Paper Menu >>
Journal Menu >>
 Vol.2, No.12, 1345-1348 (2010) Health doi: 10.4236/health.2010.212200 Copyright © 2010 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ Filaggrin mutations are associated with ichthyosis vulgaris in the Southern Chinese population* Chang-Xing Li1, Quan Luo2, Xue-Mei Li2, Xi-Bao Zhang2#, Chun-Lei Han1, Ze-Lin Ma1, Dong-Zi Lin1 1Department of Dermatology, Dongguan Institute of Dermatology, Dongguan, China; 2Department of Dermatology, Guangzhou Institute of Dermatology, Guangzhou, China; #Corresponding Author: lilichangxing@163.com; Authors Chang-Xing Li, and Quan Luo contributed equally to this work. Received 13 October 2010; revised 1 November 2010; accepted 5 November 2010 ABSTRACT Filaggrin (FLG) plays an important role in the epidermal barrier function, which identified in patients with ichthyosis vulgaris(IV).To study the genetics of FLG mutations in Southern Chinese patients with IV. We evaluated the in- fluence of five mutations (3321 delA, 441delA, 1249 insG, E1795X and S 3296X) in a c ohort of 65 IV Chinese patients and in 100 control individu- als using the Sequenom® MassARRAY® sys- tem. The null allele frequency of 3321delA was 52.31%(34/65). FLG mutation 441delA was only fo u n d i n o n e I V p a t i e n t s . F L G m u t a t i o n s 1249in sG, E1795X and S3296X were not found in these patients. These findings show that the mutation 3321delA represent the most frequent genetic cause in Southern Chinese IV patients. Our findings confirm and extend the knowledge of the influence of FLG mutations in IV. Keywords: Ichthyosis Vulgaris; Southern Chinese Population; Filaggrin; Mutation 1. INTRODUCTION Ichthyosis vulgaris (OMIM 146700) is the most com- mon ichthyosis with an estimated prevalence of 1:250- 1000, and inherited in a semidominant pattern [1]. Pa- tients have light grey scaling, keratosis pilaris, increased palmoplantar markings and associated atopic manifesta- tions. Involvement is generally mild and may vary greatly with climate and humidity. There is reduced or absence of keratohyalin granules in the epidermis [2]. In addition, there is a decrease or absence of profilaggrin, a major component of keratohyalin granules [3,4]. FLG is coding gene located in the epidermal differen- tiation complex on chromosome 1q21 [5]. Recently, mu- tations in the FLG have been shown to be a major risk factor for IV and atopic dermatitis(AD) [5,6]. The asso- ciation of FLG mutations with IV in Northern European, Japanese, Bangladeshi, Korean, Taiwanese and Singapo- rean Chinese populations has been confirmed in subse- quent studies [7-17]. To date, more than different 20 mu- tations have been identified in different populations, all leading to a filaggrin deficiency. FLG mutations 3321delA, 441delA, 1249insG, E1795X and S3296X have been previously reported, from Asia including China, Japan, Korea, Taiwan [8]. In this study, we identified five FLG mutations 3321delA, 441delA, 1249insG, E1795X and S3296X in Southern Chinese IV patients. 