Paper Menu >>
Journal Menu >>
 Advances in Bioscience and Biotechnology, 2010, 1, 372-383 ABB doi:10.4236/abb.2010.15050 Published Online December 2010 (http://www.SciRP.org/journal/abb/). Published Online December 2010 in SciRes. http://www.scirp.org/journal/ABB Morphological characteristics and identification of new monosomic stocks for cotton (Gossypium hirsutum L.) Marina F. Sanamyan1, Julia E. Petlyakova1, Elnora A. Sharipova1, Ibrokhim Y. A bdurakhmonov2* 1Cotton Genetics Laboratory, National University of Uzbekistan, Tashkent, Uzbekistan; 2Center of Genomic Technologies, Institute of Genetics and Plant Experimental Biology, Academy of Sciences of Uzbekistan, Yuqori Yuz, Tashkent, Uzbekistan. Email: *genomics@uzsci.net Received 28 July 2010; revised 24 August 2010; accepted 25 August 2010. ABSTRACT The presence of distinct morphological markers in monosomics is important for selection and mainte- nance of the monosomic plants in subsequent genera- tions and for a well-targeted chromosome substitu- tions. Here we present cytological and morphological features of the cotton (Gossypium hirsutum L.) mono- somic lines developed in Uzbekistan, and their identi- fication by means of translocation tests. We report “reduced” stigma as a new phenotypic marker for cotton monosomics, which makes it possible to dis- tinguish cytotypes without cytological analyses. We identified eleven cotton monosomes by translocation tests using our 28 translocation cotton lines. We de- termined such features of the cotton monosomic lines as significant lowering of the pollen fertility, genetic determination of variation in pollen fertility in dif- ferent flowers of the same monosomic plants and variation of both meiotic index and tetrads with mi- cronuclei in different buds. New features of cotton monosomic lines, described herein, should be useful for future cotton genome investigation and develop- ment of new chromosome substitution lines. Keywords: cotton Monosomic Stocks; Morphological Markers; Translocation Test; Identification of Monosome; Reduced Stigma 1. INTRODUCTION The development of monosomic stocks for one of the widely-grown fiber crops, cotton (Gossypium hirsutum L.), has taken place over many years. The current inven- tory of monosomics lacks deficiencies for five chromo- somes 8, 11, 13, 19 and 24. Therefore, development of monosomics for one or more of these aforementioned chromosomes is a task of high priority. Although the aneuploid lines provide incomplete cotton genome cov- erage [1], the chromosome assignments of many mo- lecular markers and candidate genes have been success- fully accomplished [2-7]. Use of F1 hypoaneuploid hy- brids resulting from the crosses of G. hirsutum ane- uploids and G. barbadense L. species in molecular ge- netic analyses has facilitated the localization of different molecular markers on specific cotton chromosomes [8-11]. However, some loci were not assigned using the aneuploids due to the lack of a full set of cotton ane- uploids [11-14]. During the past several decades, we extensively worked on the development of aneuploid cotton lines from common genetic background of highly inbred line L-458 of G. hirsutum using radioactive irra- diation techniques that resulted in creation of novel sets of monosomic and translocation lines for cotton. The preliminary cytogenetic and morphological characteris- tics of this new collection were partially reported previ- ously [15-21]. Cytogenetic details of this new mono- somic collection are also studied (Sanamyan et al. 2010, unpublished, submitted for publication elsewhere). Here we report the details of morphological characteristics of the cotton monosomic stocks and the results of identifi- cation of some of our monosomic line by using translo- cation lines. 2. MATERIALS AND METHODS 2.1. Morphological Analyses All aberrant plants were analyzed morphologically. Ve- getative and generative plant organs were studied to re- veal new morphological markers. We studied plant ar- chitecture, brunching type, leaf plate, stem and leaf pu- bescence, detailed flower morphology including number of stamens and ovules, as well as structural features of all plant organs. 