- ies and hundreds to thousands of partial copies [24]. Pre- viously, the degenerated inactive copies were found to be more abundant in heterochromatic regions than in the euchromatin [30,31]. Potential negative effects of MGE on the fitness of their hosts necessitate the development of strategies for transposon control. One of the most important mecha- nisms to regulate MGE expression rates and transposi- tion frequencies is RNA interference (RNAi) pathway which recognizes intracellular double-stranded RNA (dsRNA). MGE may represent a source of dsRNA and thus represent the RNAi target [32]. RNAi as an impor- tant tool to elucidate gene function has been identified in S. mansoni and S. japonicum [33,34]. RNA silencing system is likely to be common in other flatworms, in- cluding avian schistosomes. Recently, analysis of S. japonicum endogenous short interfering RNAs (siRNAs) detected that the majority of them are transposable-elements-derived [35,36]. It was discovered that such siRNAs in Drosophila were derived from heterochromatic genomic loci which have previ- ously been identified as master regulators of transposon activity [37]. These loci contain numerous defective clus- tered MGE copies (fragments of MGE as well as nested copies). Likewise, described in this study T. szidati DNA fragments homological to retroelements may act as a source of siRNA to put down MGE expansion. The most part of these regions (86.4%) contains frameshifts, stop- codons and gaps. Nevertheless, these sequences possess a genetic memory of MGE bursts in the evolutionary history. Taking into account that transposable element activity facilitates emergence of new genes, modifies gene expression patterns and promotes chromosomal rearrangements, MGE bursts can contribute to the evolu- tion of lineage-specific traits that increase adaptability of the species [38,39]. Recently, it was suggested that the higher nLTRs content in S. mansoni (15.4%) in relation Copyright © 2013 SciRes. OPEN ACCESS  A. Korsunenko et al. / Open Journal of Genetics 3 (2013) 141-158 147 to S. japonicum (8.3%) can be attributed to higher repre- sentation of two retrotransposon families (SR2 and Per- ere-3/SR3) of the RTE clade compared with the repre- sentation of their closest relative families in S. japonicum [21]. Considering the model of origin of African schis- tosomes from a migrating ancestral species dwelling in Asia it is proposed that bursts of SR2 and Perere-3/SR3 in S. mansoni would be a consequence of the selection of parasite populations in a new environment during the migration and speciation of blood flukes in Africa. Likewise, the bursts of MGE transpositions may have