2. MATERIALS AND METHODS 2.1 Cases and Controls DNA samples from 65 IV were of Southern China ori- gin and recruited with ethical approval and had given their informed consent. The study was conducted according to the principles of the Declaration of Helsinki and all appli- cable local regulations related to protection of human subjects. The institutional review boards of the respective institutions approved the study. DNA samples from 100 healthy volunteers were available to serve as controls. Controls had never been diagnosed with IV or AD and information from medical records where available. 2.2 Genotyping Genomic DNA was extracted from peripheral blood using standard techniques. All SNPs were genotyped by MassARRAY™ system (Sequenom, San Diego, CA, http://www.sequenom.com/ ). The iPLEX™ assay was followed according to manufacturers instructions (http:// www.sequenom.com/ ) using 10 ng of genomic DNA. The primer sequences are available upon request (primer sequences in Table 1). *Supported by Medical Scientific Research Foundation of Guangdong Province, China  C.-X. Li et al. / Health 2 (2010) 1345-1348 Copyright © 2010 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 1346 Table 1. Primer sequences for all 5 FLG mutations using iPlex chemistry on a MassARRAY platform. Mutations Polymerase chain reaction primer 1 sequence Polymerase chain reaction primer 2 sequence Extension primer se- quence 3321del E1795X 1249 insG 441 delA S3296X 5’-ACGTTGGATGATCAGCAGAGCC ACCAAGAG-3’ 5’-ACGTTGGATGAGCACTGGAGGA AGACAAAG-3’ 5’-ACGTTGGATGGGCAAGCTTCATC TGCAGTC-3’ 5’-ACGTTGGATGCCCTCAAGTCTG GAAAGAAG-3’ 5’-ACGTTGGATGTGCATCATCCCAT GAACAGG-3’ 5’-ACGTTGGATGACCTGGTAGAT GAAAGACCC-3’ 5’-ACGTTGGATGATGGTGTCCTG ACCCTCTTG-3’ 5’-ACGTTGGATGTGAATGTCCCT CACTGTCAC-3’ 5’-ACGTTGGATGTTCAGAACTAG ATTCATGCC-3’ 5’-ACGTTGGATGTGTCTGCTGAC TGCTGGTG-3’ 5’-TCCAGACCTTCCCC C-3’ 5’-TGTCTCGTGCCTGCT -3’ 5’-GCCTGACTACCGCT AG-3’ 5’-TCATGCCTTTTCCCC CC-3’ 5’-GTGTCTCTCTCCTGG ACTT-3’ 2.3 Statistical Analysis Genotype frequencies between cases and controls were using the continuity-adjusted chi-square test. The reported P-values are the results of two-sided tests. P-values ≤ 0.05 were considered to be statistically sig- nificant. All computations have been performed using SAS software version 8.01. 3. RESULTS A total of 5 polymorphic SNPs across the FLG gene locus were analysed (Figures 1,2). The minor allele fre- quencies of the two functional polymorphisms (7638G/G and 7717.9A/A) located in the FLG gene were 52.31% and 1.54% in the IV cases and 5.17% and 0% in controls, respectively. The assay showed good clusters and mass spectra (Figures 1,2).The null allele frequency of FLG mutation 3321delA were 52.31% (34/65) and 4% (4/100) in cases and controls respectively. FLG mutation 441delA was only found in one IV patients. FLG muta- tion 441delA was not found in the controls. FLG muta- tions 1249insG, E1795X and S3296X were not found in these patients and controls. When comparing IV patients with controls, only 3321delA mutation showed a statis- tically significant association with IV (χ2 = 49.1718 P < 0.0001). 