2.2. Identification of the Monosomics Identification of the monosomes was carried out using 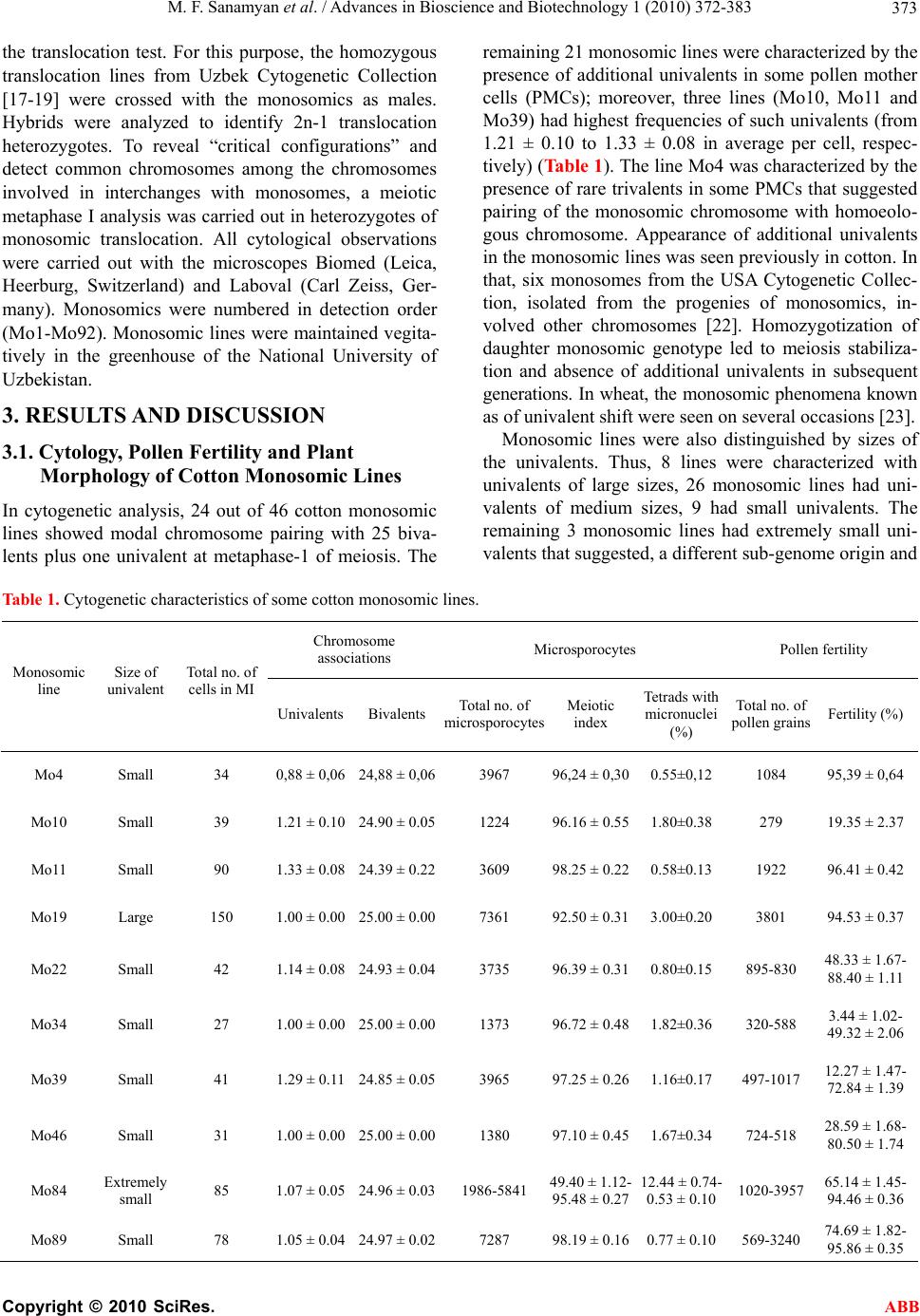 M. F. Sanamyan et al. / Advances in Bioscience and Biotechnology 1 (2010) 372-383 Copyright © 2010 SciRes. ABB 373 the translocation test. For this purpose, the homozygous translocation lines from Uzbek Cytogenetic Collection [17-19] were crossed with the monosomics as males. Hybrids were analyzed to identify 2n-1 translocation heterozygotes. To reveal “critical configurations” and detect common chromosomes among the chromosomes involved in interchanges with monosomes, a meiotic metaphase I analysis was carried out in heterozygotes of monosomic translocation. All cytological observations were carried out with the microscopes Biomed (Leica, Heerburg, Switzerland) and Laboval (Carl Zeiss, Ger- many). Monosomics were numbered in detection order (Mo1-Mo92). Monosomic lines were maintained vegita- tively in the greenhouse of the National University of Uzbekistan. 3. RESULTS AND DISCUSSION 3.1. Cytology, Pollen Fertility and Plant Morphology of Cotton Monosomic Lines In cytogenetic analysis, 24 out of 46 cotton monosomic lines showed modal chromosome pairing with 25 biva- lents plus one univalent at metaphase-1 of meiosis. The remaining 21 monosomic lines were characterized by the presence of additional univalents in some pollen mother cells (PMCs); moreover, three lines (Mo10, Mo11 and Mo39) had highest frequencies of such univalents (from 1.21 ± 0.10 to 1.33 ± 0.08 in average per cell, respec- tively) (Table 1). The line Mo4 was characterized by the presence of rare trivalents in some PMCs that suggested pairing of the monosomic chromosome with homoeolo- gous chromosome. Appearance of additional univalents in the monosomic lines was seen previously in cotton. In that, six monosomes from the USA Cytogenetic Collec- tion, isolated from the progenies of monosomics, in- volved other chromosomes [22]. Homozygotization of daughter monosomic genotype led to meiosis stabiliza- tion and absence of additional univalents in subsequent generations. In wheat, the monosomic phenomena known as of univalent shift were seen on several occasions [23]. Monosomic lines were also distinguished by sizes of the univalents. Thus, 8 lines were characterized with univalents of large sizes, 26 monosomic lines had uni- valents of medium sizes, 9 had small univalents. The remaining 3 monosomic lines had extremely small uni- valents that suggested, a different sub-genome origin and Table 1. Cytogenetic characteristics of some cotton monosomic lines. Chromosome associations Microsporocytes Pollen fertility Monosomic line Size of univalent Total no. of cells in МI Univalents BivalentsTotal no. of microsporocytes Meiotic index Tetrads with micronuclei (%) Total no. of pollen grains Fertility (%) Mo4 Small 34 0,88 ± 0,06 24,88 ± 0,063967 96,24 ± 0,300.55±0,12 1084 95,39 ± 0,64 Мо10 Small 39 1.21 ± 0.10 24.90 ± 0.051224 96.16 ± 0.551.80±0.38 279 19.35 ± 2.37 Mo11 Small 90 1.33 ± 0.08 24.39 ± 0.223609 98.25 ± 0.220.58±0.13 1922 96.41 ± 0.42 Мо19 Large 150 1.00 ± 0.00 25.00 ± 0.007361 92.50 ± 0.313.00±0.20 3801 94.53 ± 0.37 Мо22 Small 42 1.14 ± 0.08 24.93 ± 0.043735 96.39 ± 0.310.80±0.15 895-830 48.33 ± 1.67- 88.40 ± 1.11 Мо34 Small 27 1.00 ± 0.00 25.00 ± 0.001373 96.72 ± 0.481.82±0.36 320-588 3.44 ± 1.02- 49.32 ± 2.06 Мо39 Small 41 1.29 ± 0.11 24.85 ± 0.053965 97.25 ± 0.261.16±0.17 497-1017 12.27 ± 1.47- 72.84 ± 1.39 Мо46 Small 31 1.00 ± 0.00 25.00 ± 0.001380 97.10 ± 0.451.67±0.34 724-518 28.59 ± 1.68- 80.50 ± 1.74 Мо84 Extremely small 85 1.07 ± 0.05 24.96 ± 0.031986-5841 49.40 ± 1.12- 95.48 ± 0.27 12.44 ± 0.74- 0.53 ± 0.10 1020-3957 65.14 ± 1.45- 94.46 ± 0.36 Мо89 Small 78 1.05 ± 0.04 24.97 ± 0.027287 98.19 ± 0.160.77 ± 0.10 569-3240 74.69 ± 1.82- 95.86 ± 0.35  M. F. Sanamyan et al. / Advances in Bioscience and Biotechnology 1 (2010) 372-383 Copyright © 2010 SciRes. ABB 374 genetic non-uniformity. In three monosomic lines (Mo1, Mo9 and Mo46), the sizes of univalents differed various in the parental and daughter monosomics, underlining the possibility of univalent shifts in