taken place in the evolutionary history of T. szidati stud- ied in the present work. As it was mentioned above, the germinal cells of parthenitae (daughter sporocysts and rediae) undergo mitotic division instead meiotic, which may have sug- gested their genetic identity. The main cause of genetic instability in this case is homologous recombination during mitosis. Mitotic recombination can lead to either reciprocal (mitotic crossing over) or non-reciprocal (gene conversion) transfer of genetic material. The frequency of mitotic crossing over in somatic cells of the most of eukaryotes is quite low. For example, spontaneous homologous recombination occurs at a rate of 10−6 to 10−5 per cell cycle between repeated DNA se- quences in mammalian cells [40]. In contrast, the fre- quency of homologous recombination can be very high in fertilized eggs (1/500) and embryonic cells (up to 10−1) [41,42]. Likewise, the frequency of homologous recom- bination in proliferating germinal cells of digenean tre- matodes appears to be significantly high. The longevity of the germinal cell proliferation period was found to vary in different groups of trematodes [4]. Generative func- tion is absent or weakly expressed in the mother sporo- cysts of the most archaic and primitive trematodes (Fas- ciolidae, Philophthalmidae, Cyclocoelidae, Notocotyli- dae, Halipegidae, and many Echinostomatidae). In con- trast, the long proliferation period of germinal cells dur- ing the parthenitae development was found in more spe- cialized groups (Sanguinicolidae, Schistosomatidae, Bucephalidae, Diplostomatidae, Strigeidae, Plagiorchii- dae) [3]. These findings are well supported by our data obtained earlier [15] in that we found that species which have more germinal cell proliferation cycles tend to exhibit higher levels of clonal variability. Based on RAPD markers we revealed significant genetic hetero- geneity (the percentage of polymorphic loci (P) ranged from 17.8% - 29.4%) between cercariae within daughter sporocysts of five specialized forms of digenean trema- todes (Schistosomatidae, Gorgoderidae, Bucephalidae, Diplostomatidae, Plagiorchiidae). Additionally, the high level of clonal variability between cercariae within dau- ghter sporocysts of Bucephalidae representative was confirmed using several snail-hosts in another study [43]. In contrast, the low level of clonal variability (P values ranged in 5.2% - 6.5%) was determined between cer- cariae within rediae of four archaic forms of digenean trematodes (Halipegidae, Notocotylidae, and Echinosto- matidae). The average percentage of polymorphic loci for specialized forms was four times higher (25.4%) compared with