4. DISCUSSION IV and AD are relatively common heritable skin dis- eases. Lack of expression of the protein filaggrin has been shown to predispose to the development of IV and atopic eczema or dermatitis [5,16]. The filaggrin gene resides on human chromosome 1q21 within the epider- mal differentiation complex, a region that also harbours genes for several other proteins that are important for the normal barrier function of the epidermis [5]. Recent studies showed that both diseases are closely related to loss-of-function mutations in the FLG. The homozygous or compound heterozygous loss-of-function mutations p.R501X and c.2282del4 in FLG have been identified in moderate or severe IV patients from Ireland, Figure 1. FLG mutation 3321del and 441delA genotyped in 65 IV cases and 100 controls using Sequenom Mas- sARRAY platform. The assay showed good clusters. No obvious genotyping errors were apparent. 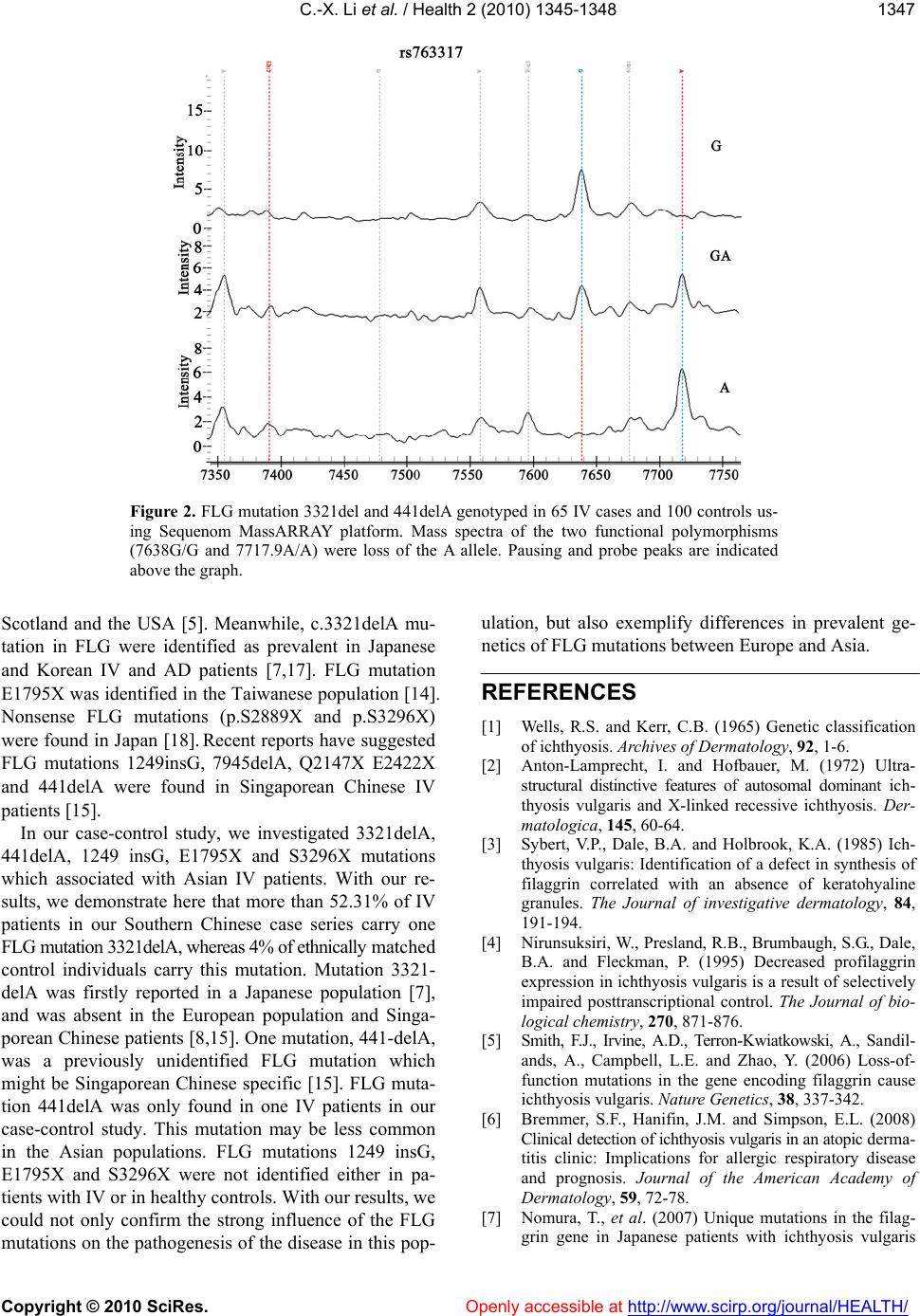 C.-X. Li et al. / Health 2 (2010) 1345-1348 Copyright © 2010 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 1347 Figure 2. FLG mutation 3321del and 441delA genotyped in 65 IV cases and 100 controls us- ing Sequenom MassARRAY platform. Mass spectra of the two functional polymorphisms (7638G/G and 7717.9A/A) were loss of the A allele. Pausing and probe peaks are indicated above the graph. Scotland and the USA [5]. Meanwhile, c.3321delA mu- tation in FLG were identified as prevalent in Japanese and Korean IV and AD patients [7,17]. FLG mutation E1795X was identified in the Taiwanese population [14]. Nonsense FLG mutations (p.S2889X and p.S3296X) were found in Japan [18]. Recent reports have suggested FLG mutations 1249insG, 7945delA, Q2147X E2422X and 441delA were found in Singaporean Chinese IV patients [15]. In our case-control study, we investigated 3321delA, 441delA, 1249 insG, E1795X and S3296X mutations which associated with Asian IV patients. With our re- sults, we demonstrate here that more than 52.31% of IV patients in our Southern Chinese case series carry one FLG mutation 3321delA, whereas 4% of ethnically matched control individuals carry this mutation. Mutation 3321- delA was firstly reported in a Japanese population [7], and was absent in the European population and Singa- porean Chinese patients [8,15]. One mutation, 441-delA, was a previously unidentified FLG mutation which might be Singaporean Chinese specific [15]. FLG muta- tion 441delA was only found in one IV patients in our case-control study. This mutation may be less common in the Asian populations. FLG mutations 1249 insG, E1795X and S3296X were not identified either in pa- tients with IV or in healthy controls. With our results, we could not only confirm the strong influence of the FLG mutations on the pathogenesis of the disease in this pop- ulation, but also exemplify differences in prevalent ge- netics of FLG mutations between Europe and Asia. REFERENCES [1] Wells, R.S. and Kerr, C.B. (1965) Genetic classification of ichthyosis. Archives of Dermatology, 92, 1-6. [2] Anton-Lamprecht, I. and Hofbauer, M. (1972) Ultra- structural distinctive features of autosomal dominant ich- thyosis vulgaris and X-linked recessive ichthyosis. Der- matologica, 145, 60-64. [3] Sybert, V.P., Dale, B.A. and Holbrook, K.A. (1985) Ich- thyosis vulgaris: Identification of a defect in synthesis of filaggrin correlated with an absence of keratohyaline granules. The Journal of investigative dermatology, 84, 191-194. [4] Nirunsuksiri, W., Presland, R.B., Brumbaugh, S.G., Dale, B.A. and Fleckman, P. (1995) Decreased