Paper Menu >>
Journal Menu >>
 American Journal of Plant Sciences, 2010, 1, 69-76 doi:10.4236/ajps.2010.12009 Published Online September 2010 (http://www.SciRP.org/journal/ajps) Copyright © 2010 SciRes. AJPS 69 Differential Expression of microRNAs in Maize Inbred and Hybrid Lines during Salt and Drought Stress Yeqin M. Kong1*, Axel A. Elling1,2*, Beibei Chen1, Xing Wang Deng1 1Department of Molecular, Cellular and Developmental Biology, Yale University, New Haven, CT, USA; 2Current affiliation: De- partment of Plant Pathology, Washington State University, Pullman, WA, USA. Email: * xingwang.deng@yale.edu Received August 14th, 2010; revised November 7th, 2010; accepted November 23rd, 2010. ABSTRACT Here, we analyzed whether the microRNA (miRNA) expression levels differ between maize inbred lines B73 and Mo17 and their reciprocal hybrids under salt and drought stress. We found that miR156, miR164, miR166, miR168, miR171 and miR319 are differentially expressed under abiotic stress. Interestingly, Mo17 × B73 showed the strongest change in miRNA expression in response to sa lt or drought stress, and was also the most resilient line when under abiotic stress in terms of water loss. In summary, our findings open the possibility that differential miRNA expression levels might be involved in heightened stress tolerance in ma ize hybrids. Keywords: microRNA, Maize, Hybrid, Inbred, Salt Stress, Drough 1. Introduction MicroRNAs (miRNAs) are non-coding RNAs appro- ximately 20-24 nucleotides in length that act as negative post-transcriptional regulators [1,2]. In plants, single- stranded primary miRNAs are transcribed from miRNA loci and are processed by Dicer-like 1 (DCL1) to yield mature single-stranded miRNAs, which are loaded into the RNA-induced silencing complex (RISC). miRNA- loaded RISC targets cognate transcripts and induces their cleavage [3]. To date, about 1000 miRNAs have been identified in various plant species, with 20 miRNA families that are well conserved between dicots and monocots [4]. As one of the world’s most important crop species, significant progress has been made in characterizing and analyzing miRNAs in the maize (Zea mays) genome [5,6]. While a significant proportion of known miRNA target genes regulates plant development [1,7,8], recent studies have shown that miRNAs are also involved in abiotic and bi- otic stress responses [2,9]. Abiotic stress, in particular drought and salt stress, is a significant yield-limiting factor for agriculture in many regions of the world. Thus, understanding plant responses to abiotic stress is vital for improving crop productivity. It is well documented that the F1 hybrid progeny of inbred parental lines shows superior performance and stress tolerance compared to either parent [10-12]. This effect is called heterosis and is widely exploited in plant breeding. In the present study, we determined whether seedlings of maize inbred lines B73 and Mo17 and their reciprocal F1 hybrids show dif- ferential miRNA expression patterns in response to salt and drought stress and whether heightened stress toler- ance in F1 hybrids correlates with changes in miRNA abundance. 