archaic forms (6.2%). So, the numerous proliferation events of germinal cells in parthenitae of the studied specialized digeneans may lead to increased recombination frequency and consequent accumulation of different genome rearrangements, while the low level of clonal variability observed using RAPD markers in studied archaic digeneans reflects the limited multiplica- tion of germinal cells and reduced frequency of recom- bination. The recombination frequency is also influenced by the content and chromosome localization of recom- bined sequences. Taking into account the specificities of RAPD tech- nique [11,12] and sequence analysis results obtained here we supposed that rearrangements occur in moderate and highly repetitive genome fraction of T. szidati. The clonal heterogeneity detected previously in mammalian schistosomes [6-9] was shown to be based on the repeti- tive DNA variability. To compare sequence analysis re- sults of T. szidati (the specialized digenean representa- tive) for one of the archaic digenean representative (Echinoparyphium aconiatum) two polymorphic RAPD fragments which reveal clonal variability in cercariae within rediae were cloned and sequenced by us previ- ously (unpublished data). The obtained sequences of 441 bp and 667 bp contained regions homological at amino acid level to pol products of S. mansoni retrotransposons. One of these sequences (GenBank KC902786) revealed homology with Saci-3, the member of Gypsy/Ty3 clade of LTRs. The other one (GenBank KC902787) was homological to Perere-5 from CR1 clade of nLTRs. So, in comparison to T. szidati clonal variability in E. aco- niatum cercariae also seemed to be associated with rear- rangements in repetitive DNA including retroelement- like sequences. Since an important consequence of mi- totic recombination is homozygosis of heterozygous markers, the increase of recombination frequency may be considered as the evolutionary adaptation of more spe- cialized and evolutionary advanced digeneans to prevent the spatial distribution of MGE compared to the archaic forms. However, comparative analysis of MGE abun- dance in both specialized and archaic groups of digene- ans is required to support this hypothesis. 5. ACKNOWLEDGEMENTS This work received financial support from the Russian Foundation for Basic Research (12-04-01153-a, 12-04-90034-Bel_a), President RF Pro- gram of Leading Scientific Schools (S.S. -5233.2012.4), the Russian Academy of Sciences Programs (Molecular and Cell Biology) of the Copyright © 2013 SciRes. OPEN ACCESS  A. Korsunenko et al. / Open Journal of Genetics 3 (2013) 141-158 148 Presidium of the Russian Academy of Sciences and President Grants for Government Support of Young Russian Scientists (MK-1026. 