progeny. Analysis of tetrads of microspores showed a high meiotic index in the majority of the monosomic lines with the exception of the line Mo84 which varied in both meiotic index (from 49.90 ± 1.12% to 95.48 ± 0.27%) and tetrads with micronuclei (from 12.44 ± 0.74 to 0.53 ± 0.10%) in different buds (Tabl e 1 ). The meiotic index variation led to variation in pollen fertility (from 65.14 ± 1.45% to 94.46 ± 0.36%) within individual flowers of the same plant. It should be noted that lower meiotic index was recorded in wheat monosomic lines and a high percentage of tetrads with micronuclei confirmed that univalents frequently lagged during chromosome disjunction [24]. Pollen fertility analysis of cotton monosomic lines af- ter acetocarmine staining showed high pollen fertility in the majority of the lines. Only line Mo10 was character- ized with strong lowering of the character (to 19.35 ± 2.37%) that suggested its partial sterility with chromo- some deficient pollen (Table 1). Six other monosomic lines (Mo22, Mo34, Mo39, Mo46, Mo84 and Mo89) showed variation in pollen fertility in different flowers within the same monosomic plants (Table 1). In the three parental monosomics (Mo22, Mo39 and Mo46), varia- tion in pollen fertility among different flowers within the same plants also was observed (13.43-91.37%; 2.53- 34.21%; 2.10-92.46%, respectively). The ranges of varia- tion in pollen fertility were wider in two monosomics (Mo22 and Mo46). A similar effect, detected in daughter monosomics, confirmed the genetic determination of such variation and suggested chromosome localization of the gene(s) for male gametophyte viability in the de- ficient chromosomes. It is known that the majority of cotton chromosome deficiencies are not transmissible via pollen due to non-functionality of chromatin-defi- cient pollen [25]. Besides, Kakani et al. [6] indicated that gene(s) responsible for pollen spine development were located on long arm of chromosome 12 using the advanced technique of confocal laser scanning micros- copy and substitution lines. A study of the morphology of cotton monosomic plants revealed the specific influence of monosomy on many characters that differentiated them from disomic sibs. Such characters were thin stem, feeble leafing, small leaves, short internodes, crooked sympodia, small flow- ers and bolls, as well as deformed and obligospermous bolls. At the same time, 4 monosomic lines (Mo35, Mo36, Mo40 and Mo50 (e.g. Figure 1(b)) looked like disomic sibs. Although the majority of the monosomic lines had a compact bush, 10 lines (Mo3, Mo7, Mo11, Mo31 (e.g. Figure 1(c)), Mo35, Mo39, Mo60, Mo69, Mo73 and Mo89) were characterized by a scattered bush. Two lines (Mo7 and Mo56) differed by having a crooked sympodia and 3 other lines (Mo75, Mo76 (e.g. Figure 1(d)) and Mo82) had elongated internodes. In three lines (Mo13, Mo34 and Mo66), a dense stem pubescence was observed whereas leaf pubescence was feeble. Three monosomic lines (Mo16, Mo31 and Mo48) had differ- ence in leaf sizes within the same plant and two other lines (Mo9 and Mo76) had leaf folding in the area of the main rib or lobe division, respectively (Figure 2). Four monosomic lines (Mo4, Mo10, Mo46 and Mo67) differed by having feeble budding and flowering (to 10-15 flowers during the summer) whereas three other lines (Mo22, Mo39 and Mo56) had strong budding and flowering (to 40-60 flowers during the summer) but low seed and boll set (from 10.10 ± 0.78 to 20.71 ± 0.52 per one boll). Many monosomic lines were characterized by small flowers and bracts; however, six lines (Mo4, Mo10, Mo16, Mo34, Mo46 and Mo48) were distinguished by a strong reduction in flower sizes (from 38 mm to 48 mm). Taken together, seven monosomic lines (Mo9, Mo31, Mo39, Mo71, Mo72, Mo73 and Mo76) had large bracts (to 65x67 mm for Mo9) and 5 monosomics (Mo4, Mo10, Mo34, Mo46 and Mo80) had small bracts (to 25x21mm for Mo10). Some chromosome deficient lines (Mo31, Mo72 and Mo76) differed by having a large number of bract teeth (from 14 to 18) whereas other lines had small number of bract teeth (Mo4, Mo10, Mo19, Mo34, Mo46 and Mo80) (from 8 to 12) (Figure 3). In the