2. Materials and Methods 2.1. Plant Material and Stress Treatment Seeds of maize (Zea mays) inbred lines B73 and Mo17, and their reciprocal hybrids B73 × Mo17 and Mo17 × B73 were individually planted in pots containing a 3:2 soil:vermiculite mixture. Plants were grown under con- trolled environmental conditions (15 h light/25℃, 9 h dark/20℃) in a growth room, and watered with 0.7 mM Ca(NO3)2 for 13 days. Salt or drought stress treatments began at the onset of day 14 by either watering with 200 mM NaCl, or by carefully removing plants from potted soil and dehydrating them on filter paper following previ- ously described methods [13]. Control plants continued to grow in pots watered with 0.7 mM Ca(NO3)2. Stress *These authors contributed equally to this work.  Differential Expression of microRNAs in Maize Inbred and Hybrid Lines during Salt and Drought Stress Copyright © 2010 SciRes. AJPS 70 treatment lasted for 24 h, after which all stress- treated and control seedlings were harvested, separated into shoots and roots and stored at –80℃. Each treatment was set up in three replicates with five to seven seedlings per geno- type each. Water content of 14-day-old shoot tissues was assayed by measuring fresh and dry weight of shoots of salt- treated, drought-treated, and control seedlings at 0, 2, 12, and 24 h after onset of treatment following previously described methods [14]. 2.2. RNA Isolation and Northern Blot Analysis of miRNA Expression Total RNA was extracted using TRIzol reagent (Invitro- gen). Northern blot analysis of miRNA was performed as described [15]. Briefly, 20 μg of total RNA was loaded per lane and resolved on a denaturing 12% polyacryla- mide gel, electrophoretically transferred to Hybond-N+ membranes (GE Healthcare) and UV cross-linked. We selected one miRNA each of 20 miRNA families that were known in maize at the time of this study (miR156, miR159, miR160, miR162, miR164, miR166, miR167, miR168, miR169, miR171, miR172, miR319, miR390, miR393, miR394, miR396, miR397, miR398, miR399, miR408. Membranes were hybridized with DNA oli- gonucleotide probes end-labeled with γ-32P-ATP, which were comple- mentary to the mature miRNA sequences. Membranes were exposed on BioMax MR film (Kodak) for 18 h, which were scanned to quantify miRNA abun- dance using ImageJ [16]. tRNA was used for normaliza- tion. Only those six miRNAs that showed a change in expression level of 25% or more in at least one compari- son between different genotypes and treatments were included in our subsequent analyses and are shown; miR172 was included as a representative example of a miRNA that was not responsive to the stress treatment in this study (Tab le 1). Due to the technique chosen, calcu- lating statistically significant differences for miRNAs was not possible here. 2.3. Real-Time Quantitative PCR Analysis of miRNA Target Gene Expression Aliquots of TRIzol-purified total RNA used for northern blots were reverse transcribed using RETROscript kit (Ambion). After DNase treatment, miRNA target gene expression was determined by quantitative real-time PCR on an ABI 7900HT real-time PCR system (Applied Bio- systems) using Quantitect SYBR Green PCR kit (Qiagen). To ensure quantification of only 3’ cleavage products of microRNA target genes, primers for target genes were designed such that they either span the miRNA cleavage site or are downstream from it [17]. 