2012.4). REFERENCES [1] Bell, G. (1982) The masterpiece of nature: The evolution and genetics of sexuality. University of California Press, Berkeley. [2] Galaktionov, K. and Dobrovolskij, A. (1998) The origin and evolution of trematode life cycles. Nauka, Saint-Pe- tersburg. [3] Galaktionov, K. and Dobrovolskij, A. (2003) The biology and evolution of trematodes. Kluwer Academic Publish- ers, Boston, Dordrecht, London. [4] Cort, W.W., Ameel, D.J. and Van Der Woude, A. (1954) Germinal development in the sporocysts and rediae of the digenetic trematodes. Experimental Parasitology, 3, 185- 225. doi:10.1016/0014-4894(54)90008-9 [5] Lushai, G. and Loxdale, H.D. (2002) The biological im- probability of a clone. Genetics Research, 79, 1-9. doi:10.1017/S0016672301009582 [6] Bayne, C.J. and Grevelding, C.G. (2003) Cloning of Schistosoma mansoni sporocysts in vitro and detection of genetic heterogeneity among individuals within clones. Journal of Parasitology, 89, 1056-1060. doi:10.1645/GE-3186RN [7] Grevelding, C.G. (1999) Genomic instability in Schisto- soma mansoni. Molecular and Biochemical Parasitology, 101, 207-216. doi:10.1016/S0166-6851(99)00078-X [8] Gower, C.M., Shrivastava, J., Lamberton, P.H., et al. (2007) Development and application of an ethically and epidemiologically advantageous assay for the multi-locus microsatellite analysis of Schistosoma mansoni. Parasi- tology, 134, 523-536. doi:10.1017/S0031182006001685 [9] Yin, M., Hu, W., Mo, X., et al. (2008) Multiple near- identical genotypes of Schistosoma japonicum can occur in snails and have implications for population-genetic analyses. International Journal for Parasitology, 38, 1681-1691. doi:10.1016/j.ijpara.2008.05.015 [10] Khalturin, K.V., Mikhailova, N.A. and Granovich, A.I. (2000) Genetic heterogeneity in natural populations of Microphallus piriformes and M. pygmaeus parthenites (Trematoda: Microphallidae). Parazitologiia, 34, 486-501. [11] Welsh, J. and McClelland, M. (1990) Fingerprinting ge- nomes using PCR with arbitrary primers. Nucleic Acids Research, 18, 7213-7218. doi:10.1093/nar/18.24.7213 [12] Williams, J.G., Kubelik, A.R., Livak, K.J., Rafalski, J.A. and Tingey, S.V. (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nu- cleic Acids Research, 18, 6531-6535. doi:10.1093/nar/18.22.6531 [13] Zarlenga, D.S. and Higgins, J. (2001) PCR as a diagnos- tic and quantitative technique in veterinary parasitology. Veterinary Parasitology, 101, 215-230. doi:10.1016/S0304-4017(01)00568-4 [14] Korsunenko, A.V., Chrisanfova, G.G., Ryskov, A.P., Movsessian, S.O., Vasilyev, V.A. and Semyenova, S.K. (2010) Detection