Mo39 line additional bracts were present, in the Mo17 the bracts were asymmetrical and in the Mo27 the bracts were de- formed with feebly expressed teeth. The most variability was observed for the character “presence/absence of nectary” where in 15 monosomic lines not all bracts had nectarines (Figure 4), and Mo66 lacked any external nectaries. Nectaries of different sizes within a single flower were presented in 6 monosomic lines (Mo9, Mo27, Mo31, Mo39, Mo84 and Mo89). Monosomy had an influence on the stigma structure and sizes in a flower. Thus, there were shorter stigmata in 3 lines (Mo17, Mo19 and Mo28) and a broad “revert- ing” stigma in Mo39. A new phenotypic marker for cot- ton monosomy—“reduced” stigma was detected in Mo62. Analysis of Mo62 progeny revealed the presence of reduced stigma only in monosomic cytotypes whereas disomic ones had normal stigmas as did the control (Figure 5). This trait makes it possible to distinguish cytotypes within the progeny without cytological analy- sis. However, stigma reduction rate was varied in differ- ent flowers within the same plant (Figure 6). Thus, there were three basic reduction ranges: a little reduction (stigma to 7-9 mm), medium reduction (stigma to 2-6  M. F. Sanamyan et al. / Advances in Bioscience and Biotechnology 1 (2010) 372-383 Copyright © 2010 SciRes. ABB 375 (a) (b) (c) (d) Figure 1. Some examples of morphology of cotton monosomic plants compared to original parental line: (a) paren- tal line L-458; (b) Mo50; (c) Mo31; (d) Mo76.  M. F. Sanamyan et al. / Advances in Bioscience and Biotechnology 1 (2010) 372-383 Copyright © 2010 SciRes. ABB 376 Figure 2. Changed leafs in cotton monosomic lines with leaf folding in the area of the main rib: (a) Mo9 or lobe di- vision (b) Mo76. Figure 3. The bracts in the different cotton monosomic lines: (a) parental line L-458; (b) Mo39; (c) Mo72; (d) Mo31; (d) Mo66; and (f to k) Mo84, Mo89, Mo81, Mo88, Mo4, Mo92.  M. F. Sanamyan et al. / Advances in Bioscience and Biotechnology 1 (2010) 372-383 Copyright © 2010 SciRes. ABB 377 Figure 4. Outside view of nectaries in some cotton monosomics: (a) L-458 parental line; (b) Mo13; (c) Mo39; (d) Mo71; (e) Mo72. (a) (b) (c) (d) Figure 5. The flowers of the different cytotypes from progeny the monosomic line Mo62: (a) L-458 - parental line; (b) disomic cytotype with normal stigma; (c) monosomic cytotype with medium reduc- tion of the stigma; (d) monosomic cytotype with strong reduction of the stigma. 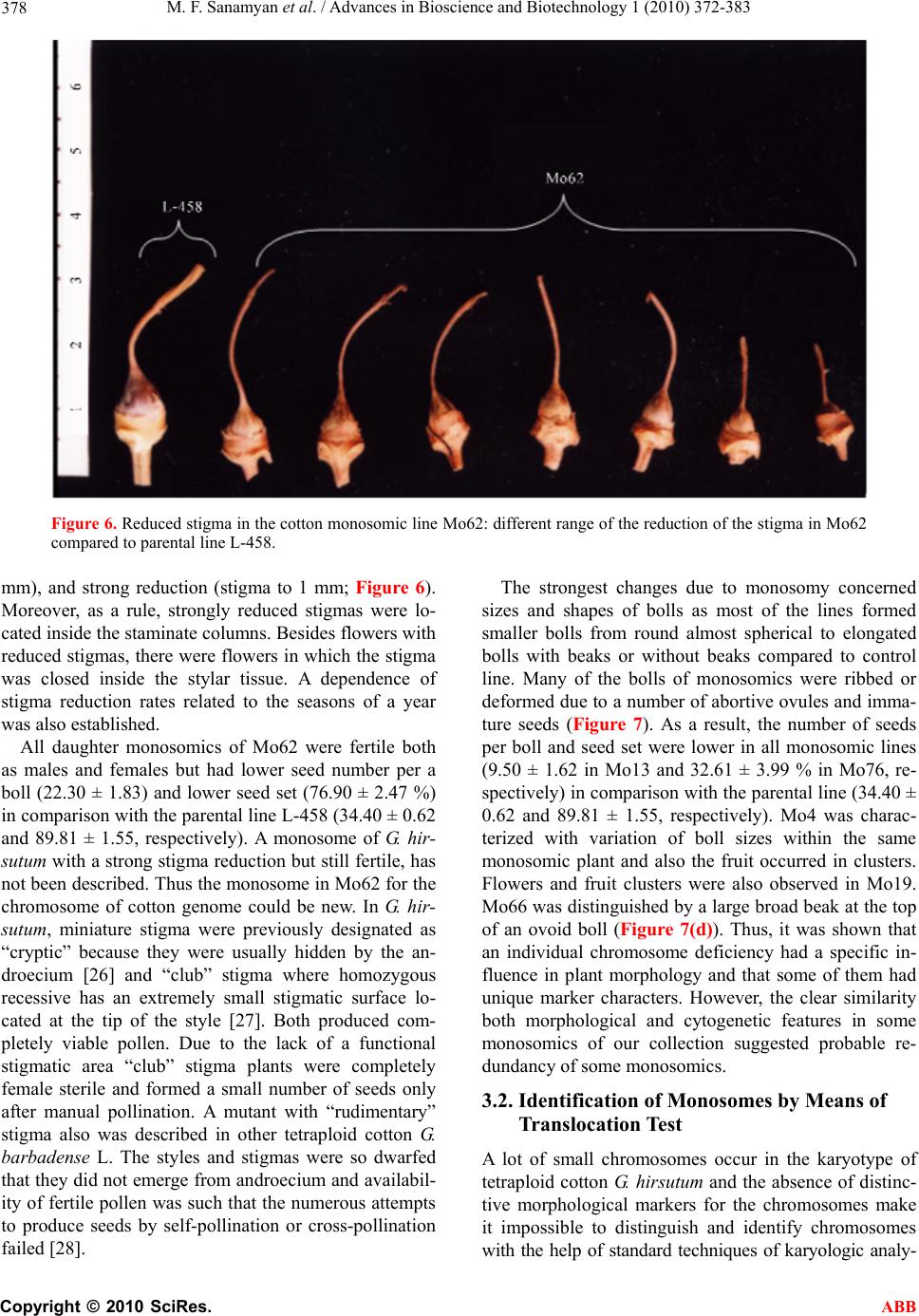 M. F. Sanamyan et al. / Advances in Bioscience and Biotechnology 1 (2010) 372-383 Copyright © 2010 SciRes. ABB 378 Figure 6. Reduced stigma in the cotton monosomic line Mo62: different range of the reduction of the stigma in Mo62 compared to parental line L-458. mm), and strong reduction (stigma to 1 mm; Figure 6). Moreover, as a rule, strongly reduced stigmas were lo- cated inside the staminate columns. Besides flowers with reduced stigmas, there were flowers in which the stigma was closed inside the stylar tissue. A dependence of stigma reduction rates related to the seasons of a year was also established. All daughter monosomics of Mo62 were fertile both as males and females but had lower seed number per a boll (22.30 ± 1.83) and lower seed set (76.90 ± 2.47 %) in comparison with the parental line L-458 (34.40 ± 0.62 and 89.81 ± 1.55, respectively). A monosome of G. hir- sutum with a strong stigma reduction but still fertile, has not been described. Thus the monosome in Mo62 for the chromosome of cotton genome could be new. In G. hi r- sutum, miniature stigma were previously designated as “cryptic” because they were usually hidden by the an- droecium [26] and “club” stigma where homozygous recessive has an extremely small stigmatic surface lo- cated at the tip of the style [27]. Both produced com- pletely viable pollen. Due to the lack of a functional stigmatic area “club” stigma plants were completely female sterile and formed a small number of seeds only after manual pollination. A mutant with “rudimentary” stigma also was described in other tetraploid cotton G. barbadense L. The styles and stigmas were so dwarfed that