18S rRNA served as reference gene. Quantification of gene expression was performed by 2-CT method [18]. All reactions were per- formed in duplicates for three biological replicates. The results are shown by their mean ± standard deviation (S.D). Fisher’s exact test was used to test for statistical significance in the statistical software package R. We used the mRNA medium expression values of three bio- logical replicates for control and treated tissue and used the non-treated control tissue to determine the standard expression value. The two-dimensional contingency ma- trix consisted of conditional expression values and stan- dard expression values. Using 0.05 as significance level (FDR = 0.01), all testing pairs with a p-value less than 0.05 were considered significant (only statistically sig- nificant comparisons within the same treatment group are marked with an asterisk). Table 1. Probes and forw ard and reverse primers used in northern blots and qrt-pcr. miRNA Probe Target Gene Primer miR156 TGTGCTCTCTCTCTTCTGTCA SPL5 TGATGAACTGATGCGGTGTCAGGAG TTAATGCATCGCGAGCAAAGTCCAC miR164 TGCACGTGCCCTGCTTCTCCA NAC1 TCGTGGACCTCAGCTACGACGACAT GGAGACGCGAAGAGCGAGGAGTAGA miR166 GGGGAATGAAGCCTGGTCCGA RLD1 ACCAAGCTGTAGCGTGGAAGGTGCT TGCATGCAACATATGCCTTTTGTCA miR168 GTCCCGATCTGCACCAAGCGA AGO1 TTGCTCCCATCTGCTACGCACATCT CACGGCTCAGCAAAAGAACATCGAG miR171 GATATTGGCACGGCTCAATCA SCL1 CAGTCAGCTTGTGCTTCTGCGAGGT CACTAACGCGGATGCTGCCAGTAAG miR172 ATGCAGCATCATCAAGATTCC GL15 AAGTGACGCGTCCTCTGTGCTTCTG TAGCTCTGGGCATCGAAGTTGGTCA miR319 GGGAGCACCCTTCAGTCCAA TCP1 AGGGCAGGAGCTGATTGCACATTCT TCTGACAAGTCGTCACCGCAACAAA 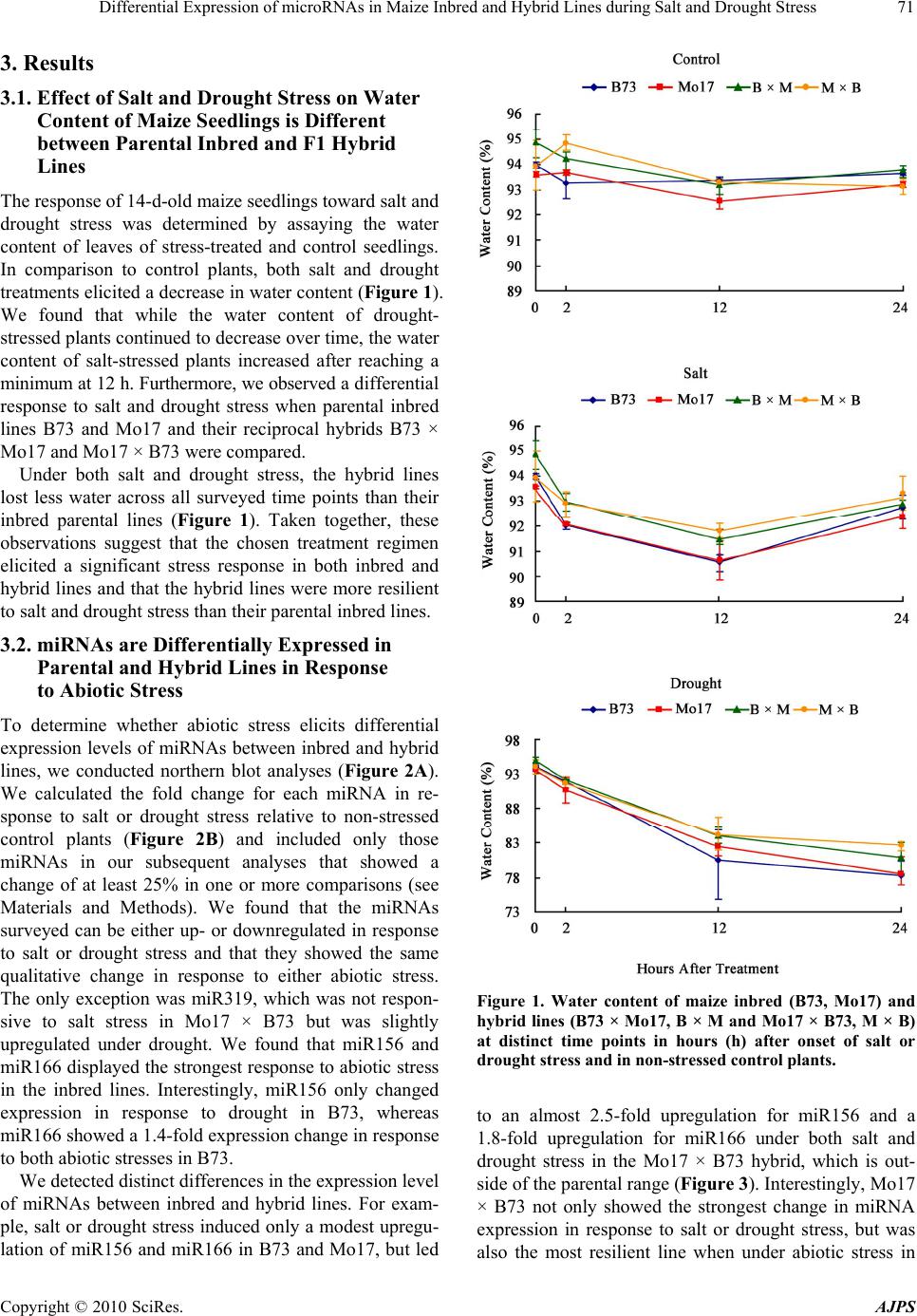 Differential Expression of microRNAs in Maize Inbred and Hybrid Lines during Salt and Drought Stress Copyright © 2010 SciRes. AJPS 71 3. Results 3.1. Effect of Salt and Drought Stress on Water Content of Maize Seedlings is Different between Parental Inbred and F1 Hybrid Lines The response of 14-d-old maize seedlings toward salt and drought stress was determined by assaying the water content of leaves of stress-treated and control seedlings. In comparison to control plants, both salt and drought treatments elicited a decrease in water content (Figure 1). We found that while the water content of drought- stressed plants continued to decrease over time, the water content of salt-stressed plants increased after reaching a minimum at 12 h. Furthermore, we observed a differential response to salt and drought stress when parental inbred lines B73 and Mo17 and their reciprocal hybrids B73 × Mo17 and Mo17 × B73 were compared. Under both salt and drought stress, the hybrid lines lost less water across all surveyed time points than their inbred parental lines (Figure 1). Taken together, these observations suggest that the chosen treatment regimen elicited a significant stress response in both inbred and hybrid lines and that the hybrid lines were more resilient to salt and drought stress than their parental inbred lines. 3.2. miRNAs are Differentially Expressed in Parental and Hybrid Lines in Response to Abiotic Stress To determine whether abiotic stress elicits differential expression levels of miRNAs between inbred and hybrid lines, we conducted northern blot analyses (Figure 2A). We calculated the fold change for each miRNA in re- sponse to salt or drought stress relative to non-stressed control plants (Figure 2B) and included only those miRNAs in our subsequent analyses that showed a change of at least 25% in one or more comparisons (see Materials and Methods). We found that the miRNAs surveyed can be either up- or downregulated in response to salt or drought stress and that they showed the same qualitative change in response to either abiotic stress. The