of European Trichobilharzia schisto- somes (T. franki, T. szidati, and T. regenti) based on novel genome sequences. Journal of Parasitology, 96, 802-806. doi:10.1645/GE-2297.1 [15] Semyenova, S.K., Khrisanfova, G.G., Korsunenko, A.V., et al. (2007) Multilocus variation in cercariae, partheno- genetic progeny of different species of the class Trema- toda. Doklady Biological Sciences, 414, 235-238. doi:10.1134/S0012496607030192 [16] Dvorak, J., Vanacova, S., Hampl, V., Flegr, J. and Horak, P. (2002) Comparison of European Trichobilharzia spe- cies based on ITS1 and ITS2 sequences. Parasitology, 124, 307-313. [17] Korsunenko, A., Chrisanfova, G., Lopatkin, A., Beer, S. A., Voronin, M., Ryskov, A.P. and Semyenova, S.K. (2012) Genetic differentiation of cercariae infrapopula- tions of the avian schistosome Trichobilharzia szidati based on RAPD markers and mitochondrial cox1 gene. Parasitology Research, 110, 833-841. doi:10.1007/s00436-011-2562-6 [18] Benson, G. (1999) Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Research, 27, 573-580. doi:10.1093/nar/27.2.573 [19] Warburton, P.E., Giordano, J., Cheung, F., Gelfand, Y. and Benson, G. (2004) Inverted repeat structure of the human genome: The X-chromosome contains a prepon- derance of large, highly homologous inverted repeats that contain testes genes. Genome Research, 14, 1861-1869. doi:10.1101/gr.2542904 [20] DeMarco, R., Machado, A.A., Bisson-Filho, A.W. and Verjovski-Almeida, S. (2005) Identification of 18 new transcribed retrotransposons in Schistosoma mansoni. Biochemical and Biophysical Research Communications, 333, 230-240. doi:10.1016/j.bbrc.2005.05.080 [21] Venancio, T.M., Wilson, R.A., Verjovski-Almeida, S. and DeMarco, R. (2010) Bursts of transposition from non-long terminal repeat retrotransposon families of the RTE clade in Schistosoma mansoni. International Jour- nal for Parasitology, 40, 743-749. doi:10.1016/j.ijpara.2009.11.013 [22] Berriman, M., Haas, B.J., LoVerde, P.T., et al. (2009) The genome of the blood fluke Schistosoma mansoni. Nature, 460, 352-358. doi:10.1038/nature08160 [23] Young, N.D., Jex, A.R., Li, B., et al. (2012) Whole-ge- nome sequence of Schistosoma haematobium. Nature Ge - netics, 44, 221-225. doi:10.1038/ng.1065 [24] Zhou, Y., Zheng, H., Chen, Y., et al. (2009). The Schis- tosoma japonicum genome reveals features of host-para- site interplay. Nature, 460, 345-351. doi:10.1038/nature08140 [25] Berg, D. and Howe, M. (1989). Mobile DNA. American Society of Microbiology, Washington DC. [26] Kidwell, M.G. and Lisch, D. (1997) Transposable ele- ments as sources of variation in animals and plants. Pro- ceedings of the National Academy of Sciences of the United of America, 94, 7704-7711. doi:10.1073/pnas.94.15.7704 Copyright © 2013 SciRes. OPEN ACCESS  A. Korsunenko et al. / Open Journal of Genetics 3 (2013) 141-158 Copyright © 2013 SciRes. 