they did not emerge from androecium and availabil- ity of fertile pollen was such that the numerous attempts to produce seeds by self-pollination or cross-pollination failed [28]. The strongest changes due to monosomy concerned sizes and shapes of bolls as most of the lines formed smaller bolls from round almost spherical to elongated bolls with beaks or without beaks compared to control line. Many of the bolls of monosomics were ribbed or deformed due to a number of abortive ovules and imma- ture seeds (Figure 7). As a result, the number of seeds per boll and seed set were lower in all monosomic lines (9.50 ± 1.62 in Mo13 and 32.61 ± 3.99 % in Mo76, re- spectively) in comparison with the parental line (34.40 ± 0.62 and 89.81 ± 1.55, respectively). Mo4 was charac- terized with variation of boll sizes within the same monosomic plant and also the fruit occurred in clusters. Flowers and fruit clusters were also observed in Mo19. Mo66 was distinguished by a large broad beak at the top of an ovoid boll (Figure 7(d)). Thus, it was shown that an individual chromosome deficiency had a specific in- fluence in plant morphology and that some of them had unique marker characters. However, the clear similarity both morphological and cytogenetic features in some monosomics of our collection suggested probable re- dundancy of some monosomics. 3.2. Identification of Monosomes by Means of Translocation Test A lot of small chromosomes occur in the karyotype of tetraploid cotton G. hirsutum and the absence of distinc- tive morphological markers for the chromosomes make it impossible to distinguish and identify chromosomes with the help of standard techniques of karyologic analy-  M. F. Sanamyan et al. / Advances in Bioscience and Biotechnology 1 (2010) 372-383 Copyright © 2010 SciRes. ABB 379 Figure 7. The bolls of the different cotton monosomic lines: (a) L-458 - parental line; (b to h) Mo72, Mo31, Mo66, Mo60, Mo50, Mo39, Mo16 and (I to r) Mo80, Mo4, Mo92, Mo89, Mo81, Mo76, Mo62, Mo75, Mo87, Mo88. sis. Therefore, we identified monosomes to be specific chromosomes of the cotton genome using the transloca- tion tests on hybrids of monosomics with translocation lines from the Uzbek Cytogenetic Collection. Analysis of hybrid chromosome pairing was used to reveal monosomic translocation F1 hybrids and to study “criti- cal configurations”. The recently developed 28 translo- cation lines (Tr1-Tr28) from our collection [17,18] were used for monosome identification according to the method described previously [29]. Our initial efforts to monosome identification were presented in previous article [16]. Here we present identification of 15 new monosomics in addition to 20 the monosomics identified previously. According to Table 2 eleven monosomics from our collection (Mo3, Mo10, Mo11, Mo19, Mo27, Mo39, Mo48, Mo53, Mo56, Mo73 and Mo85) were associated with the chromosomes of seven translocation lines (Tr1, Tr3, Tr5, Tr8, Tr11, Tr12 and Tr16) as chromosome pairing of 24 bivalents plus one trivalent was observed in PMCs of the F1 monosomic hybrid plants (Figure 8a). In this study, we also identified four monosome pairs (Mo10 and Mo73; Mo39 and Mo56; Mo48 and Mo53; Mo11 and Mo19) that were associated with the translo- cation lines Tr3, Tr5, Tr12 and Tr16, respectively. Thus, three of the above-mentioned monosome pairs (Mo10 and Mo73, Mo39 and Mo56, Mo 11 and Mo19) involved the same chromosomes with the each pair. In future analyses, hybrids from the crosses of the monosomics and other translocation lines, involving the same chro- mosomes, will confirm our interpretation and identifica- tion. However, there is evidence for monosomes Mo48 and Mo53 that nonhomologous as the chromosomes from two different sub-genomes all involved with trans- location line Tr12. According to the preliminary numera- tion that was used in this investigation, differed from the numeration published by Brown [30], chromosome 5 is from the At -genome and chromosome 14 from Dt-ge- nome [31]. In nonhomology of the monosome and chromosomes involved in interchange of cross of Mo48 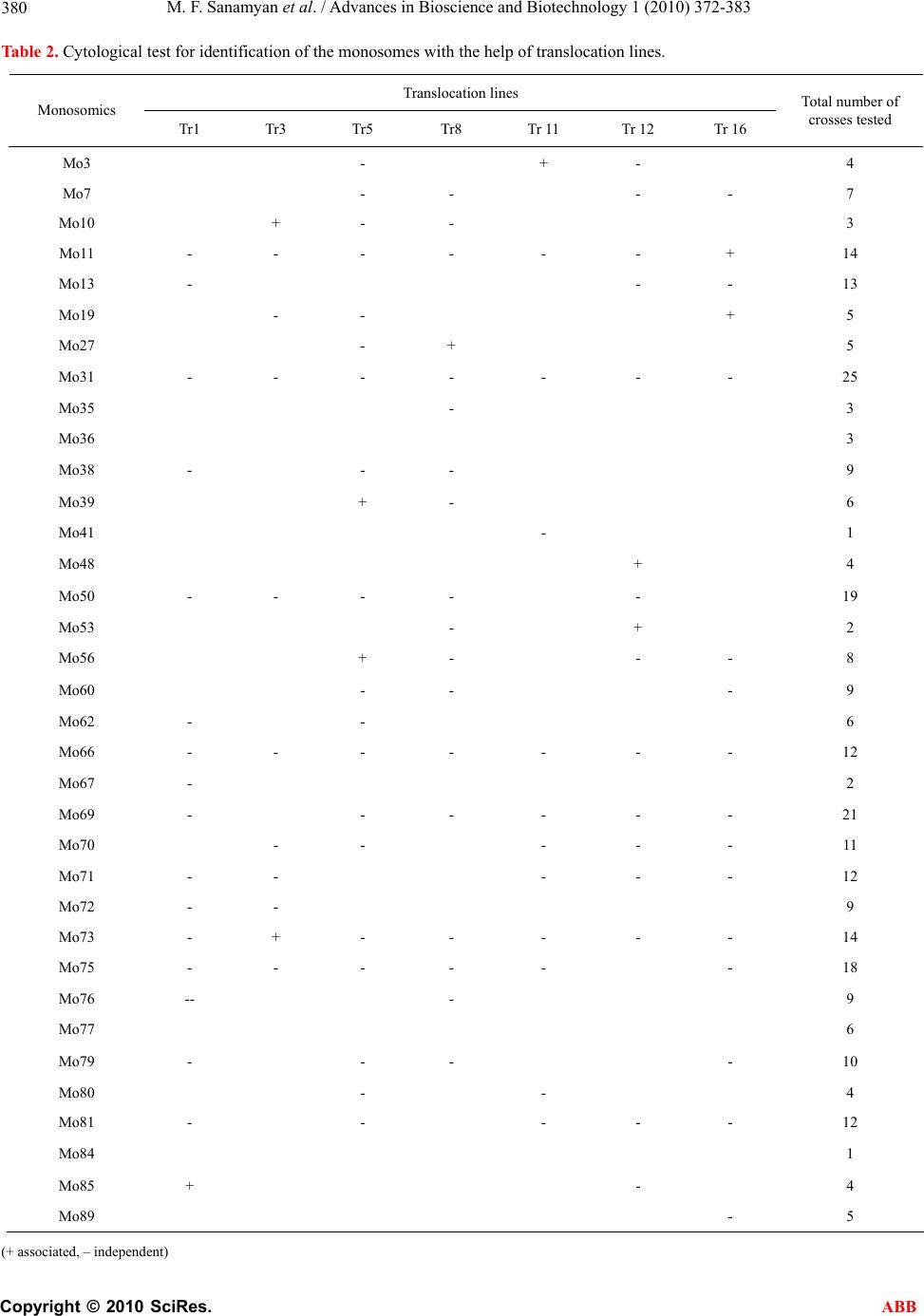 M. F. Sanamyan et al. / Advances in