only exception was miR319, which was not respon- sive to salt stress in Mo17 × B73 but was slightly upregulated under drought. We found that miR156 and miR166 displayed the strongest response to abiotic stress in the inbred lines. Interestingly, miR156 only changed expression in response to drought in B73, whereas miR166 showed a 1.4-fold expression change in response to both abiotic stresses in B73. We detected distinct differences in the expression level of miRNAs between inbred and hybrid lines. For exam- ple, salt or drought stress induced only a modest upregu- lation of miR156 and miR166 in B73 and Mo17, but led Figure 1. Water content of maize inbred (B73, Mo17) and hybrid lines (B73 × Mo17, B × M and Mo17 × B73, M × B) at distinct time points in hours (h) after onset of salt or drought stress and in non-stressed c o ntr ol plants. to an almost 2.5-fold upregulation for miR156 and a 1.8-fold upregulation for miR166 under both salt and drought stress in the Mo17 × B73 hybrid, which is out- side of the parental range (Figure 3). Interestingly, Mo17 × B73 not only showed the strongest change in miRNA expression in response to salt or drought stress, but was also the most resilient line when under abiotic stress in  Differential Expression of microRNAs in Maize Inbred and Hybrid Lines during Salt and Drought Stress Copyright © 2010 SciRes. AJPS 72 terms of water loss (Figure 1). In contrast, the reciprocal hybrid B73 × Mo17 showed a modest increase that was close to the range found in the parental inbred lines. Even though miR164, miR171 and miR319 showed differen- tial expression in the absence of abiotic stress (Figures 2(a), (b)), they only displayed modest differences be- tween inbred and hybrid lines in the presence of salt or drought stress (Figure 3). Interestingly, abiotic stress induced downregulation of miR171 and miR319 in Mo17 and B73 × Mo17 when compared to B73 and Mo17 × B73, which might indicate parental effects. The other 14 miRNAs we examined were not responsive to salt or drought stress or showed no differential stress response between inbred parents and hybrid offspring. Taken together, our results show that miRNAs in maize respond differentially to abiotic stress depending on whether they reside in an inbred or hybrid genome. 3.3. Expression Pattern of miRNA Target Genes To determine the expression level of miRNA target genes in maize, one bona fide target of each miRNA was selected based on previously validated target genes in maize or homology to validated targets in other plant species. Previous studies confirmed that homologous genes are targeted by the respective miRNAs in other plant species such as Arabidop sis, including the squamosa promoter binding protein-like transcription factor SPL4 (homologue of maize SPL5) for miR156 [19], NAM/ ATAF/CUC (NAC) domain-encoding gene NAC1 for miR164 [20], the PHABULOSA gene (homologue of maize rolled leaf1) for miR166 [21], ARGONAUTE 1 (AGO1) gene for miR168 [22], Scarecrow-like transcrip- tion factor 1 (SCL1) for miR171 [23], the APETALA2- like gene glossy15 (GL15) for miR172 [24], and the TCP (TEOSINTE BRANCHED/CYCLOIDEA/PCF) transcrip- tion factor 1 (TCP1) for miR319 [25]. As with miRNA accumulation, we determined the fold change of the target genes in each line of stress-treated plants relative to controls (Figure 3). Apart from a few exceptions, we found that the expression changes of most target