149 OPEN ACCESS [27] Thomas, M.C., Macias, F., Alonso, C. and Lopez, M.C. (2010) The biology and evolution of transposable ele- ments in parasites. Trends in Parasitology, 26, 350-362. doi:10.1016/j.pt.2010.04.001 [28] Laten, H.M., Mogil, L.S. and Wright, L.N. (2009) A shotgun approach to discovering and reconstructing con- sensus retrotransposons ex novo from dense contigs of short sequences derived from Genbank Genome Survey Sequence database records. Gene, 448, 168-173. doi:10.1016/j.gene.2009.06.011 [29] Drew, A.C., Minchella, D.J., King, L.T., Rollinson, D. and Brindley, P.J. (1999) SR2 elements, non-long termi- nal repeat retrotransposons of the RTE-1 lineage from the human blood fluke Schistosoma mansoni. Molecular Bi- ology and Evolution, 16, 1256-1269. doi:10.1093/oxfordjournals.molbev.a026216 [30] Dimitri, P., Junakovic, N. and Arca, B. (2003) Coloniza- tion of heterochromatic genes by transposable elements in Drosophila. Molecular Biology and Evolution, 20, 503- 512. doi:10.1093/molbev/msg048 [31] Mandrioli, M. (2000) Mariner-like transposable elements are interspersed within the rDNA-associated heterochro- matin of the pufferfish Tetraodon fluviatilis (Osteich- thyes). Chromosome Research, 8, 177-179. doi:10.1023/A:1009254805686 [32] Sijen, T. and Plasterk, R.H. (2003) Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature, 426, 310-314. doi:10.1038/nature02107 [33] Boyle, J.P., Wu, X.J., Shoemaker, C.B. and Yoshino, T.P. (2003) Using RNA interference to manipulate endoge- nous gene expression in Schistosoma mansoni sporocysts. Molecular and Biochemical Parasitol ogy , 128, 205-215. doi:10.1016/S0166-6851(03)00078-1 [34] Zhao, Z.R., Lei, L., Liu, M., Zhu, S.C., Ren, C.P., Wang, X.N. and Shen, J.J. (2008) Schistosoma japonicum: Inhi- bition of Mago nashi gene expression by shRNA-medi- ated RNA interference. Experimental Parasitology, 119, 379-384. doi:10.1016/j.exppara.2008.03.015 [35] Hao, L., Cai, P., Jiang, N., Wang, H. and Chen, Q. (2010) Identification and characterization of microRNAs and endogenous siRNAs in Schistosoma japonicum. BMC Genomics, 11, 55. doi:10.1186/1471-2164-11-55 [36] Wang, Z., Xue, X., Sun, J., et al. (2010) An “in-depth” description of the small non-coding RNA population of Schistosoma japonicum schistosomulum. PLoS Neglected Tropical Diseases, 4, e596. doi:10.1371/journal.pntd.0000596 [37] Brennecke, J., Aravin, A.A., Stark, A., Dus, M., Kellis, M., Sachidanandam, R. and Hannon, G.J. (2007) Discrete