Bioscience and Biotechnology 1 (2010) 372-383 Copyright © 2010 SciRes. ABB 380 Table 2. Cytological test for identification of the monosomes with the help of translocation lines. Translocation lines Monosomics Tr1 Tr3 Tr5 Tr8 Tr 11 Tr 12 Tr 16 Total number of crosses tested Мо3 - + - 4 Мо7 - - - - 7 Мо10 + - - 3 Мо11 - - - - - - + 14 Мо13 - - - 13 Мо19 - - + 5 Мо27 - + 5 Мо31 - - - - - - - 25 Мо35 - 3 Мо36 3 Мо38 - - - 9 Мо39 + - 6 Мо41 - 1 Мо48 + 4 Мо50 - - - - - 19 Мо53 - + 2 Мо56 + - - - 8 Мо60 - - - 9 Мо62 - - 6 Мо66 - - - - - - - 12 Мо67 - 2 Мо69 - - - - - - 21 Мо70 - - - - - 11 Мо71 - - - - - 12 Мо72 - - 9 Мо73 - + - - - - - 14 Мо75 - - - - - - 18 Мо76 -- - 9 Мо77 6 Мо79 - - - - 10 Мо80 - - 4 Мо81 - - - - - 12 Мо84 1 Мо85 + - 4 Мо89 - 5 (+ associated, – independent) 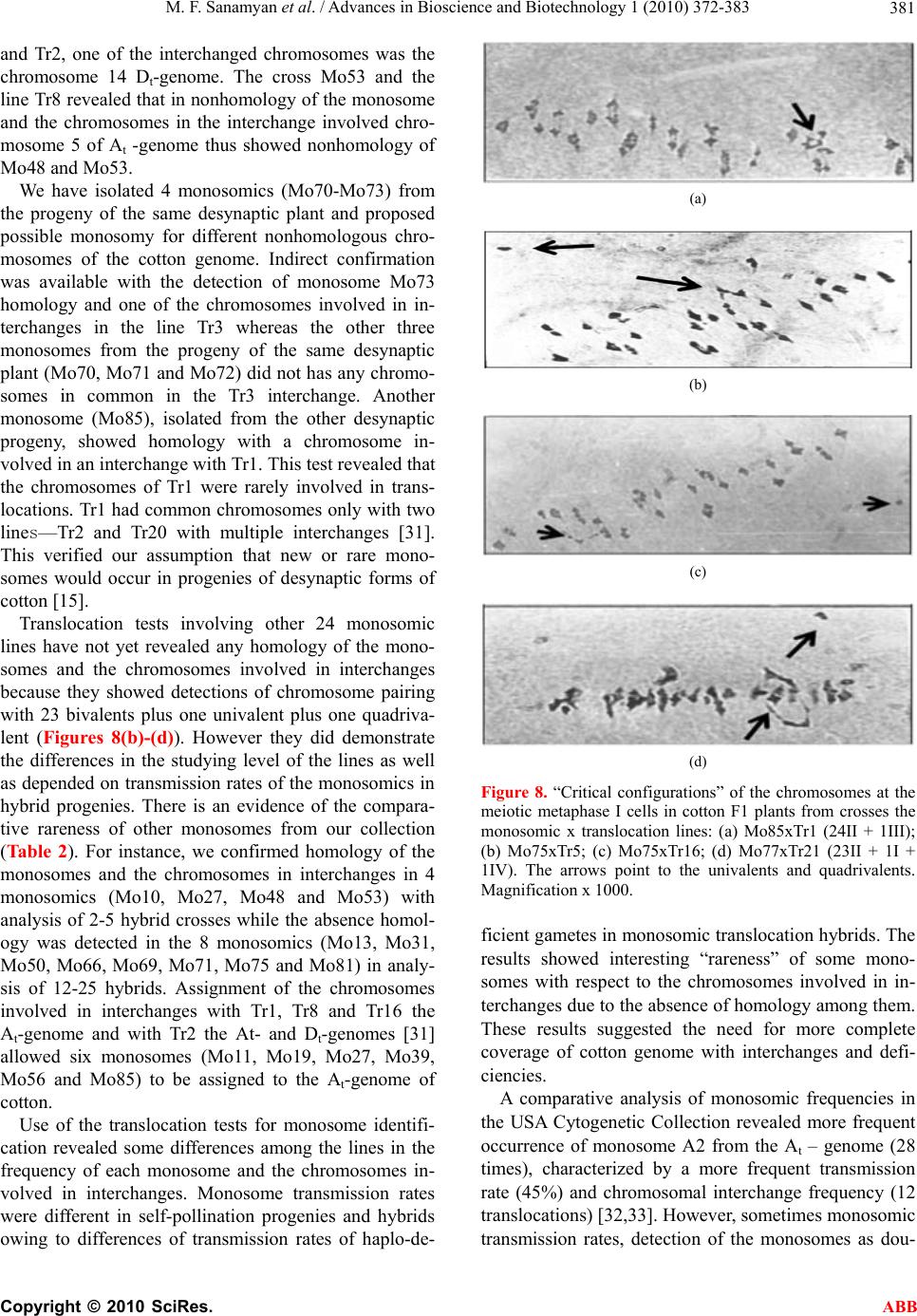 M. F. Sanamyan et al. / Advances in Bioscience and Biotechnology 1 (2010) 372-383 Copyright © 2010 SciRes. ABB 381 and Tr2, one of the interchanged chromosomes was the chromosome 14 Dt-genome. The cross Mo53 and the line Tr8 revealed that in nonhomology of the monosome and the chromosomes in the interchange involved chro- mosome 5 of At -genome thus showed nonhomology of Mo48 and Mo53. We have isolated 4 monosomics (Mo70-Mo73) from the progeny of the same desynaptic plant and proposed possible monosomy for different nonhomologous chro- mosomes of the cotton genome. Indirect confirmation was available with the detection of monosome Mo73 homology and one of the chromosomes involved in in- terchanges in the line Tr3 whereas the other three monosomes from the progeny of the same desynaptic plant (Mo70, Mo71 and Mo72) did not has any chromo- somes in common in the Tr3 interchange. Another monosome (Mo85), isolated from the other desynaptic progeny, showed homology with a chromosome in- volved in an interchange with Tr1. This test revealed that the chromosomes of Tr1 were rarely involved in trans- locations. Tr1 had common chromosomes only with two lines—Tr2 and Tr20 with multiple interchanges [31]. This verified our assumption that new or rare mono- somes would occur in progenies of desynaptic forms of cotton [15]. Translocation tests involving other 24 monosomic lines have not yet revealed any homology of the mono- somes and the chromosomes involved in interchanges because they showed detections of chromosome pairing with 23 bivalents plus one univalent plus one quadriva- lent (Figures 8(b)-(d)). However they did demonstrate the differences in the studying level of the lines as well as depended on transmission rates of the monosomics in hybrid progenies. There is an evidence of the compara- tive rareness of other monosomes from our collection (Ta bl e 2). For instance, we confirmed homology of the monosomes and the chromosomes in interchanges in 4 monosomics (Mo10, Mo27, Mo48 and Mo53) with analysis of 2-5 hybrid crosses while the absence homol- ogy was detected in the 8 monosomics (Mo13, Mo31, Mo50, Mo66, Mo69, Mo71, Mo75 and Mo81) in analy- sis of 12-25 hybrids. Assignment of the chromosomes involved in interchanges with Tr1, Tr8 and Tr16 the At-genome and with Tr2 the At- and Dt-genomes [31] allowed six monosomes (Mo11, Mo19, Mo27, Mo39, Mo56 and Mo85) to be assigned to the At-genome of cotton. Use of the translocation tests for monosome identifi- cation revealed some differences among the lines in the frequency of each monosome and the chromosomes in- volved in interchanges. Monosome transmission rates were different in self-pollination progenies and hybrids owing to differences of transmission rates of haplo-de- (a) (b) (c) (d) Figure 8. “Critical configurations” of the chromosomes at the meiotic metaphase I cells in cotton F1 plants from crosses the monosomic x translocation lines: (a) Mo85xTr1 (24II + 1III); (b) Mo75xTr5; (c) Mo75xTr16; (d) Mo77xTr21 (23II + 1I + 1IV). The arrows point to the univalents and quadrivalents. Magnification x 1000. ficient gametes in monosomic translocation hybrids. The results showed interesting “rareness” of some mono- somes with respect to the chromosomes involved in in- terchanges due to the absence of homology among them. These results suggested the need