genes were statistically significant when compared to controls. Furthermore, we observed that most differ- ences in expression levels between inbred and hybrid lines under stress conditions were statistically significant, as were a large number of comparisons among hybrid lines (Figure 3). Some genes showed a more dramatic response to abiotic stress than the miRNA targeting them. For example, NAC1 was upregulated over five fold, (a) (b) Figure 2. Northern blots for miRNAs in response to salt and drought stress and non-stressed control plants in maize inbreds B73, Mo17 and hybrids B73 × Mo17 (B × M), Mo17 × B73 (M × B). (b) Relative miRNA expression level in non-stressed con- trol plants calculated from northern blot assays (a). miR172 is shown as an example for a miRNA that did not show a differ- ential response to stress treatment.  Differential Expression of microRNAs in Maize Inbred and Hybrid Lines during Salt and Drought Stress Copyright © 2010 SciRes. AJPS 73 Figure 3. miRNA expression change determined by northern blots (left panel) and miRNA target gene expression changes determined by qRT-PCR (right two panels) in response to salt and drought stress relative to non-stressed control plants in maize inbreds B73, Mo17 and hybrids B73 × Mo17 (B × M), Mo17 × B73 (M × B). p < 0.05 was considered significant and is indicated by an asterisk. miR172 is shown as an example of a miRNA that did not respond differentially to stress.  Differential Expression of microRNAs in Maize Inbred and Hybrid Lines during Salt and Drought Stress Copyright © 2010 SciRes. AJPS 74 whereas its cognate miR164 showed only modest changes (Figure 3). In general, all miRNA-target gene pairs studied showed opposite expression patterns in re- sponse to abiotic stress, confirming the role of miRNAs as negative regulators. However, miR172, which showed no significant differential expression across inbred and hybrid lines in the absence or presence of stress (Figure 2), displayed a remarkable contrast to its target gene, GL15, which was strongly downregulated in response to drought, but not salt (Figure 3). Taken together, the target gene expression patterns we observed in maize inbred and hybrid lines in response to abiotic stress corroborate our finding that stress-respon- sive miRNAs show differential expression in inbred and hybrid lines. 4. Discussion In addition to being key regulators in several develop- mental pathways in plants, there is increasing evidence that miRNAs are implicated in other mechanisms such as plant adaptive response to stress or environmental changes [9]. Besides specific miRNAs whose target genes are involved directly in stress responses, such as miR398, which targets Cu/Zn superoxide dismutases important for oxidative stress tolerance [26], the expres- sion of many highly conserved plant miRNAs has been found to be responsive to various abiotic stresses such as dehydration and ABA treatments in Arabidopsis [27], cold stress in poplar [17], salt and drought stress in rice [28,29], and salt stress in maize roots [30]. Here, we provide evidence that miRNAs show a dif- ferential expression pattern in more stress-tolerant maize hybrids compared to less stress-tolerant inbred lines. Apart from miR168, which targets AGO1, the core component of RISC [31], all other miRNAs surveyed have highly conserved roles in important overlapping plant developmental processes: miR156 [19], miR166 [20], and miR171 [23] are involved in leaf and shoot development, miR172 in flower development [32], and miR164 [21] and miR319 [25] in hormone signaling. It is well-documented that abiotic stress elicits large-scale transcriptome responses in maize of early acting genes involved in the immediate sensing and responding to stress, and of genes that produce long lasting effects by targeting growth and developmental processes [33]. miRNAs and their regulation of plant development are an important component of stress responses, because plants not only have to respond to stress to survive, but also undergo developmental reprogramming for continued productivity [30,34]. miRNAs are generally considered as negative post- transcriptional regulators of gene expression [1,2]. Our findings corroborate this hypothesis, showing that direc- tions of stress-induced change in specific miRNA accu- mulation and target gene expression were opposite of each other (Figure 3). The only exception among the miRNAs studied here was miR168, which targets AGO1. The expression level of miR168 and AGO1 showed a parallel, and not an opposite pattern (Figure 3). This can be explained by a previously described transcriptional/ translational feedback loop between miR168 and AGO1 expression [31], in which MIR168 and AGO1 are co-regulated on a transcriptional level, and AGO1 aids in posttranscriptional stabilization of miR168. In addition, GL15, the target gene of miR172, dis- played a uniform decrease in response to abiotic stress, while miR172 was non-responsive to either salt or drought stress. Prior studies have shown that miR172 acts at the translational level to suppress GL15 protein production, instead of causing the cleavage of its mRNA [32]. Therefore, the accumulation of miR172 and mRNA of GL15, which is the maize homolog of APETALA2, have no correlation with each other (Figure 3). Even though our results largely confirm the findings of recent studies aimed at characterizing the response of miRNAs and their target genes to salt and drought stress [28,30,35], our findings show that miRNA response to abiotic stresses can differ dramatically between plant species and inbred parental and their hybrid offspring lines. For example, whereas miRNA156 has been shown to be strongly downregulated upon drought and salt stress in rice [28], Ding et al. [30] reported less dramatic changes in response to salt in maize inbred lines NC286 and Huangzao4. We found that miR156 shows relatively small changes in response to salt stress in the inbred lines B73 and Mo17, but is dramatically upregulated in their reciprocal hybrids (Figure 3). Furthermore, whereas miR171 was downregulated under salt and drought stress in rice [28], it was upregulated under similar conditions in Arabidopsis [35]. Here, we observed that both miR171 and its target gene SCL1 can be either up- or downregu- lated under salt and drought stress, depending on the maize line used. These examples illustrate that the re- sponses of individual miRNAs and their target genes to stress cannot only differ among plant species, but also between different genetic lines of the same species. In summary, we found that miRNAs show differential responses to abiotic stress in maize inbred and hybrid lines, which opens the tantalizing possibility that miRNAs might be involved in the superior performance of hybrid lines. This would have a significant impact on recent approaches aimed at using miRNAs to increase crop yields [36]. 