small RNA-generating loci as master regulators of trans- poson activity in Drosophila. Cell, 128, 1089-1103. doi:10.1016/j.cell.2007.01.043 [38] Betran, E. and Long, M. (2002) Expansion of genome coding regions by acquisition of new genes. Genetica, 115, 65-80. doi:10.1023/A:1016024131097 [39] Oliver, K.R. and Greene, W.K. (2009) Transposable ele- ments: powerful facilitators of evolution. BioEssays, 31, 703-714. doi:10.1002/bies.200800219 [40] Helleday, T. (2003) Pathways for mitotic homologous recombination in mammalian cells. Mutation Research, 532, 103-115. doi:10.1016/j.mrfmmm.2003.08.013 [41] Brinster, R.L., Braun, R.E., Lo, D., Avarbock, M.R., Oram, F. and Palmiter, R.D. (1989) Targeted correction of a major histocompatibility class II E alpha gene by DNA microinjected into mouse eggs. Proceedings of the National Academy of Sciences of the United of America, 86, 7087-7091. doi:10.1073/pnas.86.18.7087 [42] Wurst, W. and Joyner, A. (1993) Production of targeted embryonic stem cell clones. In A. Joyner, Ed., Gene Tar- geting: A Practical Approach. Oxford University Press, New York, 33-61. [43] Korsunenko, A.V., Tiutin, A.V. and Semenova, S.K. (2009) Clonal and population RAPD variation of cer- cariae obtained from Bucephalus polymorphus sporocysts (Trematoda: Bucephalidae). Genetika, 45, 73-80.  A. Korsunenko et al. / Open Journal of Genetics 3 (2013) 141-158 150 SUPPLEMENTARY MATERIALS Supplementary Table 1. The list of cloned variable RAPD fragments from Trichobilharzia szidati cercariae banding patterns ob- tained with the P29, OPA09, and OPA10 primers. No. Fragment (clone) identification GenBank Acc. Num.Length, bpRandom primer Snail-host identification Significant (E ≤ 10−5) homology with protein database sequences 1 Ts04-84 JX049957 683 OPA09 Lsm2 + 2 Ts09-86 JX049976 535 OPA10 Lsm20 – 3 Ts10-87 JX049972 572 OPA10 Lsm1 – 4 Ts11-88 JX049966 792 OPA10 NLst10 – 5 Ts12-89 JX049973 549 OPA10 Lsm2 – 6 Ts27-79 JX049943 887 P29 Lsm7 – 7 Ts28-80 JX049945 510 P29 Lsm20 – 8 Ts31-81 JX049959 589 OPA09 Lsm7 – 9 Ts47-20 JX049933 1039 P29 NLst10 – 10 Ts47-26 JX049951 435 OPA09 NLst10 + 11 Ts47-78 JX049963 1243 OPA10 NLst4 – 12 Ts48-00 JX049930 428 P29 NLst9 + 13 Ts48-65 JX049944 1420 P29 Lsm7 + 14 Ts49-00 JX049950 503 OPA09 NLst9 – 15 Ts49-66 JX049937 1312 P29 Stag1 + 16 Ts50-67 JX049939 1235 P29 Lsm1 – 17 Ts51-24 JX049961 1320 OPA09 Lsm20 + 18 Ts51-49 JX049977 1137 OPA10 Lsm20 + 19 Ts51-82 JX049975 1018 OPA10 Lsm7 + 20 Ts52-50 JX049956 1082 OPA09 Lsm1 + 21 Ts53-51 JX049967 1383 OPA10 NLst10 + 22 Ts54-27 JX049942 640 P29 NLst11 – 23 Ts54-69 JX049954 739 OPA09 Stag1 – 24 Ts55-28 JX049947 1320 P29 Lsm20 – 25 Ts56-00 JX049952 522 OPA09 