for more complete coverage of cotton genome with interchanges and defi- ciencies. A comparative analysis of monosomic frequencies in the USA Cytogenetic Collection revealed more frequent occurrence of monosome A2 from the At – genome (28 times), characterized by a more frequent transmission rate (45%) and chromosomal interchange frequency (12 translocations) [32,33]. However, sometimes monosomic transmission rates, detection of the monosomes as dou-  M. F. Sanamyan et al. / Advances in Bioscience and Biotechnology 1 (2010) 372-383 Copyright © 2010 SciRes. ABB 382 bles, and/or chromosome involved in translocations were not in correspondence. For example, the transmission rate of the chromosome D18 was 43%. It was revealed 7 times from different sources and involved in only one chromosome interchange that showed differences among various chromosomes, participating in interchanges and deficiencies. 4. CONCLUSIONS The results presented in this report suggested a detection of “reduced” stigma as a new unique phenotypic marker for cotton monosomics which makes it possible to dis- tinguish cytotypes without cytological analyses. Our cotton monosomic lines are unique and should be a valuable cytogenetic tool not only for chromosome as- signment of new marker genes and genome enrichment with new chromosome deficient plants, but also for a development of new cotton chromosome substitution lines and germplasm introgression. In future, we will identify our cotton monosomic stocks using a well-de- fined tester-set of translocation lines of the USA Cyto- genetic Collection, kindly provided by Dr. D. M. Stelly, Texas A&M University, USA, under USDA germplasm exchange program. Moreover, research is underway to develop chromosome substitution lines via interspecific hybridization of monosomic stocks and G. barbadense (Pima 3-79 and 5904-I variety) for effective use of monosomics in cotton breeding programs. An effort toward identification of specific chromosomes for our collection using a priori chromosome-associated DNA markers is also in progress. 5. ACKNOWLEDGEMENTS This work was partially supported by research grants 38/96, 28/98, 26/2000 and F.4.1.15 from the Committee for Science and Technology of the Republic of Uzbekistan. We thank Dr. James McD Stewart, University of Arakansas, Fyetteville, Arkansas, Dr. Ian Dundas, Uni- versity of Adelaide, Australia, and Dr. Masoud Sheidai, Shahid Be- heshtiUniversity, Tehran, Iran for their critical reading of the manu- script and suggestions. REFERENCES [1] Stelly, D.M. (1993) Interfacing cytogenetics with the cotton genome mapping effort. Proceedings of the Belt- wide Cotton Improvement Conference, National Cotton Council, New Orleans, La, USA, 1545-1550. [2] Endrizzi, J.E. and Ray, D.T. (1991) Monosomic and monotelodisomic analysis of 34 mutant loci in cotton”, Journal of Heredity, 82(1), 53-57. [3] Endrizzi, J.E. and Ray, D.T. (1992) Mapping of the cl1, R1, yg1 and Dw loci in the long arm of chromosome 16 of cotton. Journal of Heredity, 83(1), 1-5. [4] Stelly, D.M. (1990) Localization of the Le2 locus of cot- ton (Gossypium hirsutum L.). Journal of Heredity, 81(3), 193-197. [5] Samora, P.J., Stelly, D.M. and Kohel, R.J. (1994) Local- ization and mapping of the Le1 and Gl2 loci of cotton (Gossypium hirsutum L.). Journal of Heredity, 85(2), 152-157. [6] Kakani, A., Saha, S., Sapra, V.T., Zipf, A. and Stelly, D.M. (1999) Genetic mechanism and chromosomal loca- tion of pollen-specific gene(s) in Gossypium. Crop Sci- ence, 39(3), 668-673. [7] Saha, S. and Stelly, D.M. (1994) Chromosomal location of Phosphoglucomutase 7 locus in Gossypium hirsutum. Journal of Heredity, 85(1), 35-39. [8] Kohel, R.J., Yu, J., Park, Y.