5. Acknowledgements This research was supported by grants to X. W. D. from  Differential Expression of microRNAs in Maize Inbred and Hybrid Lines during Salt and Drought Stress Copyright © 2010 SciRes. AJPS 75 the National Institutes of Health (GM047850), the Na- tional Science Foundation Plant Genome Program (DBI0421675), and the National Science Foundation 2010 program (MCB-0929100). A. A. E. was supported by a Yale University Brown-Coxe Postdoctoral Fellow- ship. REFERENCES [1] M. W. Jones-Rhoades, D. P. Bartel and B. Bartel, “Mi- croRNAs and Their regulatory Roles in Plants,” Annual Review of Plant Biology, Vol. 57, January 2006, pp. 19-53. [2] D. H. Jeong, M. A. German, L. A. Rymarquis, S. R. Thatcher and P. J. Green, “Abiotic Stress-Associated miRNAs: Detection and Functional Analysis,” Methods in Molecular Bi ology, Vol. 592, January 2009, pp. 203-230. [3] D. P. Bartel, “MicroRNAs: Genomics, Biogenesis, Mechanism, and Function,” Cell, Vol. 116, January 2004, pp. 281-297. [4] S. Griffiths-Jones, H. K. Saini, S. van Dongen and A. J. Enright, “miRBase: Tools for microRNA Genomics,” Nucleic Acids Research, Vol. 36, January 2008, D154- D158. [5] E. Mica, L. Gianfranceschi and M. E. Pe, “Characteriza- tion of Five microRNA Families in Maize,” Journal of Experimental Botany, Vol. 57, July 2006, pp. 2601-2612. [6] L. Zhang, J-M. Chia, S. Kumari, J.C. Stein, Z. Liu, et al., “A Genome-Wide Characterization of microRNA Genes in Maize,” PLoS Genetics, Vol. 5, November 2009, e716. [7] X. Chen, “MicroRNA Biogenesis and Function in Plants,” FEBS Letters, Vol. 579, August 2005, pp. 5923-5931. [8] A.C. Mallory and H. Vaucheret, “Functions of microR- NAs and Related Small RNAs in Plants,” Nature Genet- ics, Vol. 38, May 2006, S31-S36. [9] L.I. Shukla, V. Chinnusamy and R. Sunkar, “The Role of microRNAs and Other Endogenous Small RNAs in Plant Stress Responses,” Biochimica et Biophysica Acta, Vol. 1779, November 2008, pp. 743-748. [10] G. H. Shull, “The Composition of a Field of Maize,” American Breeders Association, Vol. 4, January 1908, pp. 296-301. [11] K. P. Kollipara, I. N. Saab, R. D. Wych, M. J. Lauer and G. W. Singletary, “Expression Profiling of Reciprocal Maize Hybrids Divergent for Cold Germination and Des- iccation Tolerance,” Plant Physiology, Vol. 129, June 2002, pp. 974-992. [12] P. Rhode, D. K. Hincha and A. G. Heyer “Heterosis in the Freezing Tolerance of Crosses between Two Arabidopsis Thaliana Accessions (Columbia-0 and C24) that Show Differences in Non-Acclimated and Acclimated Freezing Tolerance,” Plant Journal, Vol. 38, June 2004, pp. 790- 799. [13] K. Yamaguchi-Shinozaki and K. Shinozaki, “A Novel Cis-Acting Element in an Arabidopsis Gene is Involved in Responsiveness to Drought, Low-Temperature, or High-Salt stress,” Plant Cell, Vol. 6, February 1994, pp. 251-264. [14] C. Zhu, D. Schraut, W. Hartung and A. R. Schäffner, “Differential Responses of Maize MIP Genes to Salt Stress and ABA,” Journal of Experimental Botany, Vol. 56, November 2005, pp. 2971-2981. [15] E. Varallyay, J. Burgyan and Z. Havelda, “MicroRNA Detection by Northern Blotting Using Locked Nucleic Acid Probes,” Nature Protocol, Vol. 3, January 2008, pp. 190- 196. [16] M. D. Abramoff, P. J. Magelhaes and S. J. Ram, “Image Processing with ImageJ,” Biophotonics International, Vol. 11, 2004, pp. 26-42. [17] S. Lu, Y. H. Sun