profilaggrin expression in ichthyosis vulgaris is a result of selectively impaired posttranscriptional control. The Journal of bio- logical chemistry, 270, 871-876. [5] Smith, F.J., Irvine, A.D., Terron-Kwiatkowski, A., Sandil- ands, A., Campbell, L.E. and Zhao, Y. (2006) Loss-of- function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nature Genetics, 38, 337-342. [6] Bremmer, S.F., Hanifin, J.M. and Simpson, E.L. (2008) Clinical detection of ichthyosis vulgaris in an atopic derma- titis clinic: Implications for allergic respiratory disease and prognosis. Journal of the American Academy of Dermatology, 59, 72-78. [7] Nomura, T., et al. (2007) Unique mutations in the filag- grin gene in Japanese patients with ichthyosis vulgaris  C.-X. Li et al. / Health 2 (2010) 1345-1348 Copyright © 2010 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 1348 and atopic dermatitis. The Journal of Allergy and Clini- cal Immunology, 119, 434-440. [8] Akiyama, M. (2010) FLG mutations in ichthyosis vulga- ris and atopic eczema. Spectrum of mutations and popula- tion genetics. The British Journal of Dermatology, 162, 472-477. [9] Sandilands, A., et al. (2006) Prevalent and rare mutations in the gene encoding filaggrin cause ichthyosis vulgaris and predispose individuals to atopic dermatitis. The Journal of Investigative Dermatology, 126, 1770-1775. [10] Hamada, T., et al. (2008) De novo occurrence of the filaggrin mutation p.R501X with prevalent mutation c.3321delA in a Japanese family with ichthyosis vulgaris complicated by atopic dermatitis. The Journal of Inves- tigative Dermatology, 128, 1323-1325. [11] Sandilands, A., et al. (2007) Comprehensive analysis of the gene encoding filaggrin uncovers prevalent and rare mutations in ichthyosis vulgaris and atopic eczema. Na- ture Genetics, 39, 650-654. [12] Gruber, R., Janecke, A.R., Fauth, C., Utermann, G., Fritsch, P.O. and Schmuth, M. (2006) Filaggrin muta- tions p.R501X and c.2282del4 in ichthyosis vulgaris. European Journal of Human Genetics: EJHG, 15,179-184. [13] Sinclair, C., et al. (2009) Filaggrin mutations are asso- ciated with ichthyosis vulgaris in the Bangladeshi popu- lation. The British Journal of Derm atology, 160, 1113-1115. [14] Hsu, C.K., et al. (2009) Analysis of Taiwanese ichthyosis vulgaris families further demonstrates differences in FLG mutations between European and Asian populations. The British Journal of Dermatology, 161, 448-451. [15] Chen, H., et al. (2008) Unique and recurrent mutations in the filaggrin gene in Singaporean Chinese patients with ichthyosis vulgaris. The Journal of Investigative Derma- tology, 128, 1669-1675. [16] Palmer, C.N., et al. (2006) Common lossof-function va- riants of the epidermal barrier protein filaggrin are a ma- jor predisposing factor for atopic dermatitis. Nature Ge- netics, 38, 441-446. [17] Kang, T.W., Lee, J.S., Oh, S.W. and Kim, S.C. (2009) Filaggrin mutation c.3321delA in a Korean patient with ichthyosis vulgaris and atopic dermatitis. Dermatology, 218, 186-187. [18] Nomura, T., et al. (2008) Specific filaggrin mutations cause ichthyosis vulgaris and are significantly associated with atopic dermatitis in Japan. The Journal of Investiga- tive Dermatology, 128, 1436-1441. |