NLst10 – 26 Ts57-00 JX049953 288 OPA09 NLst11 – 27 Ts58-00 JX049955 281 OPA09 Lsm1 – 28 Ts59-00 JX049932 299 P29 NLst10 – 29 Ts59-60 JX049969 1190 OPA10 NLst11 – 30 Ts64-57 JX049940 1210 P29 Lsm2 + Copyright © 2013 SciRes. OPEN ACCESS 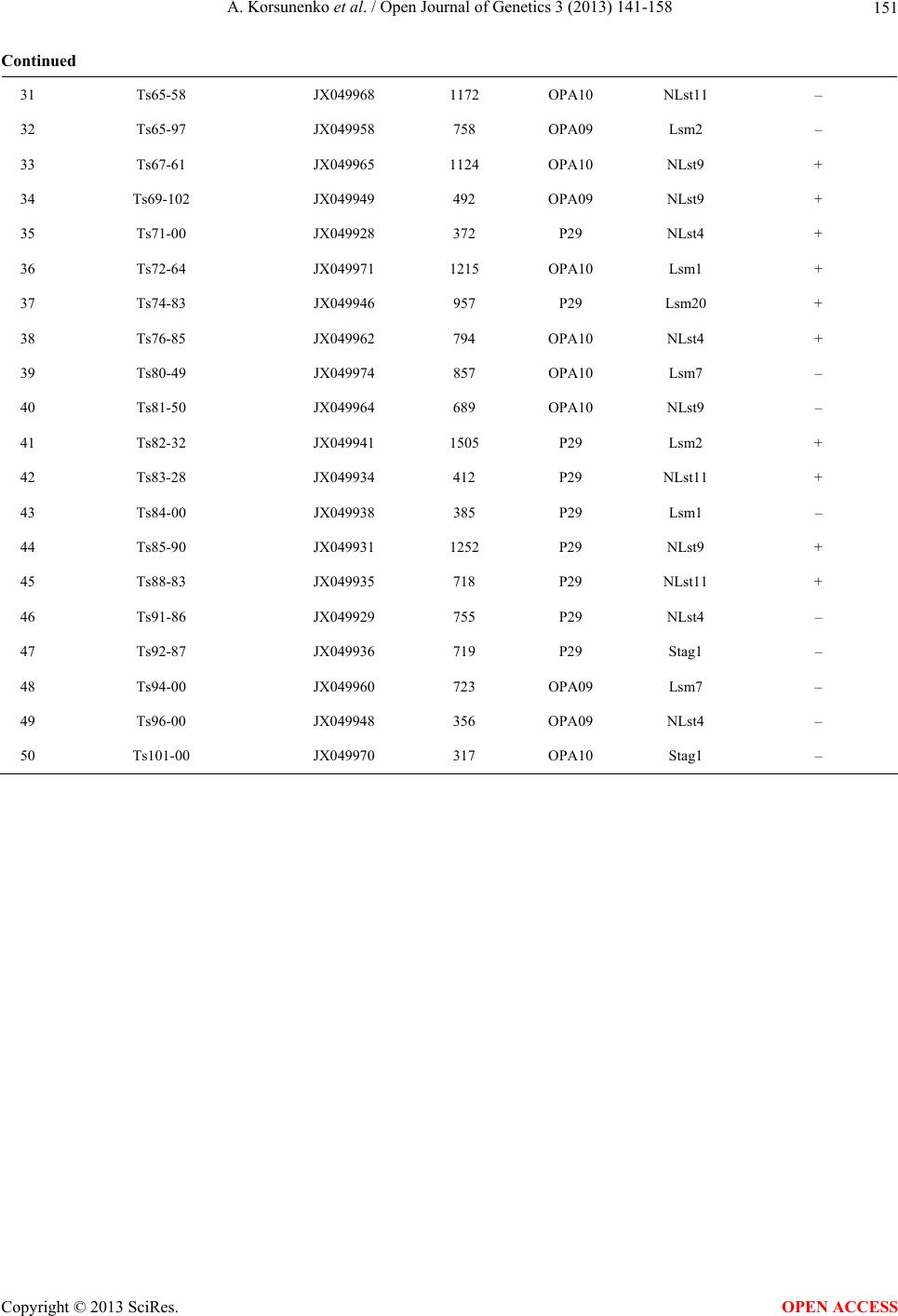 A. Korsunenko et al. / Open Journal of Genetics 3 (2013) 141-158 151 Continued 31 Ts65-58 JX049968 1172 OPA10 NLst11 – 32 Ts65-97 JX049958 758 OPA09 Lsm2 – 33 Ts67-61 JX049965 1124 OPA10 NLst9 + 34 Ts69-102 JX049949 492 OPA09 NLst9 + 35 Ts71-00 JX049928 372 P29 NLst4 + 36 Ts72-64 JX049971 1215 OPA10 Lsm1 + 37 Ts74-83 JX049946 957 P29 Lsm20 + 38 Ts76-85 JX049962 794 OPA10 NLst4 + 39 Ts80-49 JX049974 857 OPA10 Lsm7 – 40 Ts81-50 JX049964 689 OPA10 NLst9 – 41 Ts82-32 JX049941 1505 P29 Lsm2 + 42 Ts83-28 JX049934 412 P29 NLst11 + 43 Ts84-00 JX049938 385 P29 Lsm1 – 44 Ts85-90 JX049931 1252 P29 NLst9 + 45 Ts88-83 JX049935 718 P29 NLst11 + 46 Ts91-86 JX049929 755 P29 NLst4 – 47 Ts92-87 JX049936 719 P29 Stag1 – 48 Ts94-00 JX049960 723 OPA09 Lsm7 – 49 Ts96-00 JX049948 356 OPA09 NLst4 – 50 Ts101-00 JX049970 317 OPA10 Stag1 – Copyright © 2013 SciRes. OPEN ACCESS 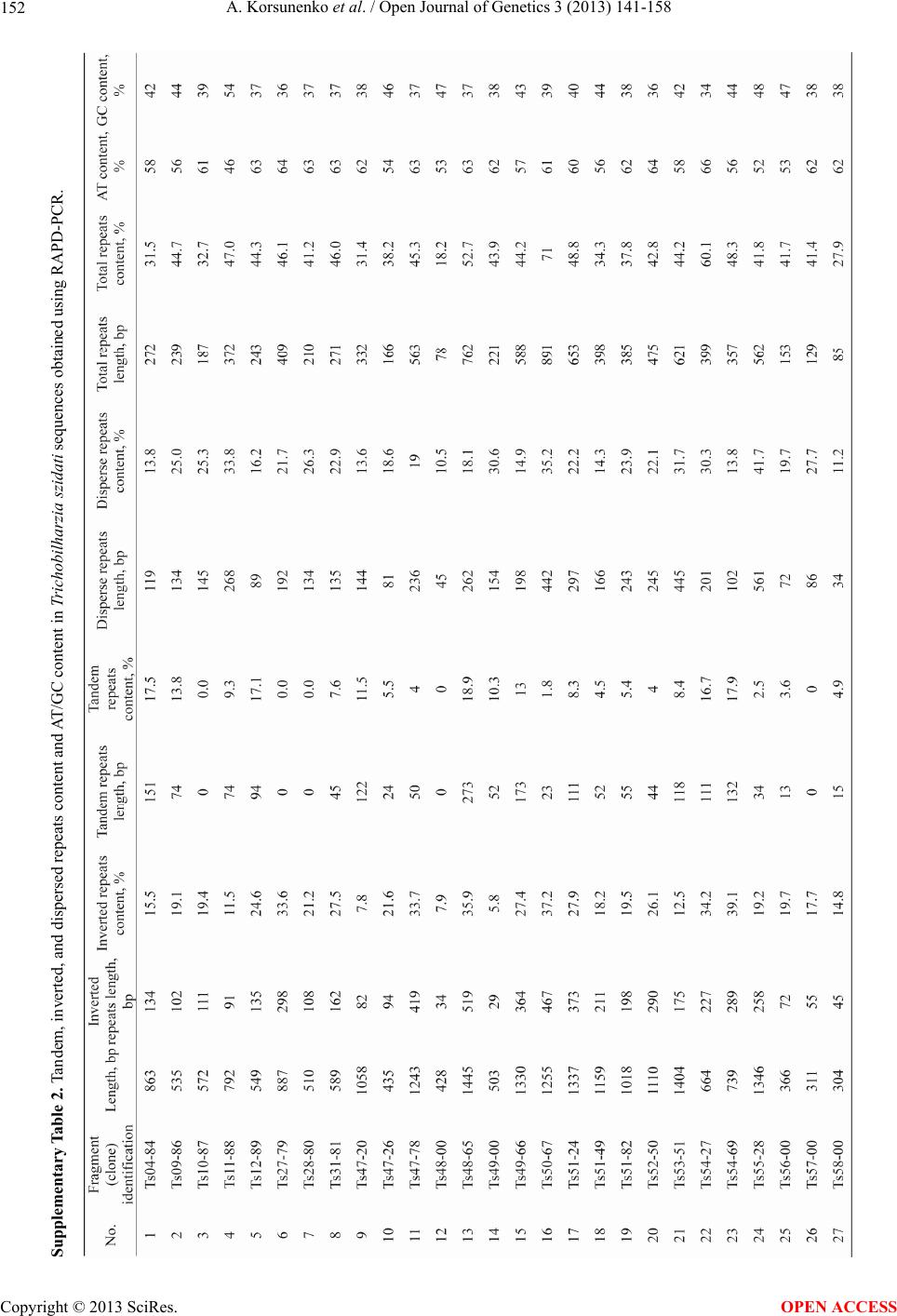 A. Korsunenko et al. / Open Journal of Genetics 3 (2013) 141-158 152 Supplementary Table 2. Tandem, inverted, and dispersed repeats content and AT/GC content in Trichobilharzia szidati sequences obtained using RAPD-PCR. Copyright © 2013 SciRes. OPEN ACCESS  A. Korsunenko et al. / Open Journal of Genetics 3 (2013) 141-158 153 Continued Copyright © 2013 SciRes. OPEN ACCESS 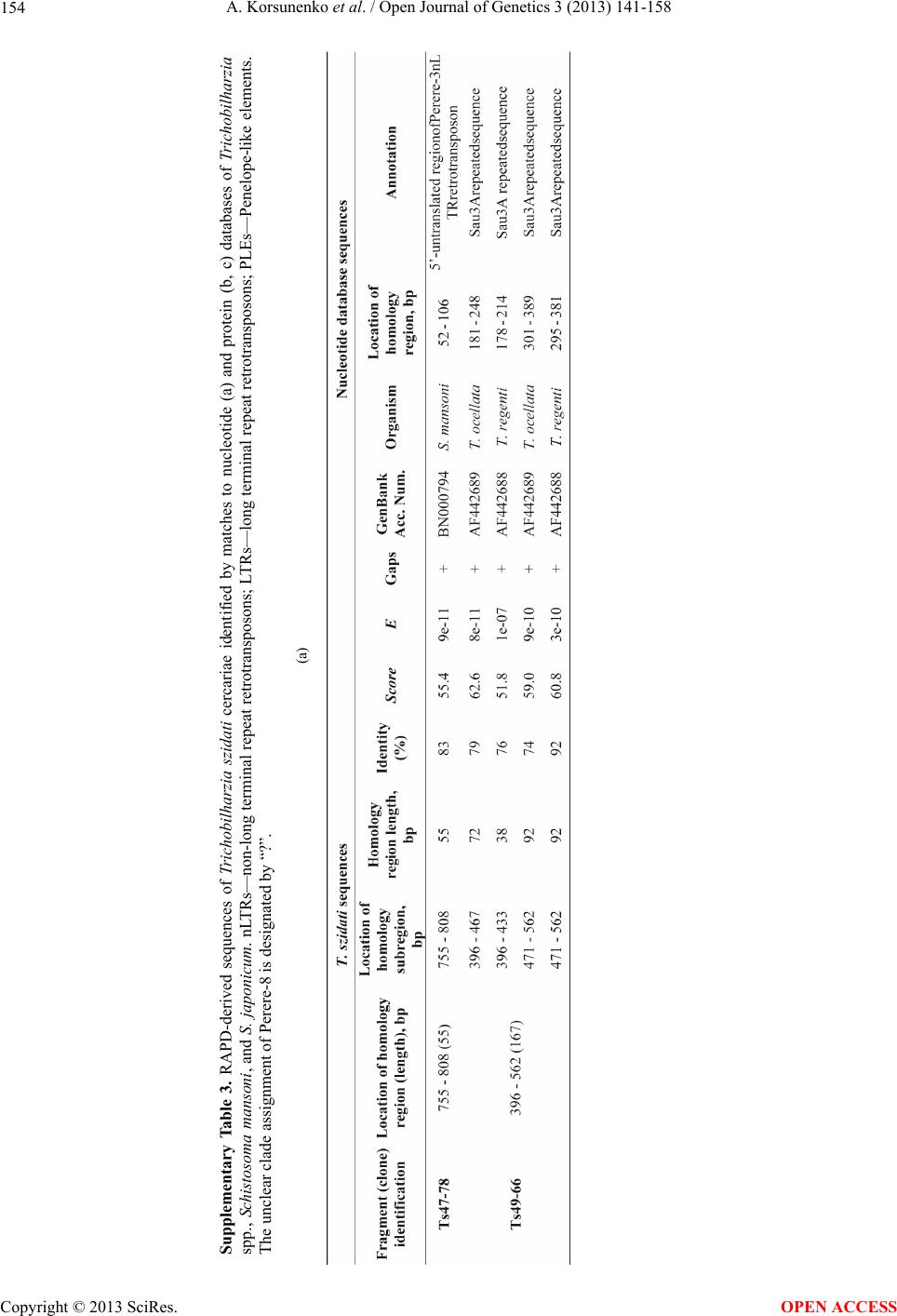 A. Korsunenko et al. / Open Journal of Genetics 3 (2013) 141-158 154 Supplementary Table 3. RAPD-derived sequences of Trichobilharzia szidati cercariae identified by matches to nucleotide (a) and protein (b, c) databases of Trichobilharzia spp., Schistosoma mansoni, and S. japonicum. nLTRs—non-long terminal repeat retrotransposons; LTRs—long terminal repeat retrotransposons; PLEs—Penelope-like elements. The unclear clade assignment of Perere-8 is designated by “?”. (a) Copyright © 2013 SciRes. OPEN ACCESS 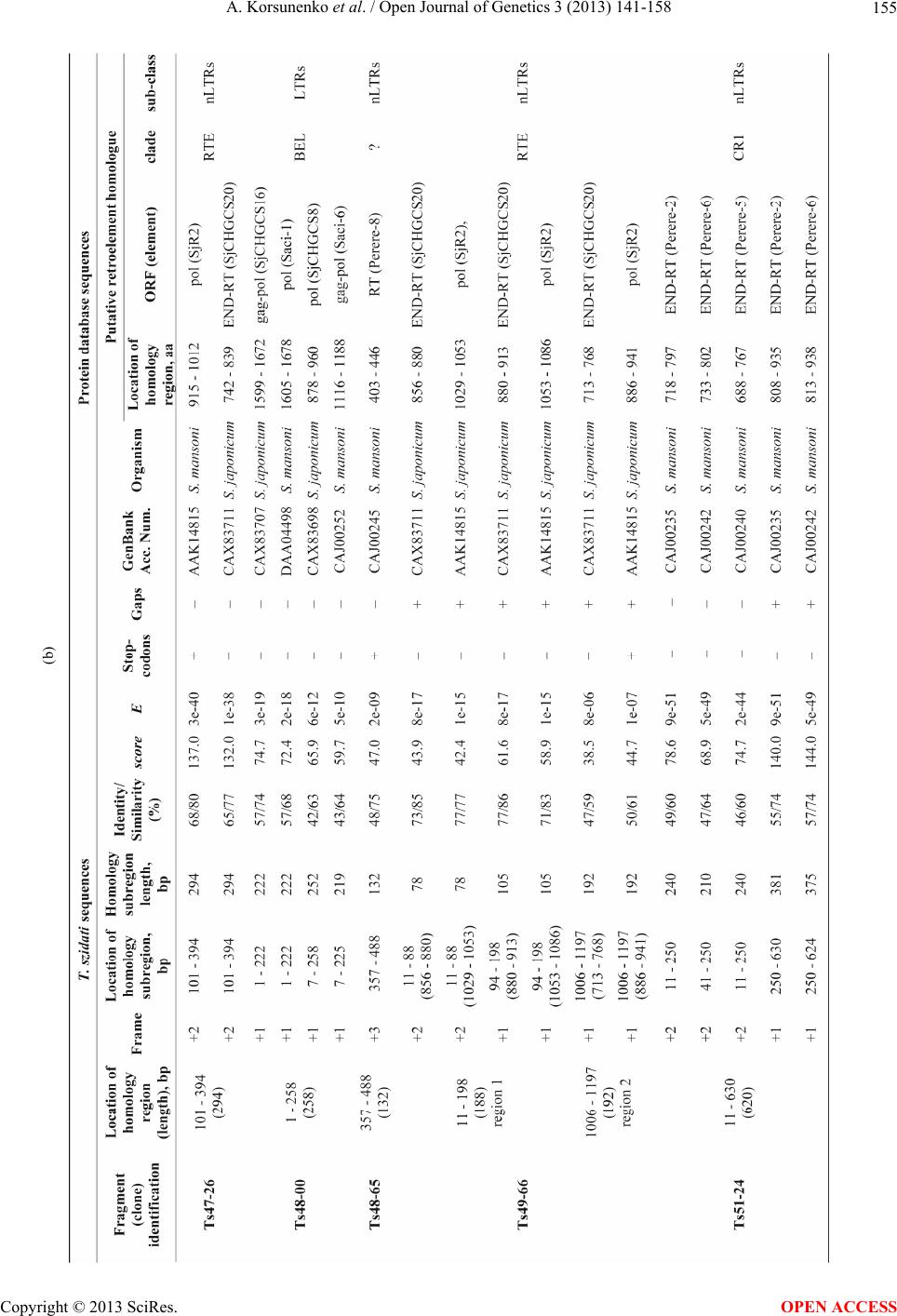 A. Korsunenko et al. / Open Journal of Genetics 3 (2013) 141-158 155 (b) Copyright © 2013 SciRes. OPEN ACCESS 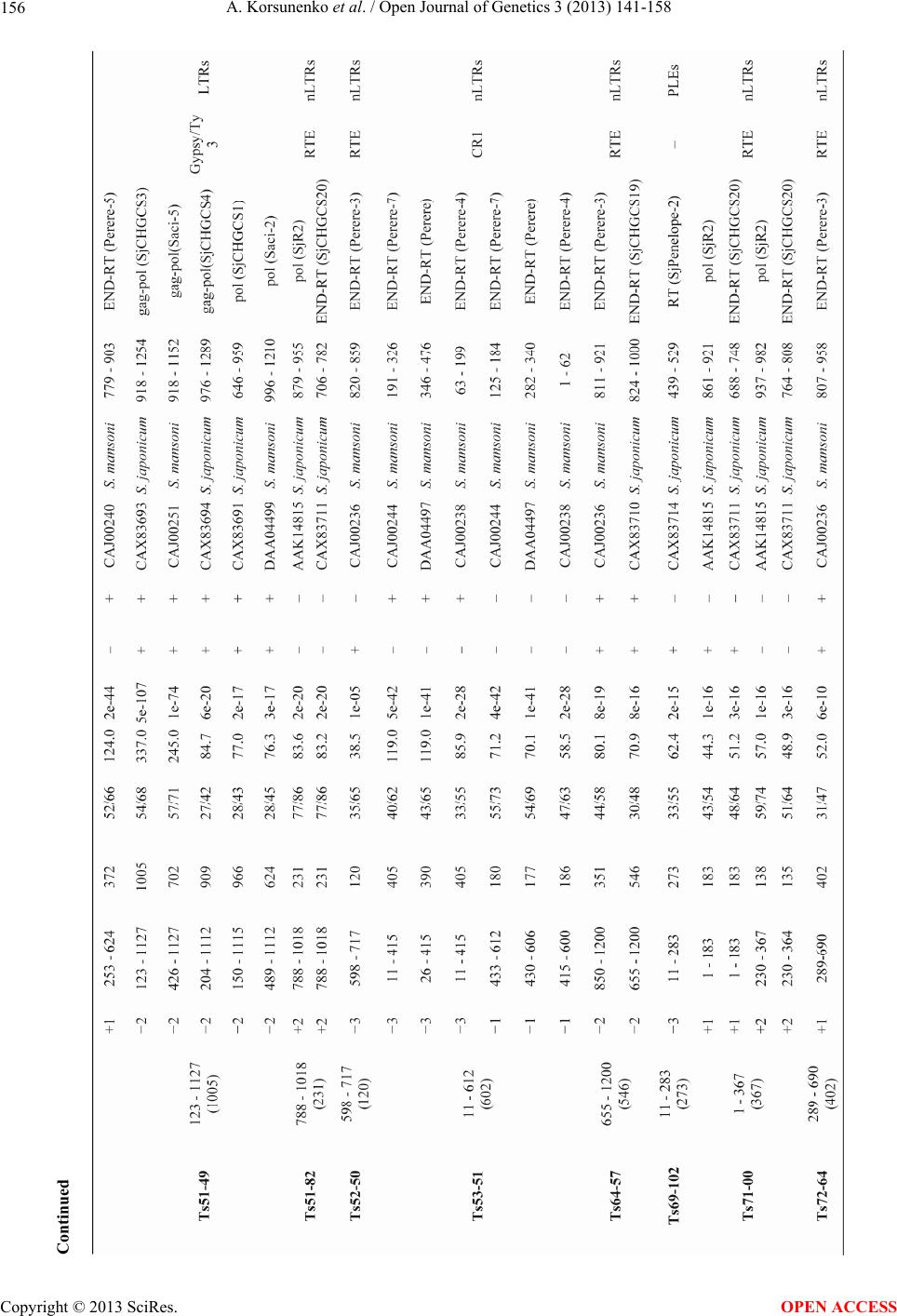 A. Korsunenko et al. / Open Journal of Genetics 3 (2013) 141-158 156 Continued Copyright © 2013 SciRes. OPEN ACCESS 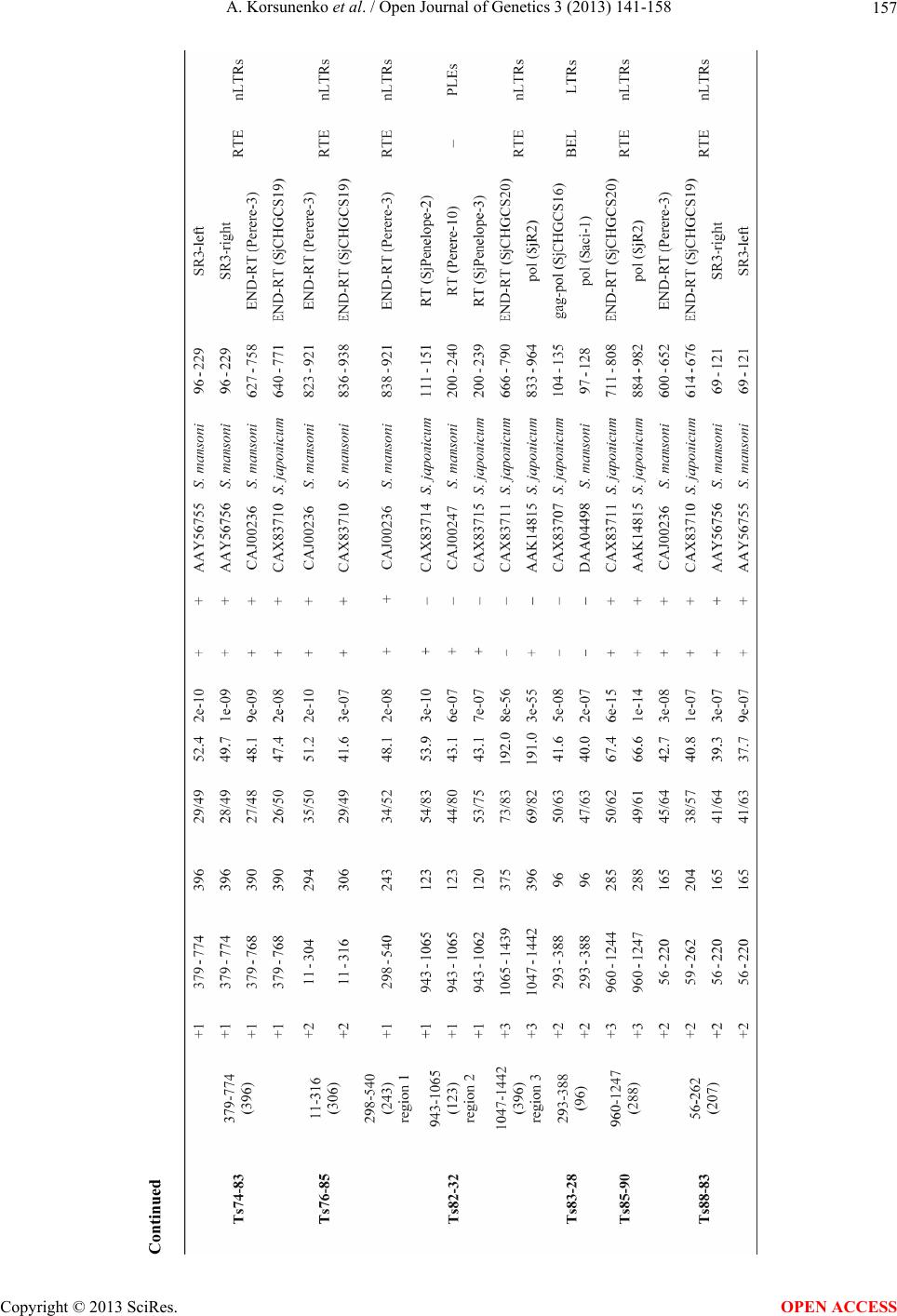 A. Korsunenko et al. / Open Journal of Genetics 3 (2013) 141-158 157 Continued Copyright © 2013 SciRes. OPEN ACCESS  A. Korsunenko et al. / Open Journal of Genetics 3 (2013) 141-158 Copyright © 2013 SciRes. 158 (c) OPEN ACCESS
|