and V. L. Chiang, “Stress-Responsive microRNAs in Populus,” Plant Journal, Vol. 55, July 2008, pp. 131-151. [18] K. J. Livak and T. D. Schmittgen, “Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-C T Method,” Methods, Vol. 25, Decem- ber 2001, pp. 402-408. [19] G. Wu and R. S. Poethig, “Temporal Regulation of Shoot Development in Arabidopsis thaliana by miR156 and Its Target SPL3,” Development, Vol. 133, August 2006, pp. 3539-3547. [20] A. C. Mallory, B. J. Reinhart, M. W. Jones-Rhoades, G. Tang, P. D. Zamore, M. K. Barton and D. P. Bartel, “Mi- croRNA Control of PHABULOSA in Leaf Development: Importance of Pairing to the microRNA 5′ Region,” EMBO Journal, Vol. 23, July 2004, pp. 3356-3364. [21] H. S. Guo, Q. Xie, J. F. Fei, and N. H. Chua, “MicroRNA Directs mRNA Cleavage of the Transcription Factor NAC1 to Downregulate Auxin Signals for Arabidopsis Lateral Root Development,” Plant Cell, Vol. 17, April 2005, pp. 1376- 1386. [22] H. Vaucheret, A. C. Mallory and D. P. Bartel, “AGO1 Homeostasis Entails Coexpression of MIR168 and AGO1 and Preferential Stabilization of miR168 by AGO1,” Mo- lecular Cell, Vol. 22, April 2006, pp. 129-136. [23] C. Llave, Z. Xie, K. D. Kasschau and J.C. Carrington, “Cleavage of Scarecrow-Like mRNA Targets Directed by a Class of Arabidopsis miRNA,” Science, Vol. 297, Sep- tember 2002, pp. 2053-2056. [24] N. Lauter, A. Kampani, S. Carlson, M. Goebel and S. Moose, “MicroRNA172 Down-Regulates Glossy15 to Promote Vegetative Phase Change in Maize,” Proceedings of the National Academies of Science USA, Vol. 102, May 2005, pp. 9412-9417. [25] C. Schommer, J. F. Palatnik, P. Aggarwal, A. Chételat, P. Cubas, E.E. Farmer, U. Nath and D. Weigel, “Control of Jasmonate Biosynthesis and Senescence by miR319 Tar- gets,” PLoS Biology, Vol. 6, September 2008, p. e230. [26] R. Sunkar, A. Kapoor and J.K. Zhu, “Posttranscriptional Induction of Two Cu/Zn Superoxide Dismutase Genes in Arabidopsis is Mediated by Downregulation of miR398 and Important for Oxidative Stress Tolerance,” Plant Cell, Vol. 18, July 2006, pp. 2051-2065.  Differential Expression of microRNAs in Maize Inbred and Hybrid Lines during Salt and Drought Stress Copyright © 2010 SciRes. AJPS 76 [27] R. Sunkar and J.K. Zhu, “Novel and Stress-Regulated microRNAs and Other Small RNAs from Arabidopsis,” Plant Cell, vol. 16, August 2004, pp. 2001-2019. [28] R. Sunkar, X, Zhou, Y. Zheng, W. Zhang and J.K. Zhu, “Identification of Novel and Candidate miRNAs in Rice by High Throughput Sequencing,” BMC Plant Biology, Vol. 8, February 2008, 25. [29] B. Zhao, R. Liang, L. Ge,W. Li, H. Xiao, H. Lin, K. Ruan and Y. Jin, “Identification of Drought-Induced microR- NAs in Rice,” Biochemical and Biophysical Research Communications, Vol. 354, March 2007, pp. 585-590. [30] D. Ding, L. Zhang, H. Wang, Z. Liu, Z. Zhang and Y. Zheng, “Differential Expression of miRNAs in Response to Salt Stress in Maize Roots,” Annals of Botany, Vol. 103, January 2009, pp. 29-38. [31] H. Vaucheret, “Plant ARGONAUTES,” Trends in Plant Science, Vol. 13, July 2008, pp. 350-358. [32] X.M. Chen, “A microRNA as a Translational Repressor of APETALA2 in Arabidopsis Flower Development,” Science, Vol. 303, March 2004, pp. 2022-2025. [33] J. Fernandes, D. J. Morrow,P. Casati and V. Walbot, “Distinctive Transcriptome Responses to Adverse Envi- ronmental Conditions in Zea mays L.” Plant Biotech- nology Journal, Vol. 6, October 2008, pp. 782-798. |