-H. and Lazo, G.R. (2001) Molecular mapping and characterization of traits control- ling fiber quality in cotton. Euphytica, 121(2), 163-172. [9] Karaca, M., Saha, S., Jenkins, J.N., Zipf, A., Kohel, R.J. and Stelly, D.M. (2002) Simple sequence repeat (SSR) markers linked to the Ligon Lintless (Li1) mutant in cot- ton. Journal of Heredity, 93(3), 221-224. [10] Mei, M., Syed, N.H., Gao, W., Thaxton, P.M., Smith, C.W., Stelly, D.M. and Chen, Z.J. (2004) Genetic map- ping and QTL analysis of fiber-related traits in cotton (Gossypium). Theoretical and Applied Genetics, 108(2), 280-291. [11] Abdurakhmonov, I.Y., Buriev, Z.T., Saha, S., Pepper, A. E., Musaev, J.A., Almatov, A., Shermatov, S.E., Ku- shanov, F.N., Mavlonov, G.T., Reddy, U.K., Yu, J.Z., Jen- kins, J.N., Kohel, R.J. and Abdukarimov, A. (2007) Mi- crosatellite markers associated with lint percentage trait in cotton, Gossypium hirsutum. Euphytica, 156(1-2), 141-156. [12] Kohel, R.J., Stelly, D.M. and Yu, J. (2002) Tests of six cotton (Gossypium hirsutum L.) mutants for association with aneuploids. Journal of Heredity, 93(2), 130-132. [13] Liu, S., Saha, S., Stelly, D.M., Burr, B. and Cantrell, R.G. (2000) Chromosomal assignment of microsatellite loci in cotton. Journal of Heredity, 91(4), 326-332. [14] Lacape, J.M., Nguyen, T.B., Thibivillers, S., Bojinov, B., Courtols, B., Cantrell, R.G., Burr, B. and Hau, B. (2003) A combined RFLP-SSR-AFLP map of tetraploid cotton based on a Gossypium hirsutum x Gossypium bar- badense backcross population. Genome, 46(4), 612-626. [15] Sanamyan, M.F., Petlyakova, J.E. and Musaev, D.A. (2000) The development and a cytogenetic study of monosomics of Gossypium hirsutum L. Biologia Plan- tarum, 43(2), 193-197. [16] Sanamyan, M.F. and Petlyakova, J.E. (2002) Cytogenetic analysis of cotton monosomics. Biologia Plantarum, 45(3), 367-373,. [17] Sanamyan, M.F. and Rakhmatullina, E.M. (2003) Cyto- genetic analysis of translocations in cotton. Plant Breed- ing, 122(6), 511-516. [18] Sanamyan, M.F. and Musaev, D.A. (1992) Production and cytogenetic study of the homozygous translocation stocks of cotton. Genetika, 28(75-82). [19] Sanamyan, M.F. and Musaev, D.A. (1995) Cytological and morphological peciliarities of the new translocated lines of cotton. Dokl. Akad. Nauk Republ. Uzbek., No. 11-12, 63-65. [20] Sanamyan, M.F. (2003) Evaluation of the effect of pollen irradiation on karyotype variability in M1 cotton plant.  M. F. Sanamyan et al. / Advances in Bioscience and Biotechnology 1 (2010) 372-383 Copyright © 2010 SciRes. ABB 383 Russian Journal of Genetics, 39(8), 909-916. [21] Sanamyan, M.F. (2003) Evaluation of the effect of pollen irradiation on karyotype variability in M2 cotton plant. Russian Journal of Genetics, 39(7), 791-798. [22] Edwards, G.A., Brown, M.S., Niles, G.A. and Naqi, S.A. (1980) Monosomics of cotton. Crop Science, 20(4), 527- 528. [23] Person, C. (1956) Some aspects of monosomic wheat breeding. Canadian Journal of Botany, 34(1), 60-70. [24] Morrison, J.W. and Unrau, J. (1952) Frequency of mi- cronuclei in pollen quartets of common wheat mono- somics. Canadian Journal of Botany, 30(4), 371-378. [25] Endrizzi, J.E., Turcotte, E.L. and Kohel, R.J. (1984) Qualitative genetics, cytology and cytogenetics. In: Kohel, R.J. and Lewis, C.F., Eds., Cotton, American So- ciety of Agronomy, Madison, Wis., USA, 81-129. [26] McMichael, S.C. (1965) Inheritance of aberrant stigmas in the flowers of Upland cotton. Journal of Heredity, 56(1), 21-22. [27] Endrizzi, J.E. and Bray, R. (1980) Cytogenetics of disomics, monotelo- and monoisodisomics and ml1st1 mutants of chromosome 4 of cotton. Genetics, 9, 979- 988. [28] Turcotte, E.L. and Feaster, C.V. (1964) Inheritance of a mutant with a rudimentary stigma and style in Pima cot- ton, Gossypium barbadense L. Crop Science, 4, 377-378. [29] Endrizzi, J.E. and Brown, M.S. (1964) Identification of monosomes for six chromosomes in Gossypium hirsutum. American Journal of Botany, 51(2), 117-120. [30] Brown, M.S. (1980) Identification of the chromosomes of Gossypium hirsutum L. by means of translocations. Journal of Heredity, 71(4), 266-274. [31] Sanamyan, M.F. (2006) Cytogenetics of translocant and monosomic plants of cotton Gossypium hirsutum L. Doctoral dissertation, National University of Uzbekistan, Tashkent. [32] Brown, M.S. (1978) The identification of the individual chromosomes of Gossypium hirsutum L. Proceedings of the Beltwide Production Research Conference, USA, Dallas, 70. [33] Endrizzi, J.E., Turcotte, E.L. and Kohel, R.J. (1985) Ge- netics, cytology and evolution of Gossypium. Advances in Genetics, 23, 271-375. |

